Copyright
©The Author(s) 2022.
World J Clin Cases. Aug 26, 2022; 10(24): 8490-8505
Published online Aug 26, 2022. doi: 10.12998/wjcc.v10.i24.8490
Published online Aug 26, 2022. doi: 10.12998/wjcc.v10.i24.8490
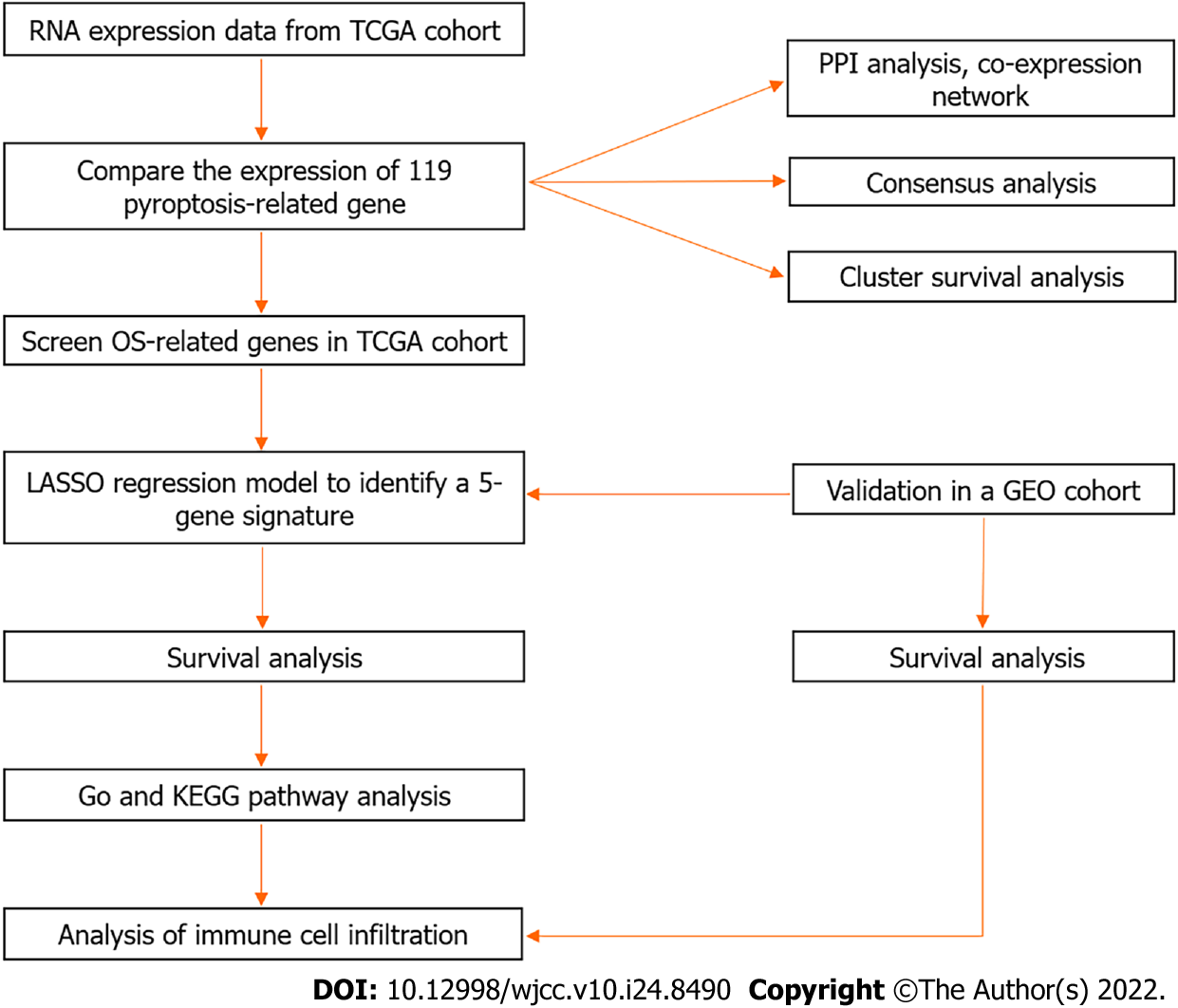
Figure 1 Workflow diagram.
TCGA: The Cancer Genome Atlas; LASSO: least absolute contraction and selection operator; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; PPI: Protein–protein interaction; GEO: Gene Expression Omnibus.
