Published online Aug 26, 2022. doi: 10.12998/wjcc.v10.i24.8490
Peer-review started: March 21, 2022
First decision: May 1, 2022
Revised: May 14, 2022
Accepted: July 16, 2022
Article in press: July 16, 2022
Published online: August 26, 2022
Processing time: 147 Days and 14.1 Hours
Pyroptosis is an inflammatory form of programmed cell death, which has been shown to be related to the prognosis of many tumors. However, its role in gastric cancer (GC) is not fully understood.
To evaluate the expression of pyroptosis-related genes in GC and its correlation with prognosis.
We constructed prognostic multigene markers of differentially expressed genes associated with pyroptosis by least absolute contraction and selection operator Cox regression. The risk model was analyzed by Kaplan-Meier curve, two-sided log-rank test and functional enrichment analysis.
Sixty-three pyroptosis-related genes were differentially expressed in tumor tissues and adjacent nontumor tissues. Based on these differentially expressed genes, 5 gene signature were constructed and all GC patients were classified into two risk groups. Kaplan-Meier survival curve showed that the overall survival (OS) of patients in the high-risk group was significantly lower than that of the low-risk group. Multivariate Cox regression analyses showed that the risk score was an independent risk factor for OS. Receiver operating characteristic curve analysis confirmed the predictive ability of the model. External validation indicated increased OS in the low-risk group. The immune function and immune cell scores of the high-risk group were generally higher than those of the low-risk group.
Pyroptosis-related genes play a significant role in tumor immune microenvironment. This novel model, which contains 5 pyroptosis-related genes, is an independent predicting factor for OS in GC patients, and may help to evaluate the prognosis of GC.
Core Tip: This study aims to evaluate the expression of pyroptosis-related genes in gastric cancer (GC) and its correlation with prognosis. Based on 63 pyroptosis differentially expressed genes, 5 gene signature were constructed and all GC patients were classified into two risk groups. Kaplan-Meier survival curve showed that the overall survival (OS) of patients in the high-risk group was significantly lower than that of the low-risk group. Multivariate Cox regression analyses showed that the risk score was an independent risk factor for OS. The immune function and immune cell scores of the high-risk group were generally higher than those of the low-risk group. Similar results were obtained in external validation.
- Citation: Guan SH, Wang XY, Shang P, Du QC, Li MZ, Xing X, Yan B. Pyroptosis-related genes play a significant role in the prognosis of gastric cancer. World J Clin Cases 2022; 10(24): 8490-8505
- URL: https://www.wjgnet.com/2307-8960/full/v10/i24/8490.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i24.8490
Gastric cancer (GC) is one of the most common malignant tumors of the digestive tract in the world. According to the data, there are approximately 1.03 million cases of GC, accounting for 5.6% of the total number of cancer cases. The incidence is approximately 11.1 per 100000, ranking fifth among all malignant tumors. There are approximately 782000 deaths from GC, accounting for 8.2% of all tumor-related deaths, with a mortality rate of approximately 8.2 per 100000; GC has the third highest death rate among cancers worldwide[1]. Almost two-thirds of GC cases occur in Asia (especially Japan, South Korea, and China). Compared with Japan and South Korea, China's early GC accounts for only 19.7%, and 40% of patients have no indications for radical surgery at the time of diagnosis[2,3]. There is significant heterogeneity in the response of GC patients to treatment. The treatment effect of advanced GC is limited, the median survival time is only about 1 year, and the prognosis has not been effectively improved[4]. Therefore, it is essential to find more targets, determine the treatment advantage population, and improve the efficiency of precise treatment of GC.
Pyroptosis is a kind of programmed cell death caused by inflammasomes, which is manifested by the continuous expansion of cells until the cell membrane ruptures, resulting in the release of cell contents and a strong inflammatory response[5]. The classic pathway of pyroptosis depends on the inflammatory caspase and gasdermines (GSDMs) protein family. The activated caspase cleaves the GSDMs protein and releases its N-terminal domain, which binds to membrane lipids and punches holes in the cell membrane. Perforating on the top causes the cell osmotic pressure to change and then swells until the cell membrane ruptures[6-8]. Pyroptosis is a crucial natural immune response of the body, which plays a vital role in fighting infection. In recent years, increasing studies have shown that it also plays a significant role in the development of tumors[9]. Rébé et al[10] found that Liver X receptor ligand-induced pyroptosis occurs in colon cancer cells. LncRNA GAS5 inhibits the glucocorticoid receptor complex and triggers the formation of inflammasomes, which in turn activates the inflammatory pathway and induces apoptosis in ovarian cancer cells[11]. Lipopolysaccharide (LPS) can induce pyroptosis and play an important role in the carcinogenesis of Barrett's esophagus. LPS activates and activates the NOD-like receptor protein 3 in Barrett cells to enhance the secretion of proinflammatory cytokines, including interleukin (IL)-18, IL-1β and lactate dehydrogenase (indicators of pyroptosis)[12].
Therefore, we conducted a systematic study on pyroptosis genes. We constructed a prognostic polygene signature of differentially expressed genes (DEGs) related to pyroptosis based on the mRNA levels of pyroptosis-related genes, and externally verified. On this basis, we further carry out functional enrichment analysis to explore its potential immune mechanism. The purpose of this study is to classify GC with pyroptosis-related regulatory factors to construct a scoring model to predict the prognosis of GC and explore the prognostic value of these genes, to provide new ideas for the clinical treatment of GC.
As of August 23, 2021 RNA sequencing (RNA-seq) data (n = 373) and clinical features data (n = 401) for GC information were downloaded from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/repository), and the RNA-seq data including 343 GC tissues and 30 adjacent nontumor tissues. A total of 432 GC patients information of the external validation cohort were downloaded from Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). Patients without survival information were excluded from further analysis. All above datasets are publicly available. Therefore, this study did not involve relevant ethics and strictly follows the relevant database access policies and guidelines.
We searched for pyroptosis-related genes from AmiGO2 (http://amigo.geneontology.org/amigo/Landing), Gene-National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/gene/) and Gene Set Enrichment Analysis (http://www.gsea-msigdb.org/gsea/index.jsp), deleted duplicate entries, and finally identified 119 pyroptosis-related genes (the detailed data are available in Supplementary material). The data in TCGA database were normalized to fragment per kilobase million (FPKM) values before comparison.
The "limma" package was used to identify DEGs with a P < 0.05. The DEGs were marked as follows:
Univariate Cox regression analysis was performed to assess the association between genes and survival status in the TCGA cohort, selecting 0.005 as the cutoff P value. To minimize the risk of overfitting, using the least absolute shrinkage and selection operator (LASSO) Cox regression to narrow down the candidate genes and develop a prognostic signature. The "glmnet" package in R software was used to select variables and shrink them by the LASSO algorithm, and the penalty parameter (λ) was decided by using the minimum criteria. The risk score was reckoned after centralization and standardization of the TCGA data. The risk score formula is as follows: Risk Score= ∑i11 Xi × Yi (X: coefficients, Y: gene expression level). The TCGA cohort were divided into high-risk and low-risk subgroups pursuant to the median risk score. The overall survival (OS) time of the two subgroups was compared by Kaplan-Meier analysis. Principal component analysis (PCA) and t-distributed stochastic neighbor embedding (t-SNE) were performed using the “prcomp” function in the “stats” R package according to the gene expression in the gene profile. The “survival”, “survminer” and “time-receiver operating characteristic (ROC)” R packages were employed to perform a 5-year ROC curve analysis. We used the GEO cohort for validation. The expressions of pyroptosis-related gene were also normalized by the “scale” function, and the risk score was measured by the same formula as the TCGA cohort. By adopting the median risk score from the TCGA cohort, patients in the GEO cohort were also divided into low-risk and high-risk subgroups, and these subgroups were compared to validate the gene model.