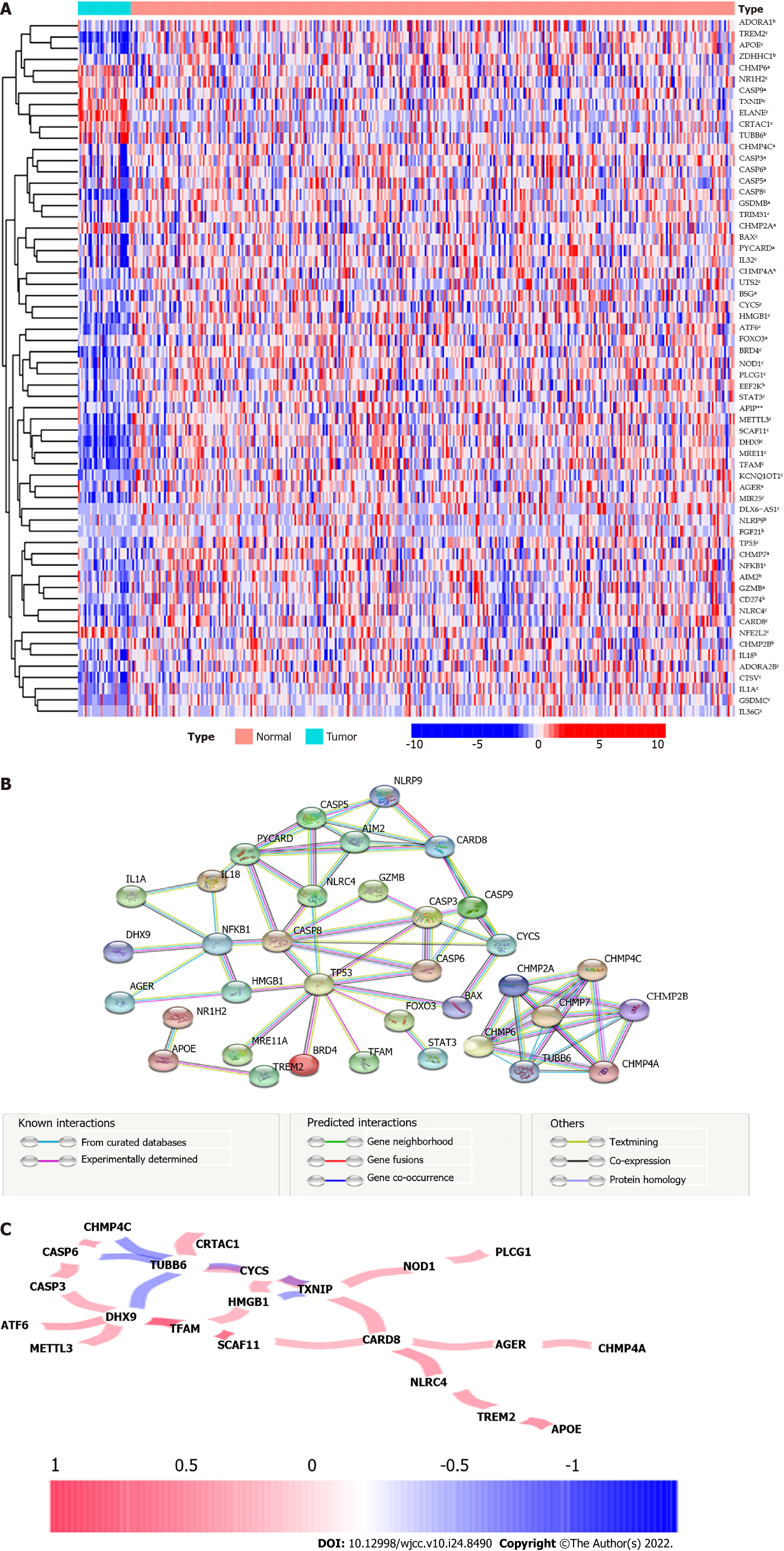
Figure 2 Interaction among 199 pyroptosis-related genes.
A: Heatmap (blue: low expression level; red: high expression level) of the pyroptosis-related genes between the adjacent nontumor tissues (brilliant blue) and the tumor tissues (red). P values were showed as: aP < 0.05, bP < 0.01, and cP < 0.001; B: Protein–protein interaction network reflecting the interactions of the differentially expressed genes (DEGs); C: The correlation network of DEGs. The correlation coefficients are represented by different colors. Red line: Positive correlation; Blue line: Negative correlation. The depth of the colors indicates the strength of the relevance.
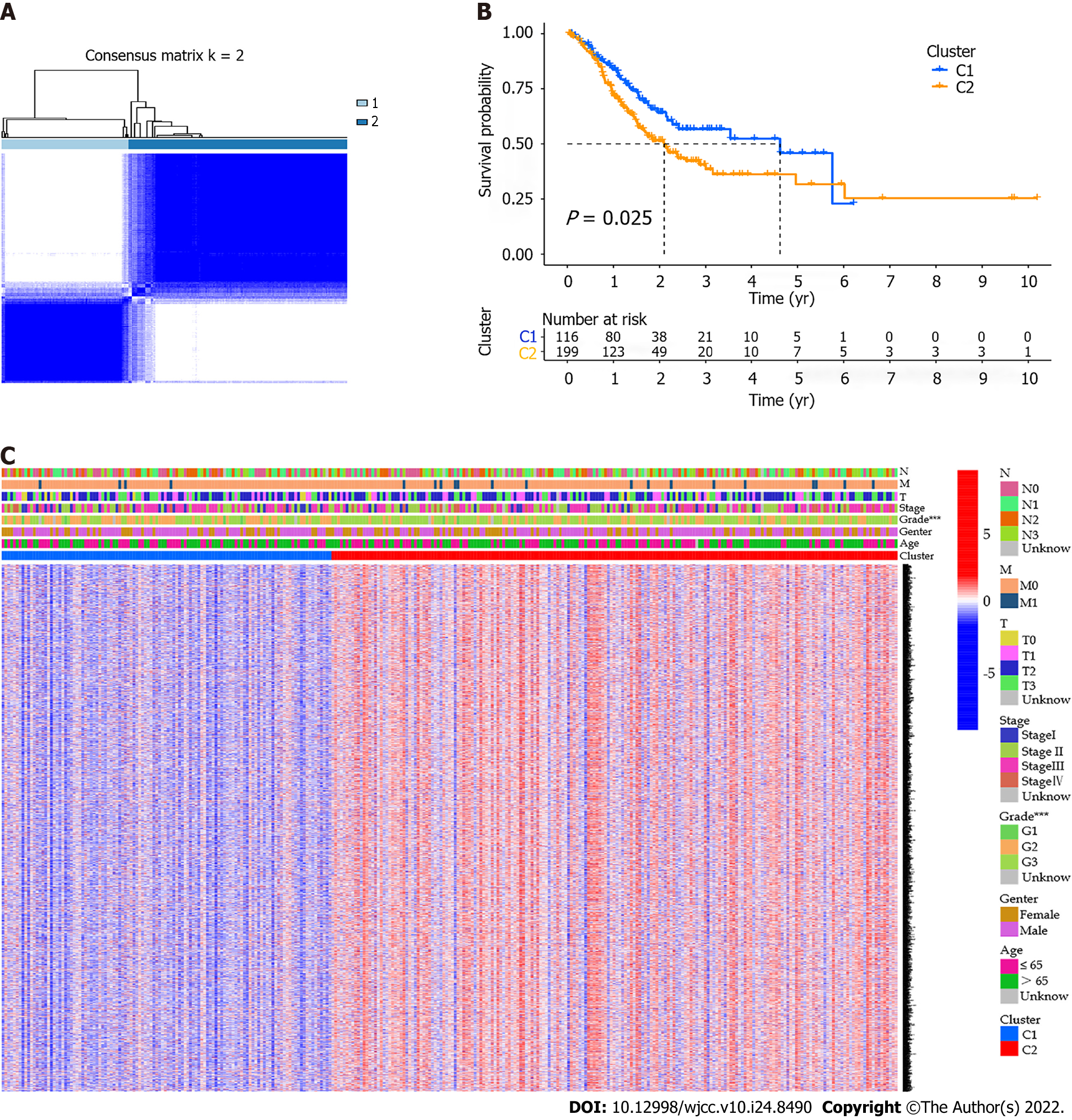
Figure 3 Tumor classification based on the pyroptosis-related differentially expressed genes.
A: Gastric cancer patients were classified into two clusters based on the consensus clustering matrix (k = 2); B: Kaplan-Meier overall survival curves for the two clusters; C: Heatmap and the clinical features of the two clusters classified by these differentially expressed genes (DEGs).