We figure out the clinical information of patients from the TCGA cohort and GEO cohort, including age, gender, tumor differentiation, depth of invasion, and lymph node metastasis. Prognostic analysis of these variables combined with risk scores. Univariate and multivariate Cox regression models were applied for analysis.
The TCGA cohort were divided into two subgroups pursuant to the median risk score. We set |log2FC| ≥ 1 and false discovery rate (FDR) < 0.05 as the criteria for screening DEGs between the low-risk group and the high-risk group. Founded on these DEGs, Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional analysis were performed by applying the “cluster Profiler” package. The “gsva” package was applied to perform single sample gene set enrichment analysis (ssGSEA) to calculate the infiltration scores of 16 kinds of immune cells and the activities of 13 immune-related pathways.
One-way analysis of variance was used to compare gene expression levels between adjacent nontumor tissues and GC tissue, while categorical variables were analyzed using Pearson chi-square test. The Kaplan-Meier method and the two-sided log-rank test were used to compare the OS of patients between subgroups. Univariate and multivariate Cox regression analysis were performed to estimate the risk model’s independent prognostic value in both TCGA cohort and GEO cohort. The immune cell infiltration and immune pathway activation between the two groups were compared by Mann–Whitney test with corrected P values. All statistical analysis was done using R software (version 4.1.0).
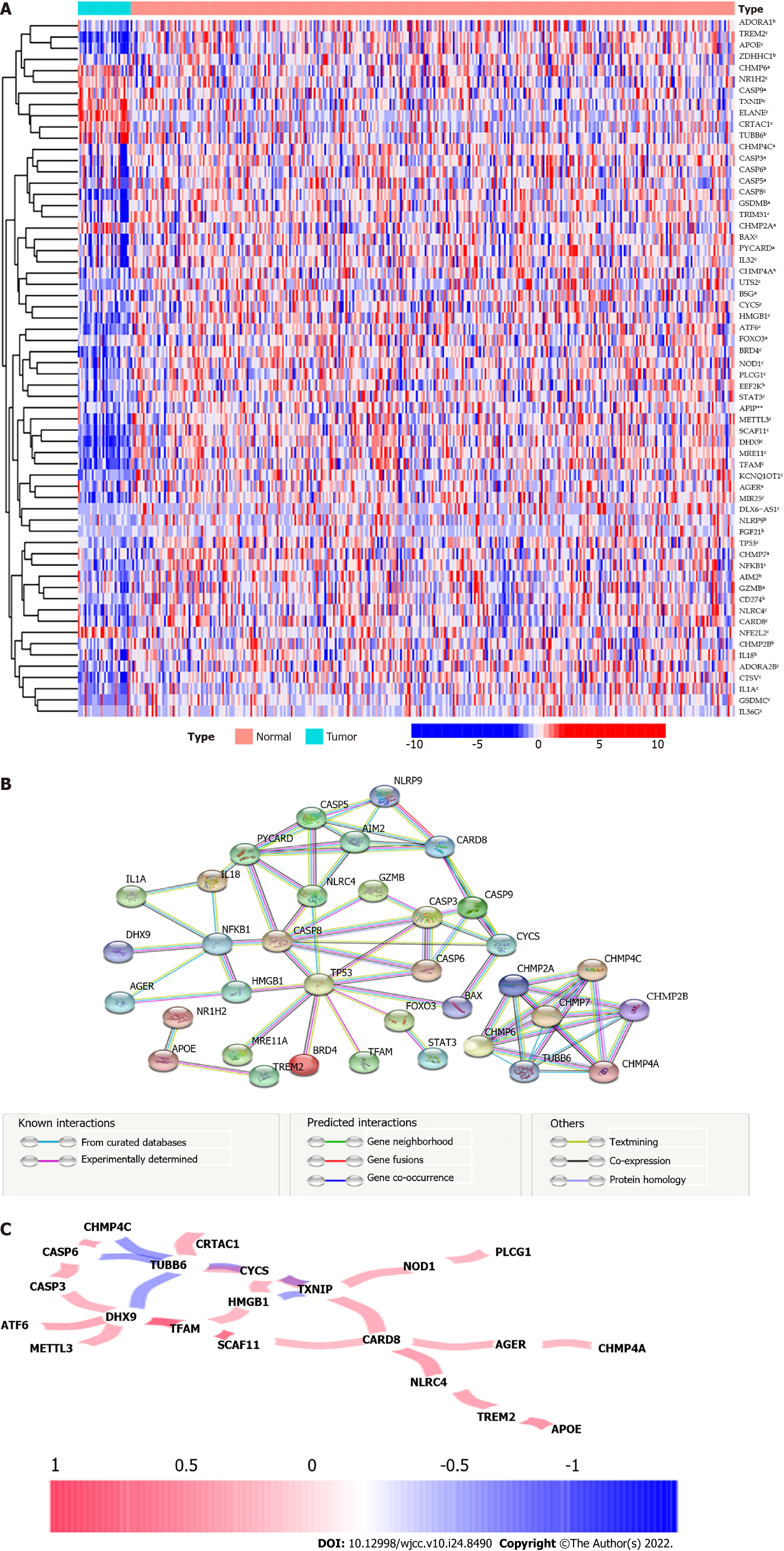
The overall flow chart is depicted in Figure 1. We compiled the genes of 30 adjacent nontumor tissues and 343 tumor tissues in the TCGA database and compared the expression levels of 118 genes related to pyroptosis. We identified 63 DEGs (all P < 0.05). The RNA levels of these genes were presented in heatmap, among them, 8 genes (CHMP6, NR1H2, CASP9, TXNIP, ELANE, CRTAC1, TUBB6 and NFE2L2) were downregulated, while the other 55 genes were abundant in the tumor group (Figure 2A). To further explore the interaction of these DEGs, we conducted a PPI analysis. The minimum interaction score required for PPI analysis is set to 0.900 (the highest confidence level), the results show that TP53, PYCARD, CASP8, NFKB1, CHMP2A, CHMP6, TUBB6, CARD8, CHMP4A and CHMP7 were hub genes, and there is a close protein interaction relationship between DEGs (Figure 2B). The correlations between these genes were presented in co-expression network (Figure 2C).