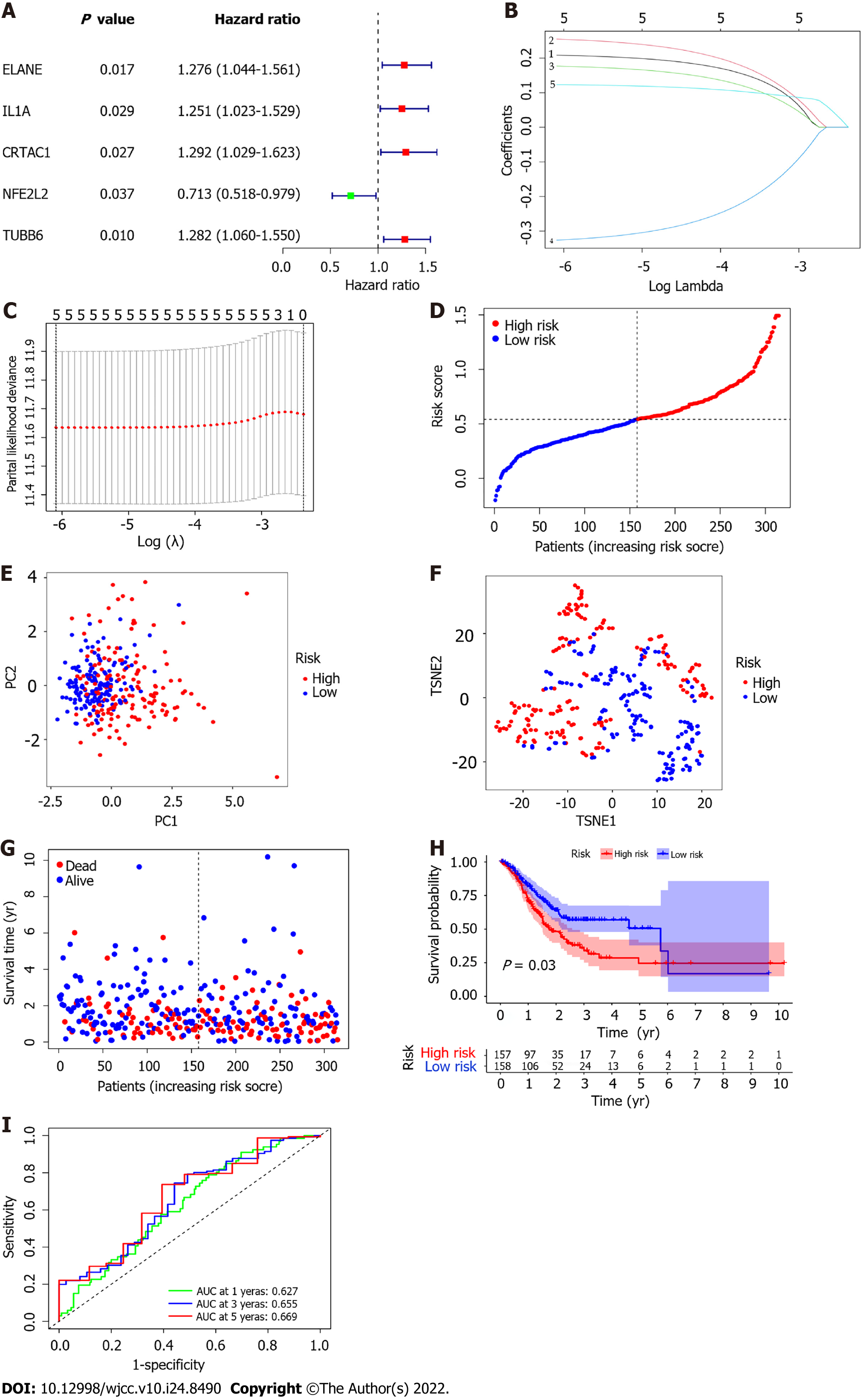
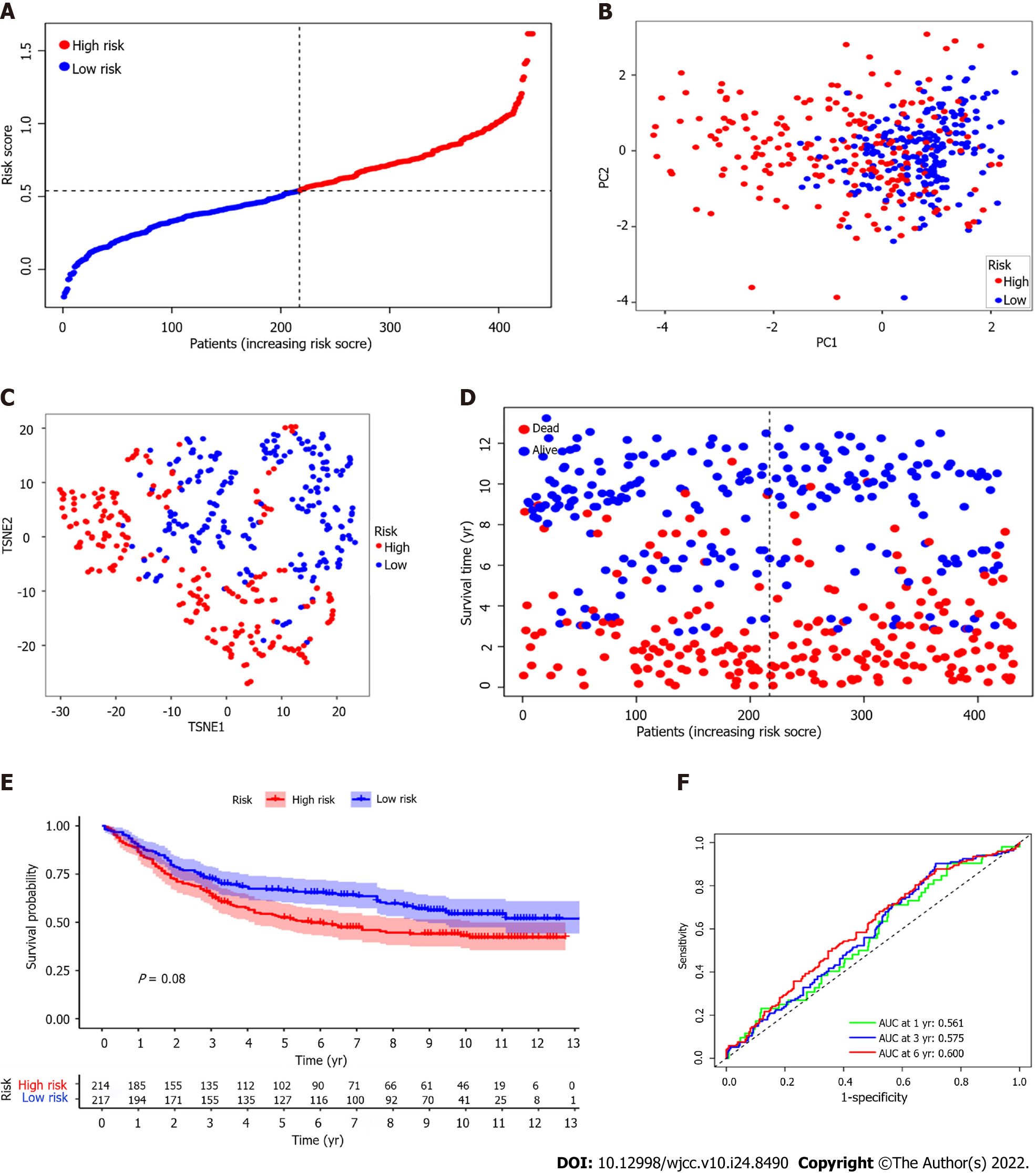
Figure 4 Construction of risk signature in The Cancer Genome Atlas cohort.
A: Univariate Cox regression analysis of overall survival (OS) for each pyroptosis-related genes, and 23 genes with P < 0.001; B and C: The results of the least absolute contraction and selection operator (LASSO) Cox regression indicated that there were 11 OS-related genes; D: Distribution of patients based on the median value of risk score; E and F: Principal component analysis (PCA) plot and t-distributed stochastic neighbor embedding (t-SNE) analysis founded on the risk score; G: The distributions of OS status, OS and risk score for each patient; H: Kaplan-Meier curves for the OS of patients in the high-risk and low-risk groups; I: Area under the curve (AUC) of time-dependent receiver operating characteristic (ROC) curves demonstrated the sensitivity and specificity of the risk score.
Figure 5 Validation of the risk model in the Gene Expression Omnibus cohort.
A: Distribution of patients in the Gene Expression Omnibus (GEO) cohort based on the median risk score in The Cancer Genome Atlas (TCGA) cohort; B and C: Principal component analysis (PCA) plot and t-distributed stochastic neighbor embedding (t-SNE) analysis for gastric cancer (GC) patients in the GEO cohort; D: The distributions of overall survival (OS) status, OS and risk score for each patient in the GEO cohort; E: Kaplan-Meier curves for comparison of the OS between low-risk and high-risk groups in the GEO cohort; F: Time-dependent receiver operating characteristic (ROC) curves for GC in the GEO cohort.