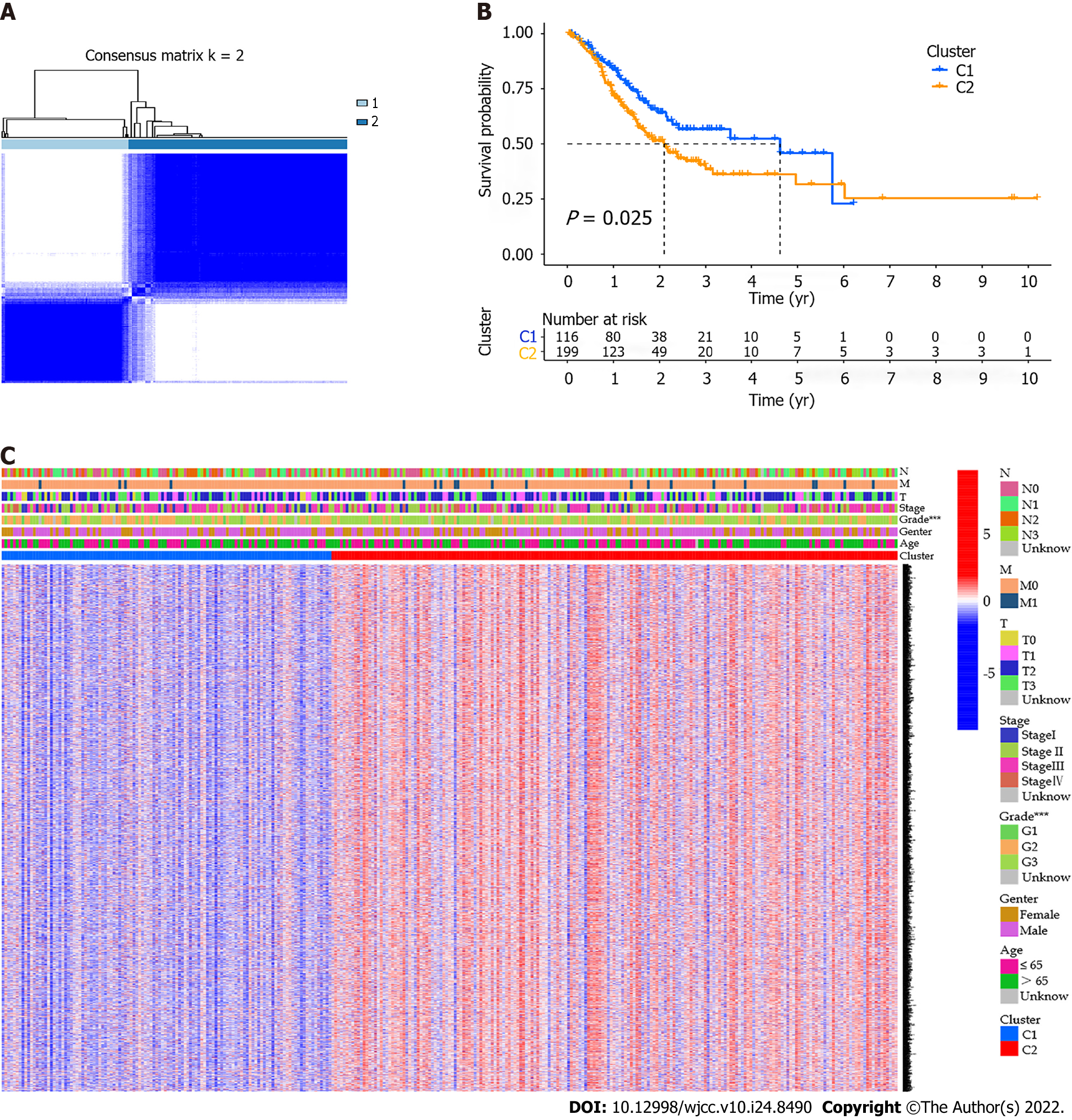
A total of 373 GC samples were matched with corresponding patients with complete survival information. In order to explore the associations between pyroptosis-related DEGs and GC subtypes, we performed a consensus clustering analysis on GC patients in the TCGA cohort. By increasing the clustering variable (k) from 2 to 10, we found that when k = 2, the intragroup correlations were the highest and the intergroup correlations were low, indicating that GC patients can be well classified into two clusters (Figure 3A). We further compared the OS time between the two clusters and found that the survival rate of the C1 cluster was higher than that of the C2 cluster, and the difference was statistically significant (P = 0.025, Figure 3B). Gene expression profile and clinical features including tumor differentiation degree, gender, age ( ≤ 65 years or > 65 years), stage, depth of tumor invasion, lymph node metastasis, and distant metastasis were all shown in a heatmap. In the figure, the difference in the degree of tumor differentiation was diversely distributed between the two clusters (Figure 3C).
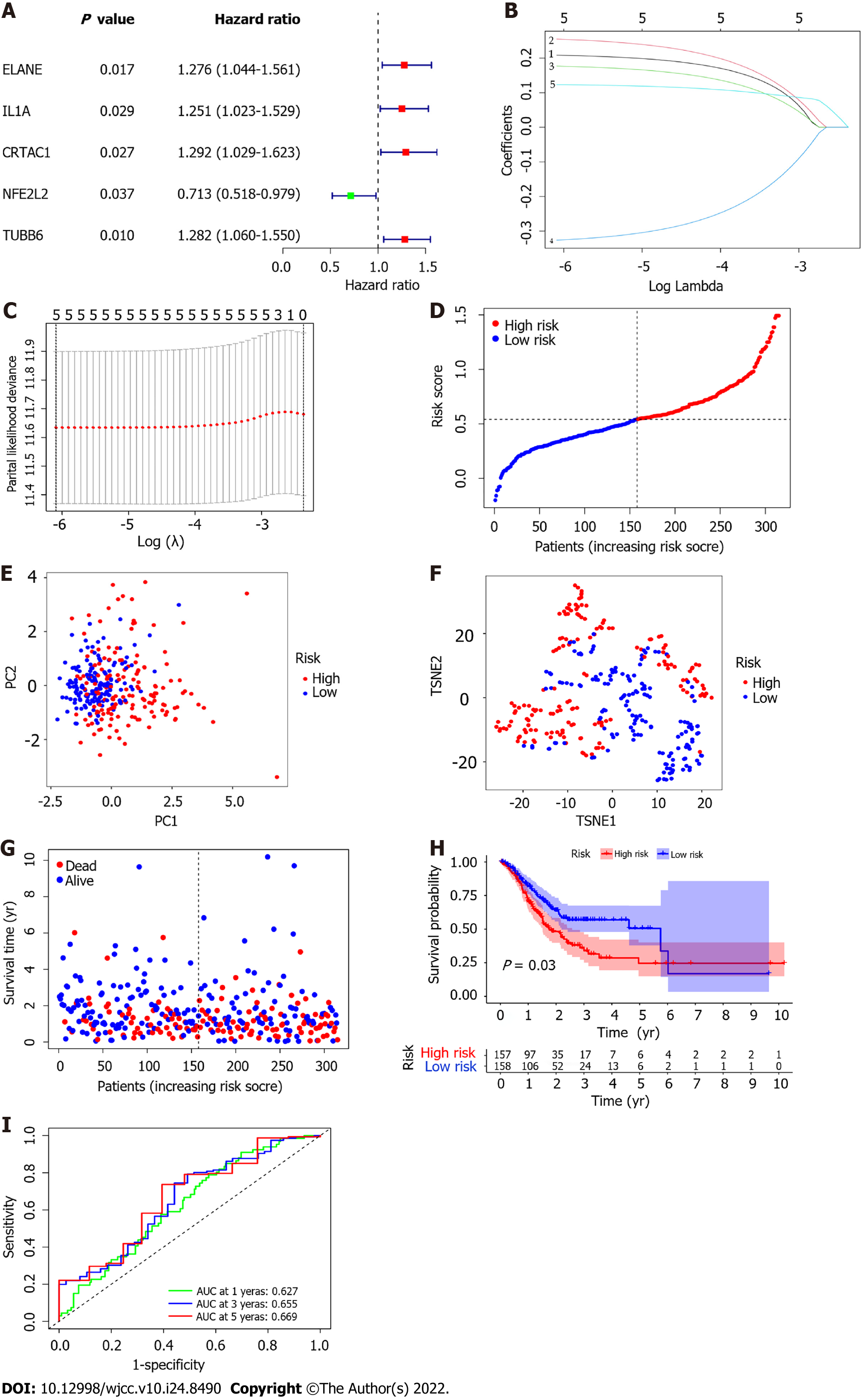
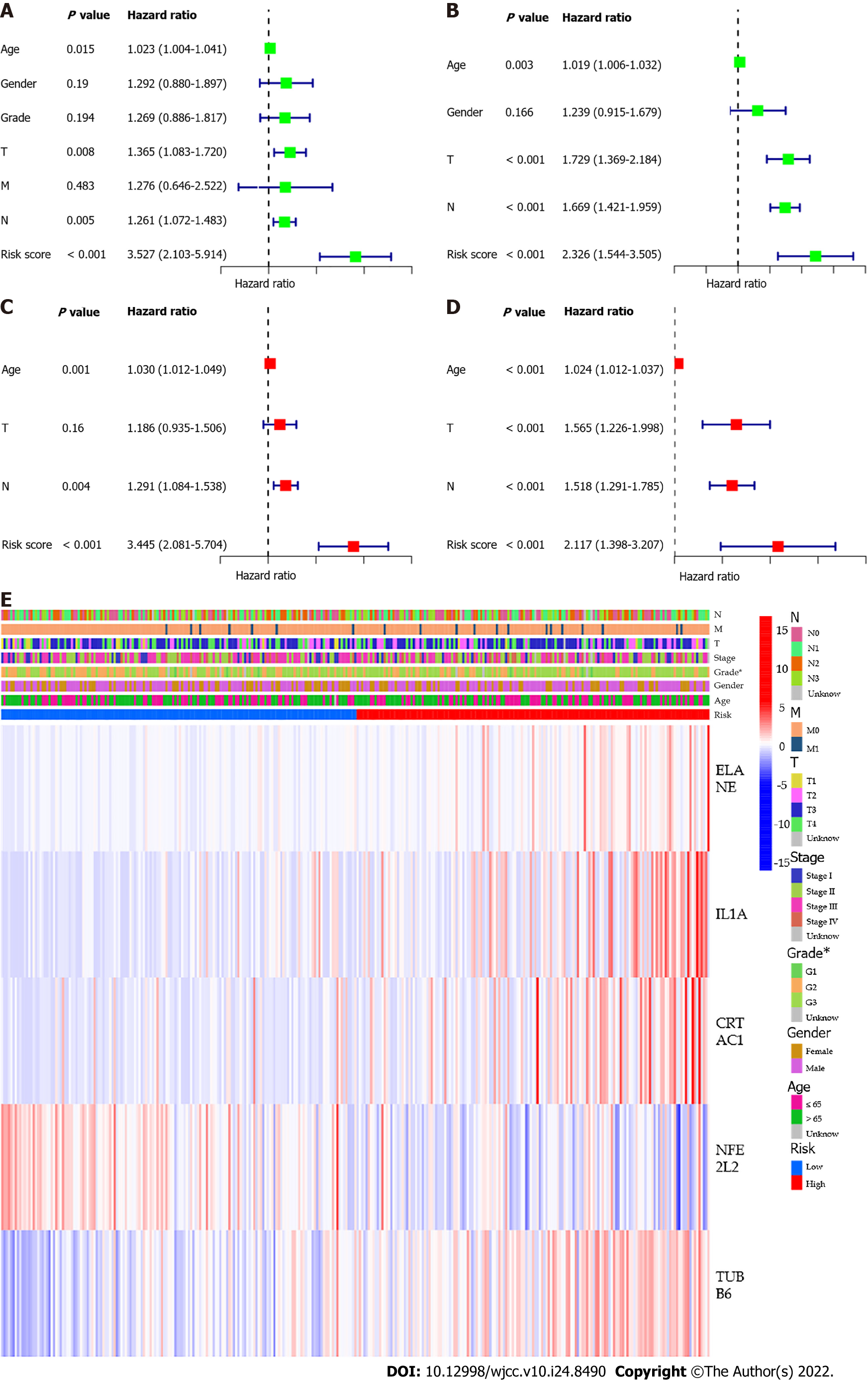
Univariate Cox regression analysis was used for preliminary screening of survival-related genes. Five genes that met the standard of P < 0.005 (ELANE, IL1A, CRTAC1, NFE2L2, TUBB6) were retained for further analysis (Figure 4A). By performing LASSO Cox regression analysis, 5 genes signature were constructed based on the optimal value of λ (Figure 4B and C). The risk score is calculated as follows: Risk score = (0.209 * ELANE exp.) + (0.255 * IL1A exp.) + (0.177 * CRTAC1 exp.) + (−0.326 * NFE2L2 exp.) + (0.123 * TUBB6 exp.). The GC patients were equally divided into low-risk and high-risk subgroups according to the median cutoff value (Figure 4D). PCA and t-SNE analysis showed the patients with different risk groups were well distributed into two clusters(Figure 4E and F). The patients with high risk had more deaths and shorter survival time than those with low risk (Figure 4G). Consistently, the Kaplan-Meier curve revealed that patients in the high-risk group had a significantly associated with poor OS (P =0.003, Figure 4H). Employing time-dependent ROC curves to evaluate the sensitivity and specificity of the prognostic model, we found that the area under the curve (AUC) was 0.627 for 1 year, 0.655 for 3 years, and 0.669 for 5 years (Figure 4I).
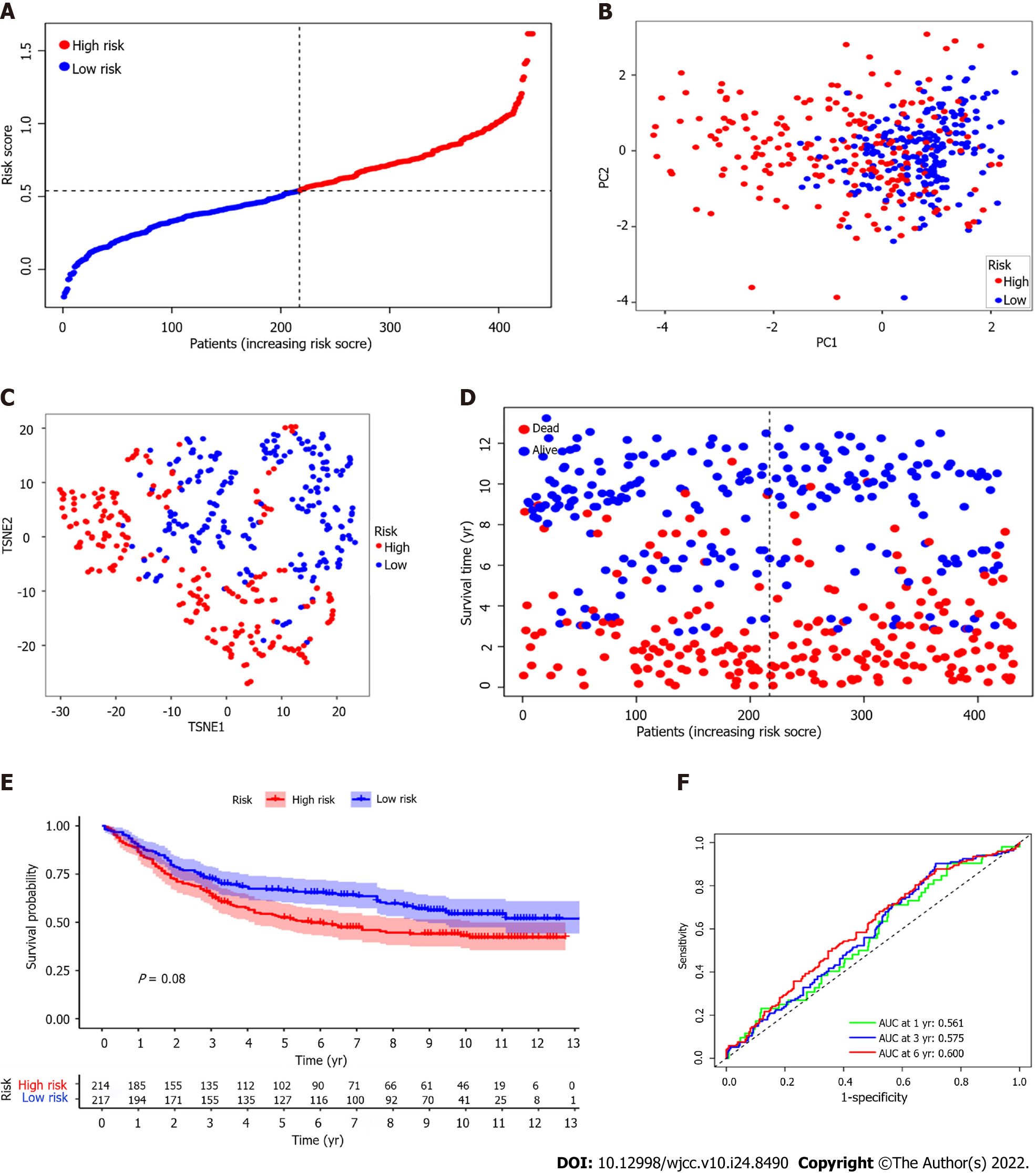
A total of 431 GC patients from the GEO cohort for external verification. According to the median risk score in the TCGA cohort, 247 patients in the GEO cohort were classified as high-risk group, while the other 184 patients were classified as low-risk group (Figure 5A). PCA and t-SNE analysis confirmed that patients in the two subgroups were distributed in discrete directions (Figure 5B and C). Similar to the results of the TCGA cohort, patients in the low-risk subgroup survived longer and lower mortality compared with those in the high-risk subgroup (Figure 5D). And the Kaplan-Meier analysis also showed that the survival time of the high-risk group was shorter (P = 0.008, Figure 5E). Besides, ROC curve analysis of the GEO cohort shows that the AUC was 0.561 for 1-year survival, 0.575 for 3-year survival, and 0.600 for 5-year survival (Figure 5F).
We used univariate and multivariate Cox regression analysis to assess whether the risk score was an independent prognostic predictor factor for OS. In univariate Cox regression analyses, the risk score was an independent factor predicting poor OS in both the TCGA and GEO cohorts (HR = 3.527, 95%CI: 2.103-5.914 and HR: 2.326, 95%CI: 1.544- 3.505, Figure 6A and B). Multivariate analysis also suggested that after correction for other confounding factors, the risk score was a prognostic factor for the two groups of GC patients (TCGA cohort: HR = 3.445, 95%CI: 2.081-5.704, GEO cohort: HR: 2.117, 95%CI: 1.398-3.207, Figure 6C and D). Besides, we generated a heatmap of clinical characteristics for the TCGA cohort and found that the degree of tumor differentiation was differently distributed between the low-risk and high-risk subgroups (P < 0.05, Figure 6E).
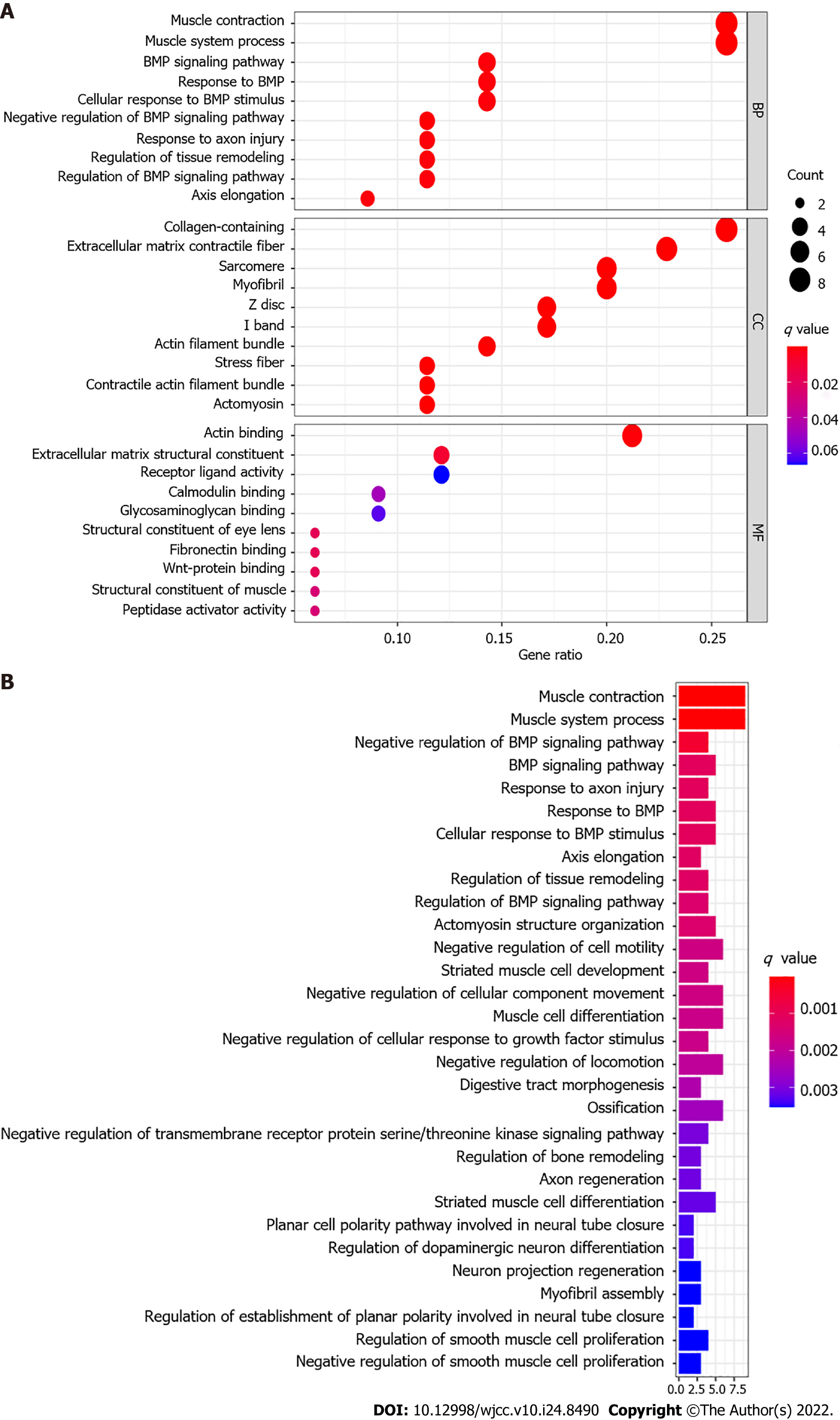
To elucidate the molecular mechanism between subgroups classified by the risk model, the DEGs between the high-risk and low-risk groups were used to perform GO enrichment and KEGG pathway analyses. GO functional enrichment analysis showed that the DEGs from the TCGA cohort were significantly enriched in muscle system, extracellular matrix, and receptor ligand activity (P adjusted < 0.05, Figure 7A). KEGG pathway analyses also indicated that DEGs are rich in some classic pathways, including bone morphogenetic protein (BMP) and regulation of transmembrane receptor protein serine/threonine kinase signaling pathway and regulation of cell motility (P adjusted < 0.05, Figure 7B).