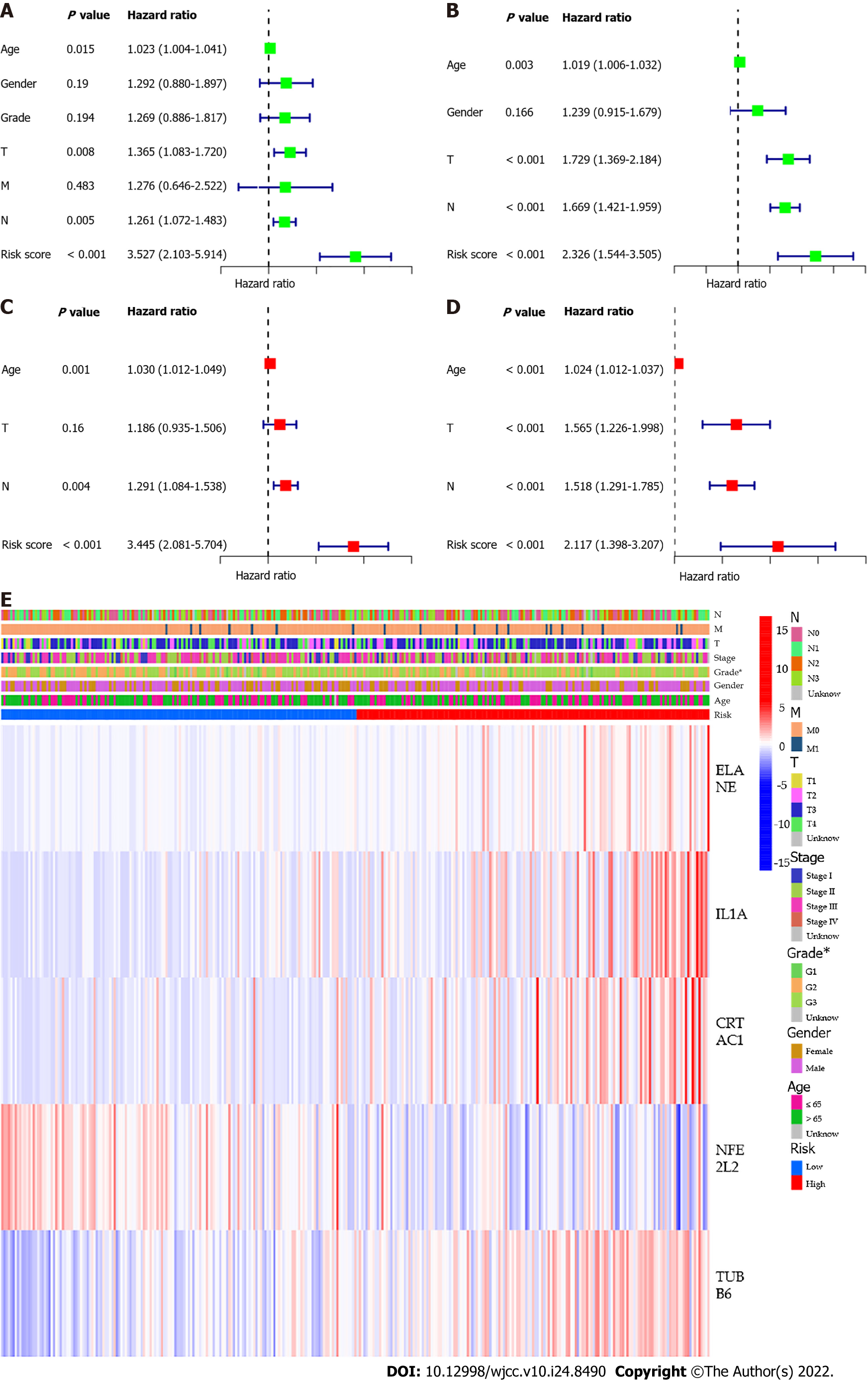
Figure 6 Univariate and multivariate Cox regression analysis of risk signature.
A: Univariate analysis for The Cancer Genome Atlas (TCGA) cohort; B: Univariate analysis for the Gene Expression Omnibus (GEO) cohort; C: Multivariate analysis for the TCGA cohort; D: Multivariate analysis for the GEO cohort; E: Heatmap for the connections between clinicopathologic features and the risk groups.
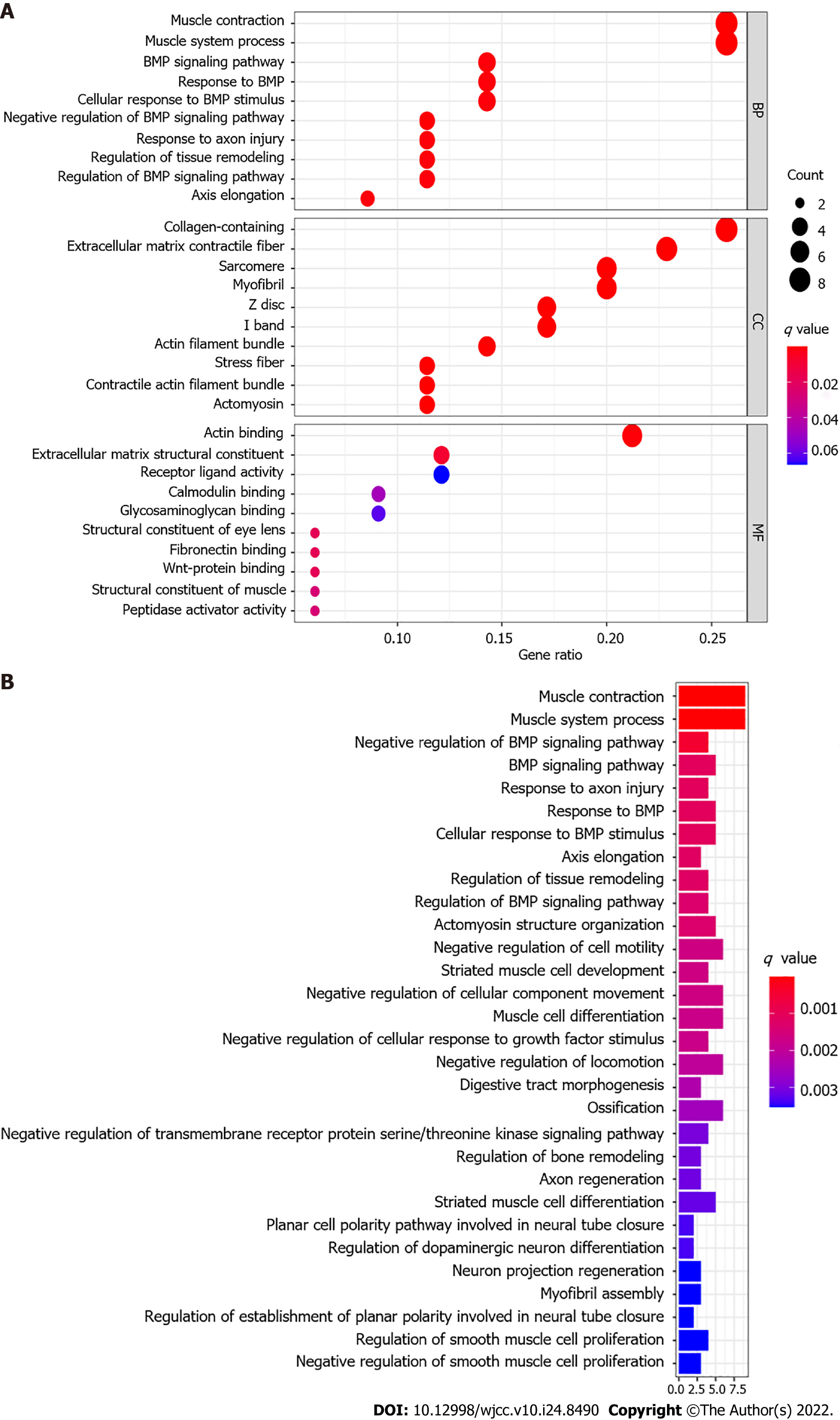
Figure 7 Functional analysis based on the differentially expressed genes in The Cancer Genome Atlas cohort.
A: The most significant Gene Ontology (GO) enrichment in The Cancer Genome Atlas (TCGA) cohort (q-value: the adjusted P value); B: The most significant Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in the TCGA cohort (q-value: the adjusted P value).
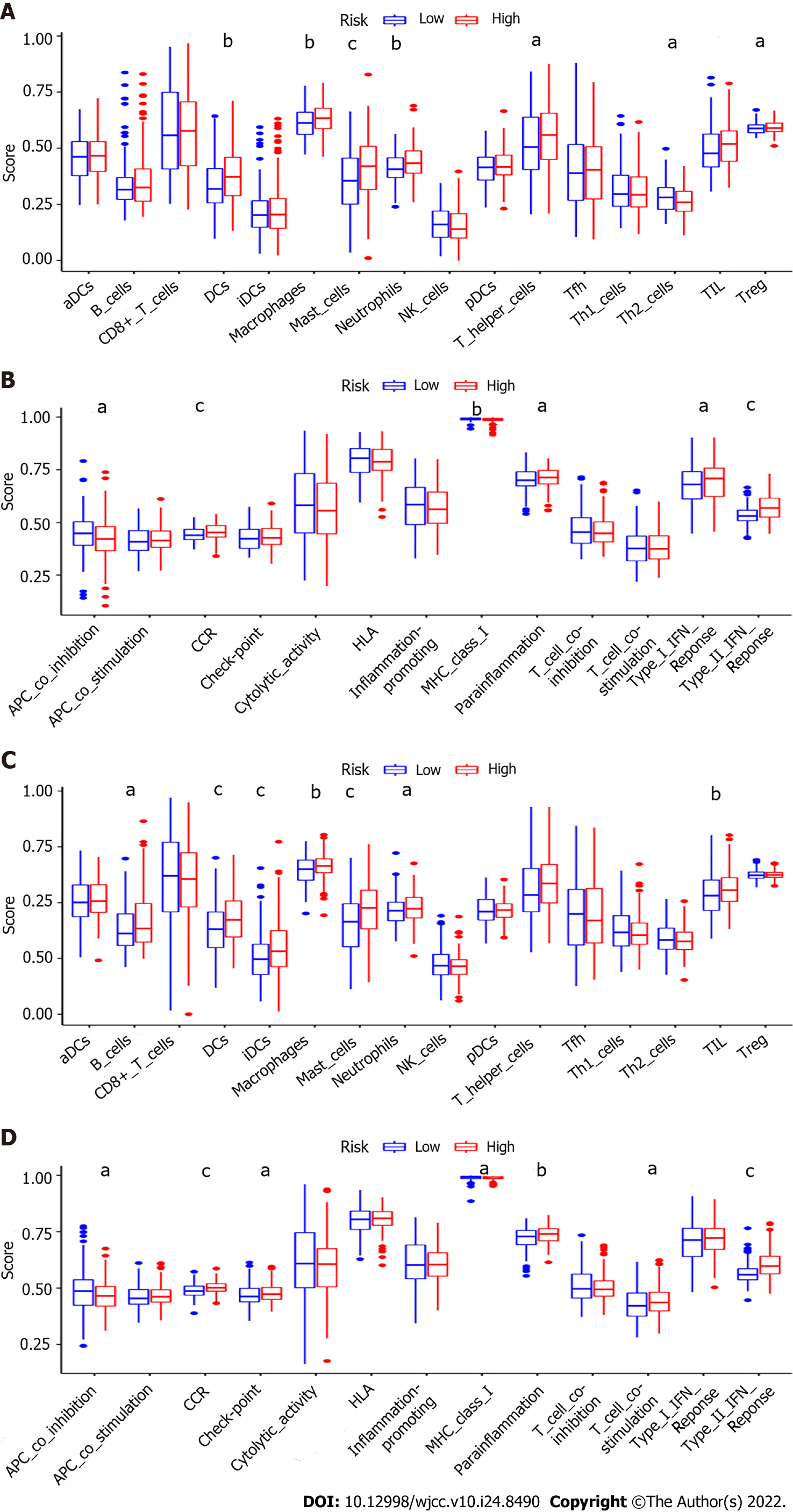
Figure 8 Comparison of the single sample gene set enrichment analysis scores for immune cells and immune pathways.
A and B: Comparison of the enrichment scores of 16 types of immune cells and 13 immune-related pathways between different risk groups in The Cancer Genome Atlas (TCGA) cohort; C and D: Comparison of the immune infiltration between different risk groups in the Gene Expression Omnibus (GEO) cohort. P values were showed as: aP < 0.05, bP < 0.01, and cP < 0.001.
- Citation: Guan SH, Wang XY, Shang P, Du QC, Li MZ, Xing X, Yan B. Pyroptosis-related genes play a significant role in the prognosis of gastric cancer. World J Clin Cases 2022; 10(24): 8490-8505
- URL: https://www.wjgnet.com/2307-8960/full/v10/i24/8490.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i24.8490