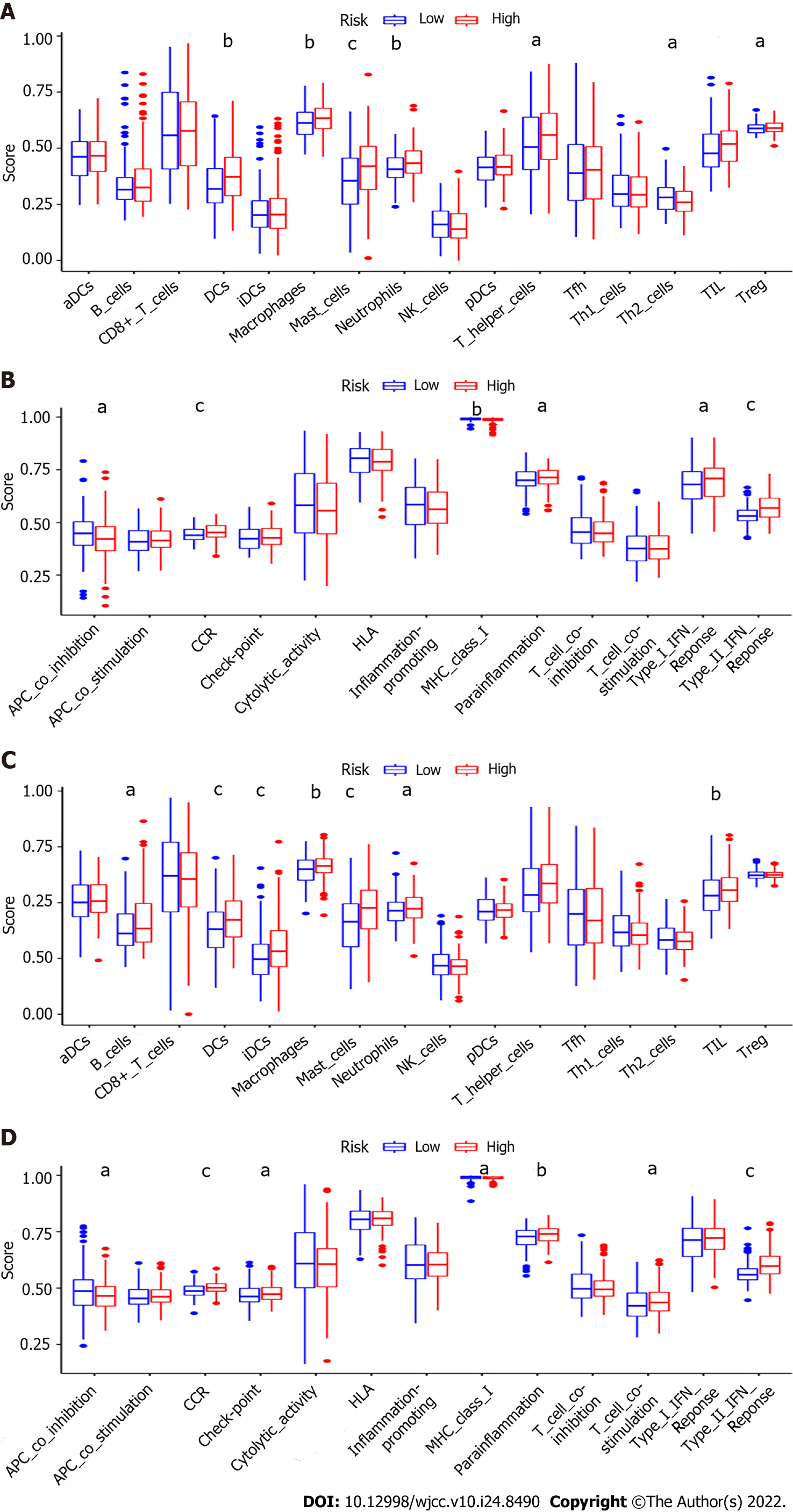
To further analyze the correlation between risk score and immune status, we used ssGSEA to compare the enrichment scores of 16 immune cells and the activities of 13 immune-related pathways in both cohorts. In the TCGA cohort, the high-risk subgroup usually has significant immune cell infiltration, especially dendritic cells(DCs), macrophages, mast cells, neutrophils, T helper 2 cells (Th2) and tumor infiltrating lymphocytes (TIL) (P adjusted < 0.05, Figure 8A). Six immune-related pathways were validated in the TCGA cohort, except for the APC co inhibition and major histocompatibility complex (MHC) class Ⅰ pathway, the activities of the other four immune pathways in the high-risk group were higher than those in the low-risk group (P adjusted < 0.05, Figure 8B). When evaluating the immune status in the GEO cohort, a similar conclusion was reached (P adjusted < 0.05, Figure 8C and D).
In the present study, we studied the expression of 118 pyroptosis-related genes in GC tissues and their association with OS. First, GC patients were classified according to the pyroptosis-related DEGs. However, the two clusters did not show significant differences in clinical characteristics. To create a better clinical application, we constructed a new prognostic signature of 5 pyroptosis-related genes through Cox univariate analysis and LASSO Cox regression analysis, and verified it in an external cohort.
Pyroptosis is a form of programmed cell death. Recent studies have shown that pyroptosis has become a new hot spot in cancer research, because it is closely related to the occurrence and development of cancer and may affect various stages of cancer. It also plays different roles in many cancers[13]. Cell pyroptosis promotes the secretion of proinflammatory factors. On the one hand, chronic inflammation caused by pyroptosis can form a microenvironment suitable for tumor cell growth, thereby promoting tumor growth, including immunosuppression, proliferation, angiogenesis and metastasis. On the other hand, it can maintain intestinal barrier integrity to suppress the occurrence of tumor development[14,15]. It has the effect of inhibiting tumor growth in ovarian cancer, colorectal cancer, and liver cancer[11,16,17], but has a bidirectional effect on breast cancer[18]. However, the effect of pyroptosis on the prognosis of GC is still unclear. Therefore, we explored as many genes as possible related to pyroptosis and found 5 pyroptosis-related genes (ELANE, IL1A, CRTAC1, NFE2L2 and TUBB6) w significantly differentially expression in GC and related to overall survival.
ELANE encodes neutrophil elastase, which is a protease packaged in the primary particles of neutrophil precursors[19]. Kambara et al[20] proved that the lysis and activation of gasdermin D(GSDMD) in neutrophils can be mediated by ELANE, and the efficiency of inducing cell pyroptosis is the same as that of gasdermin D N-terminal fragment (GSDMD-cNT). A recent study showed that ELANE proteolysis releases the CD95 death domain, which interacts with histone H1 to selectively kill cancer cells with the least toxicity to noncancer cells[21]. Pyroptosis depends on the activation of caspase-1 family members and leads to the release of interleukin-1 family cytokines. IL-1A, as an important member of the IL-1 family, is a proinflammatory cytokines, on the one hand, participates in the transformation, growth, invasion, and metastasis of malignant tumors, on the other hand, it can activate the body's immune system to limit tumor growth[22]. In the terminal differentiation process of human keratinocytes in vitro, multiple steps are involved in pyroptosis, among which the expression levels of pro-inflammatory IL-1A and IL-1B and pyroptosis pore-forming GSDMD are downregulated[23]. Recent studies have shown that genetic variants of IL-1A single nucleotide polymorphisms affect the risk of ovarian cancer, and genetic variants in the IL-1A gene region lead to susceptibility to GC[24,25]. Cartilage acid protein 1 (CRTAC1) is a novel human marker that can be used to distinguish human chondrocytes from osteoblasts and mesenchymal stem cells. Sun et al[26] found that pyroptosis markers (NLRP3, active Caspase-1, pro-Caspase-1, and GSDMD) can be induced in human lens epithelial cells by ultraviolet B (UVB) irradiation, and the downregulation of CRTAC1 significantly reversed the UVB induced cell pyroptosis. At present, there are no studies on pyroptosis and CRTAC1 in GC, but these studies may provide new findings for prognostic markers of GC. NFE2L2 is involved in the coding of injury and inflammation-related proteins, including the production of free radicals[27]. Bai et al[28] showed that reactive oxygen species (ROS) induce pyroptosis of nucleus pulposus (NPC) through the NLR family pyrin domain containing 3 (NLRP3) / PYD and CARD domain containing (PYCARD) pathway, but the increased ROS level also increases the expression of the transcription factor NFE2L2 and inhibits the pyroptosis of NPC. Activation of NFE2L2 was also found to be accompanied by inhibition of NLRP3 inflammasome in many different models of inflammatory disease. In our risk model, NFE2L2 is enriched in normal tissues and low-risk groups, which helps to prolong patient survival, indicating that NFE2L2 may play a protective role in GC. TUBB6 is a novel isoform of β-tubulin, which forms microtubules and participates in the cytoskeleton, and biological functions such as cell division, differentiation and migration, and intracellular transport[29]. The experiment of Salinas et al[30] showed that knocking down the expression of TUBB6 can increase cell pyroptosis without changing IL-1β secretion, indicating that TUBB6 only affects the cell death aspect of this pathway, and may act downstream of caspase-1 activation. TUBB6 was once considered a potential mutation hotspot gene in colorectal cancer[31]. A recent GC whole-genome and transcriptome sequencing experiment found that some nonsynonymous mutations can lead to increased gene activity and mRNA expression (including TUBB6) Up-regulation suggesting that it may contribute to the wide spread of GC cells in the abdominal cavity[32]. All in all, these five genes are involved in pyroptosis and affect the progression of cancer cells, but whether it plays a role in GC requires more exploration.
Although the underlying mechanism of tumor susceptibility to pyroptosis has been a hot research area for some time in the past, the potential adjustment between tumor immunity and pyroptosis has remained elusive. We performed functional enrichment in GO, KEGG, and ssGSEA based on DEGs between different risk groups. In GO and KEGG analysis, biological pathways such as “muscle system”, “extracellular matrix”, “actin”, “regulation of cell motility”, and “BMP signaling pathway” were significantly enriched, emphasizing the role of pyroptosis-related genes in GC. We then performed the immune function and immune cell analysis between the two groups. Antigen-presenting cells, including DCs and T helper cells, can help present pyroptosis cells to T cells and co-stimulate T cells and paracrine interferon triggers a subsequent responses. Interestingly, immune function and immune cell scores were generally higher in the high-risk group than in the low-risk group in this study. In view of the limited data from GC and the characteristics of tumor heterogeneity, our results on immune infiltration provide some insights for further research. In addition, how the immune system related to pyroptosis plays a role in GC requires more in vitro and in vivo exploration and verification.
Our research aims to identify DEGs and establish a prognostic model associating pyroptosis with the prognosis of GC patients. Although we have conducted multiangle and multidatabase verification, there are still limitations in this study that need to be considered. First, our prognostic model is constructed and verified through retrospective data from public databases. Further validation by in vitro and in vivo experiments with larger sample sizes is needed to better assess the relationship between prognostic model and pyroptosis. In addition, the association between risk score and immune activity needs to be mined or established for more GC immunotherapy data. Some single-cell sequencing results can explain the specific changes in the tumor microenvironment, which is also an aspect of our future attention.
In conclusion, our study defines a new prognostic model of 5 pyroptosis-related genes. The model was found to independently correlate with OS, providing in-depth understanding of GC prognosis prediction and providing an important basis for future studies on the association between pyroptosis-related genes and GC immunity. The underlying mechanism between pyroptosis-related genes in GC and tumor immunity is still poorly understood, and further research is needed. Such challenges motivate us to continue our efforts.
The expression of pyroptosis genes in gastric cancer (GC) has not been well analyzed for its correlation with the prognosis of GC and the immune infiltration of tumors.
This study provides some insights for a deeper understanding of GC pathogenesis.
To explore the correlation between pyroptosis genes and prognosis of GC.
The authors constructed prognostic multigene markers of differentially expressed genes associated with pyroptosis by least absolute contraction and selection operator Cox regression. The risk model was analyzed by Kaplan-Meier curve, two-sided log-rank test and functional enrichment analysis.
Based on 63 pyroptosis differentially expressed genes, 5 gene signature were constructed and all GC patients were classified into two risk groups. Multivariate Cox regression analyses showed that the risk score was an independent risk factor for overall survival (OS). The immune function and immune cell scores of the high-risk group were generally higher than those of the low-risk group. Similar results were obtained in external validation.
This novel model, which contains 5 pyroptosis-related genes, is an independent predicting factor for OS in GC patients. Pyroptosis-related genes play a significant role in tumor immune microenvironment.
The underlying mechanism between pyroptosis-related genes in GC and tumor immunity is still poorly understood, and further research is needed.
Thanks to all public databases for sharing and uploading their data.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Batyrbekov K, Kazakhstan; Mohamed SY, Egypt S-Editor: Ma YJ L-Editor: A P-Editor: Li X
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55823] [Article Influence: 7974.7] [Reference Citation Analysis (132)] |
| 2. | Johnston FM, Beckman M. Updates on Management of Gastric Cancer. Curr Oncol Rep. 2019;21:67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 316] [Article Influence: 52.7] [Reference Citation Analysis (1)] |
| 3. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12667] [Cited by in RCA: 15314] [Article Influence: 3062.8] [Reference Citation Analysis (4)] |
| 4. | Cao M, Li H, Sun D, Chen W. Cancer burden of major cancers in China: A need for sustainable actions. Cancer Commun (Lond). 2020;40:205-210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 226] [Cited by in RCA: 309] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 5. | Kovacs SB, Miao EA. Gasdermins: Effectors of Pyroptosis. Trends Cell Biol. 2017;27:673-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 706] [Cited by in RCA: 985] [Article Influence: 123.1] [Reference Citation Analysis (0)] |
| 6. | Rogers C, Fernandes-Alnemri T, Mayes L, Alnemri D, Cingolani G, Alnemri ES. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat Commun. 2017;8:14128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 578] [Cited by in RCA: 1099] [Article Influence: 137.4] [Reference Citation Analysis (0)] |
| 7. | Xia X, Wang X, Cheng Z, Qin W, Lei L, Jiang J, Hu J. The role of pyroptosis in cancer: pro-cancer or pro-"host"? Cell Death Dis. 2019;10:650. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 306] [Cited by in RCA: 610] [Article Influence: 101.7] [Reference Citation Analysis (0)] |
| 8. | Broz P, Pelegrín P, Shao F. The gasdermins, a protein family executing cell death and inflammation. Nat Rev Immunol. 2020;20:143-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 449] [Cited by in RCA: 1068] [Article Influence: 178.0] [Reference Citation Analysis (0)] |
| 9. | Kesavardhana S, Malireddi RKS, Kanneganti TD. Caspases in Cell Death, Inflammation, and Pyroptosis. Annu Rev Immunol. 2020;38:567-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 621] [Article Influence: 124.2] [Reference Citation Analysis (0)] |
| 10. | Rébé C, Raveneau M, Chevriaux A, Lakomy D, Sberna AL, Costa A, Bessède G, Athias A, Steinmetz E, Lobaccaro JM, Alves G, Menicacci A, Vachenc S, Solary E, Gambert P, Masson D. Induction of transglutaminase 2 by a liver X receptor/retinoic acid receptor alpha pathway increases the clearance of apoptotic cells by human macrophages. Circ Res. 2009;105:393-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 87] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 11. | Li J, Yang C, Li Y, Chen A, Li L, You Z. LncRNA GAS5 suppresses ovarian cancer by inducing inflammasome formation. Biosci Rep. 2018;38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 92] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 12. | Nadatani Y, Huo X, Zhang X, Yu C, Cheng E, Zhang Q, Dunbar KB, Theiss A, Pham TH, Wang DH, Watanabe T, Fujiwara Y, Arakawa T, Spechler SJ, Souza RF. NOD-Like Receptor Protein 3 Inflammasome Priming and Activation in Barrett's Epithelial Cells. Cell Mol Gastroenterol Hepatol. 2016;2:439-453. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 54] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 13. | Bedoui S, Herold MJ, Strasser A. Emerging connectivity of programmed cell death pathways and its physiological implications. Nat Rev Mol Cell Biol. 2020;21:678-695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 628] [Article Influence: 125.6] [Reference Citation Analysis (0)] |
| 14. | Zhou CB, Fang JY. The role of pyroptosis in gastrointestinal cancer and immune responses to intestinal microbial infection. Biochim Biophys Acta Rev Cancer. 2019;1872:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 119] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 15. | Karki R, Kanneganti TD. Diverging inflammasome signals in tumorigenesis and potential targeting. Nat Rev Cancer. 2019;19:197-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 468] [Article Influence: 78.0] [Reference Citation Analysis (0)] |
| 16. | Ma X, Guo P, Qiu Y, Mu K, Zhu L, Zhao W, Li T, Han L. Loss of AIM2 expression promotes hepatocarcinoma progression through activation of mTOR-S6K1 pathway. Oncotarget. 2016;7:36185-36197. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 94] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 17. | Zaki MH, Vogel P, Body-Malapel M, Lamkanfi M, Kanneganti TD. IL-18 production downstream of the Nlrp3 inflammasome confers protection against colorectal tumor formation. J Immunol. 2010;185:4912-4920. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 337] [Cited by in RCA: 329] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 18. | Chen LC, Wang LJ, Tsang NM, Ojcius DM, Chen CC, Ouyang CN, Hsueh C, Liang Y, Chang KP, Chang YS. Tumour inflammasome-derived IL-1β recruits neutrophils and improves local recurrence-free survival in EBV-induced nasopharyngeal carcinoma. EMBO Mol Med. 2012;4:1276-1293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 140] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 19. | Fu Z, Akula S, Thorpe M, Hellman L. Potent and Broad but not Unselective Cleavage of Cytokines and Chemokines by Human Neutrophil Elastase and Proteinase 3. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 20. | Kambara H, Liu F, Zhang X, Liu P, Bajrami B, Teng Y, Zhao L, Zhou S, Yu H, Zhou W, Silberstein LE, Cheng T, Han M, Xu Y, Luo HR. Gasdermin D Exerts Anti-inflammatory Effects by Promoting Neutrophil Death. Cell Rep. 2018;22:2924-2936. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 300] [Cited by in RCA: 323] [Article Influence: 46.1] [Reference Citation Analysis (0)] |
| 21. | Cui C, Chakraborty K, Tang XA, Zhou G, Schoenfelt KQ, Becker KM, Hoffman A, Chang YF, Blank A, Reardon CA, Kenny HA, Vaisar T, Lengyel E, Greene G, Becker L. Neutrophil elastase selectively kills cancer cells and attenuates tumorigenesis. Cell. 2021;184:3163-3177.e21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 205] [Article Influence: 51.3] [Reference Citation Analysis (0)] |
| 22. | Yuan J, Najafov A, Py BF. Roles of Caspases in Necrotic Cell Death. Cell. 2016;167:1693-1704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 220] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 23. | Lachner J, Mlitz V, Tschachler E, Eckhart L. Epidermal cornification is preceded by the expression of a keratinocyte-specific set of pyroptosis-related genes. Sci Rep. 2017;7:17446. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 66] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 24. | Durães C, Muñoz X, Bonet C, García N, Venceslá A, Carneiro F, Peleteiro B, Lunet N, Barros H, Lindkvist B, Boutron-Ruault MC, Bueno-de-Mesquita HB, Rizzato C, Trichopoulou A, Weiderpass E, Naccarati A, Travis RC, Tjønneland A, Gurrea AB, Johansson M, Riboli E, Figueiredo C, González CA, Capellà G, Machado JC, Sala N. Genetic variants in the IL1A gene region contribute to intestinal-type gastric carcinoma susceptibility in European populations. Int J Cancer. 2014;135:1343-1355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 25. | White KL, Schildkraut JM, Palmieri RT, Iversen ES Jr, Berchuck A, Vierkant RA, Rider DN, Charbonneau B, Cicek MS, Sutphen R, Birrer MJ, Pharoah PP, Song H, Tyrer J, Gayther SA, Ramus SJ, Wentzensen N, Yang HP, Garcia-Closas M, Phelan CM, Cunningham JM, Fridley BL, Sellers TA, Goode EL; Ovarian Cancer Association Consortium. Ovarian cancer risk associated with inherited inflammation-related variants. Cancer Res. 2012;72:1064-1069. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 40] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 26. | Sun Y, Rong X, Li D, Jiang Y, Lu Y, Ji Y. Down-regulation of CRTAC1 attenuates UVB-induced pyroptosis in HLECs through inhibiting ROS production. Biochem Biophys Res Commun. 2020;532:159-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 27. | Pajares M, Rojo AI, Arias E, Díaz-Carretero A, Cuervo AM, Cuadrado A. Transcription factor NFE2L2/NRF2 modulates chaperone-mediated autophagy through the regulation of LAMP2A. Autophagy. 2018;14:1310-1322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 160] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 28. | Bai Z, Liu W, He D, Wang Y, Yi W, Luo C, Shen J, Hu Z. Protective effects of autophagy and NFE2L2 on reactive oxygen species-induced pyroptosis of human nucleus pulposus cells. Aging (Albany NY). 2020;12:7534-7548. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 73] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 29. | Jaglin XH, Chelly J. Tubulin-related cortical dysgeneses: microtubule dysfunction underlying neuronal migration defects. Trends Genet. 2009;25:555-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 134] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 30. | Salinas RE, Ogohara C, Thomas MI, Shukla KP, Miller SI, Ko DC. A cellular genome-wide association study reveals human variation in microtubule stability and a role in inflammatory cell death. Mol Biol Cell. 2014;25:76-86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 31. | Gylfe AE, Kondelin J, Turunen M, Ristolainen H, Katainen R, Pitkänen E, Kaasinen E, Rantanen V, Tanskanen T, Varjosalo M, Lehtonen H, Palin K, Taipale M, Taipale J, Renkonen-Sinisalo L, Järvinen H, Böhm J, Mecklin JP, Ristimäki A, Kilpivaara O, Tuupanen S, Karhu A, Vahteristo P, Aaltonen LA. Identification of candidate oncogenes in human colorectal cancers with microsatellite instability. Gastroenterology. 2013;145:540-3.e22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 32. | Zhang J, Huang JY, Chen YN, Yuan F, Zhang H, Yan FH, Wang MJ, Wang G, Su M, Lu G, Huang Y, Dai H, Ji J, Zhang J, Zhang JN, Jiang YN, Chen SJ, Zhu ZG, Yu YY. Whole genome and transcriptome sequencing of matched primary and peritoneal metastatic gastric carcinoma. Sci Rep. 2015;5:13750. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |