Published online Apr 15, 2020. doi: 10.4251/wjgo.v12.i4.394
Peer-review started: December 21, 2019
First decision: January 19, 2020
Revised: February 4, 2020
Accepted: March 22, 2020
Article in press: March 22, 2020
Published online: April 15, 2020
Processing time: 116 Days and 4.1 Hours
Gastric cancer (GC) is one of the most aggressive malignancies, with a high incidence and poor prognosis worldwide. Recently, accumulating evidence has illustrated that long noncoding RNAs (lncRNAs) play pivotal roles in many cancers. It has been reported that LINC00511 contributes to tumorigenesis in various diseases. However, the role of LINC00511 in GC cell growth remains mostly unknown.
To determine whether the lncRNA LINC00511 exerted its carcinogenic function in GC via the miR-124-3p/PDK4 axis.
Cell culture and transfection, RNA extraction and quantitative real-time PCR, CCK-8 assay, Colony formation assay, Luciferase reporter assay, RIP assay, RNA pull-down assay, and Western blot analysis were used to show expression and mechanisms of LINC00511 in GC progression and apoptosis. Rescue assays were performed to verify the relationships among LINC00511, miR-124-3p and PDK4 further.
The expression of LINC00511 was remarkably upregulated in GC cells compared to that in corresponding normal cell lines. Compared to the controls, cell proliferation was inhibited, and cell apoptosis was increased upon LINC00511 knockdown, demonstrating that LINC00511 influenced GC cell growth. An exploration of the molecular mechanism revealed that LINC00511 functioned as a molecular sponge of miR-124-3p and that PDK4 was a downstream target of miR-124-3p in GC. Rescue assays showed that the overexpression of PDK4 could partly restore the inhibitory function of si-LINC00511 in GC.
These data demonstrate that LINC00511 promotes gastric cancer cell growth by acting as a ceRNA to regulate the miR-124-3p/PDK4 axis, which may be a promising therapeutic target for GC.
Core tip: In this study, we aimed to determine whether the long noncoding RNAs LINC00511 exerted its carcinogenic function in gastric cancer (GC) via the miR-124-3p/PDK4 axis. First, we investigated the expression level of LINC00511 in GC cell lines, and we then examined the biological function of LINC00511 with functional assays. Next, we predicted and confirmed the interaction between LINC00511 and miR-124-3p. Furthermore, we further found that PDK4 was a downstream target of miR-124-3p. Finally, we concluded that LINC00511 plays an oncogenic role in GC by sponging miR-124-3p and targeting PDK4, indicating that LINC00511 may be a new molecular biomarker for GC.
- Citation: Sun CB, Wang HY, Han XQ, Liu YN, Wang MC, Zhang HX, Gu YF, Leng XG. LINC00511 promotes gastric cancer cell growth by acting as a ceRNA. World J Gastrointest Oncol 2020; 12(4): 394-404
- URL: https://www.wjgnet.com/1948-5204/full/v12/i4/394.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v12.i4.394
Gastric cancer (GC), as a kind of heterogeneous disease in the digestive system, has been steadily increasing worldwide[1,2]. The overall survival rate of GC patients is poor[3]. Therefore, it is necessary to identify potential biomarkers of GC and to study their molecular regulatory mechanisms to provide patients with better therapeutic outcomes.
Long noncoding RNAs (lncRNAs), a group of RNAs that are 200 nucleotides long, do not have protein-coding and translational capacity[2,4,5]. Increasing evidence has demonstrated that the dysregulation of lncRNA expression plays a crucial role in the pathogenesis of many cancers, including in cell proliferation and apoptosis[6]. For instance, the upregulation of the long noncoding RNA SNHG6 promotes cell malignancy in esophageal squamous cell carcinoma[7]. The long noncoding RNA LINC01296 induces non-small cell lung cancer growth and progression by sponging miR-5095[8]. The long noncoding RNA NEAT1 plays an oncogenic role in triple-negative breast cancer by modulating chemoresistance and cancer stemness[9]. A recent study found that the lncRNA LINC00511 contributes to breast cancer tumorigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis[10]. The knockdown of the long noncoding RNA LINC00511 suppresses the proliferation and promotes the apoptosis of bladder cancer cells by suppressing the Wnt/β-catenin signaling pathway[11]. LINC00511 interacts with miR-765 and modulates tongue squamous cell carcinoma progression by targeting LAMC2[12]. However, the specific biological function and regulatory mechanism of LINC00511 in GC have not been extensively explored.
In this study, we aimed to determine whether the lncRNA LINC00511 exerted its carcinogenic function in GC via the miR-124-3p/PDK4 axis. First, we investigated the expression level of LINC00511 in GC cell lines, and we then examined the biological function of LINC00511 with functional assays. Next, we predicted and confirmed the interaction between LINC00511 and miR-124-3p. Furthermore, we further found that PDK4 was a downstream target of miR-124-3p. Finally, we concluded that LINC00511 plays an oncogenic role in GC by sponging miR-124-3p and targeting PDK4, indicating that LINC00511 may be a new molecular biomarker for GC.
GC cell lines (MKN-45, BGC-823, HGC-27, and MGC-803) and healthy human gastric epithelial cells (GES-1) were obtained from the American Type Culture Collection (ATCC; Manassas, VA, United States) and were cultured in Roswell Park Memorial Institute 1640 medium containing 10% fetal bovine serum (FBS; Gibco/Invitrogen Inc., Carlsbad, CA, United States), streptomycin (100 mg/ml), and penicillin (100 U/mL). All of these cells were cultivated at 37°C, with 5% CO2 in a humid atmosphere.
Short hairpin RNAs (shRNAs) specifically targeting LINC00511 were designed and synthesized to knockdown LINC00511. LINC00511 and PDK4 were overexpressed with pcDNA3.1 vectors. MiR-124-3p mimics and inhibitors were used for miR-124-3p overexpression and silencing, respectively. In addition, NC mimics and an NC inhibitor were used as negative controls. All the plasmids mentioned above were purchased from GenePharma (Shanghai, China), and they were transfected into MKN-45 and MGC-803 cells by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, United States) in accordance with the manufacturer’s recommendations.
Total RNA was extracted from MKN-45 and MGC-803 cells with Trizol reagent (Takara, Otsu, Japan). A TaqManTM Advanced miRNA cDNA Synthesis Kit (Waltham, MA, United States) or a reverse transcription kit (Takara, Otsu, Japan) was utilized to reverse transcribe the RNAs into cDNA. An quantitative real-time PCR (RT-qPCR) assay was performed with an SYBR Green PCR Kit (Takara, Otsu, Japan). GAPDH and U6 were utilized as internal controls. An Applied Biosystems Step One Plus Real-Time PCR System (Applied Biosystems, Foster City, United States) was used to analyze the results. In addition, the relative expression levels were examined with the 2−ΔΔCt method.
To investigate the proliferation of the gastric cancer cells, a Cell-Counting Kit 8 (CCK8; Dojindo Molecular Technologies) was used. The transfected cells (1 × 104 cells/well) were seeded in 96-well plates and then cultured for 24, 48, 72, and 96 h. Then, 10 μL CCK-8 solution was added to each well, and the cells were incubated for another 4 h. The absorbance at 450 nm was measured by using a Multiskan Go spectrophotometer (Thermo Fisher Scientific, Inc.).
Transfected cells (1 × 103 cells/well) were seeded into 6-well plates and maintained in Roswell Park Memorial Institute 1640 medium that was replaced every 3 d. Two weeks later, PBS was used to wash the cells, and then the cells were fixed with methanol and stained with crystal violet. Subsequently, the colonies were visualized and could be counted.
The 3′-UTR sequences of PDK4, containing the predicted binding sites for miR-124-3p, and the full-length sequences of LINC00511 were subcloned into pGL3 vectors (Promega, Madison, WI, United States); thus, the wild-type PDK4 reporter (PDK4-Wt) and the wild-type LINC00511 reporter (LINC00511-Wt) were produced. The mutant-type PDK4 reporter (PDK4-Mt) and the mutant-type LINC00511 reporter (LINC00511-Mt) were built in the same manner. The miR-124-3p mimic or miR-NC was cotransfected with LINC00511-Wt or LINC00511-Mut into MKN-45 or MGC-803 cells by using Lipofectamine 2000. MiR-124-3p mimics, or miR-NC, were also cotransfected with PDK4-Wt or PDK4-Mut into MKN-45 or MGC-803 cells. Forty-eight hours later, a luciferase reporter assay system (Promega, Madison WI, United States) was used to measure the luciferase activities.
A Magna RNA-binding protein immunoprecipitation kit (Millipore, Billerica, MA, United States) was utilized to conduct the RIP assay. Cell lysates (MKN-45 and MGC-803) were mixed with RIP buffer containing anti-Ago2-conjugated beads. Input and IgG were used as positive and negative controls, respectively. Immunoprecipitated RNAs were isolated with proteinase K, and RT-qPCR was used to detect the purified RNAs.
To further probe the binding capacity of LINC00511 and miR-124-3p, a pull-down assay was conducted. First, LINC00511 and NC were biotinylated to generate a Bio-LINC00511-Probe and Bio-NC-Probe by GenePharma Company (Shanghai, China). The Bio-LINC00511-Probe and Bio-NC-Probe were transfected into MKN-45 and MGC-803 cells. Then, biotinylated RNAs and cell lysates were cultured with streptavidin-coupled beads for 48 h. Finally, the expression of the purified RNA complexes was detected by RT-qPCR.
Total proteins were extracted by using RIPA lysis buffer (Beyotime Biotechnology, China) to which protease inhibitors (Roche, China) were added. A BCATM Protein Assay Kit (Pierce, Appleton, United States) was used to measure the protein concentration. Proteins were separated by using sodium dodecyl sulfate-polyacrylamide gel electrophoresis. After electrophoresis, the proteins were transferred onto polyvinylidene difluoride membranes, and then the membranes were blocked with skim milk and were incubated with primary antibodies at 4°C. Later, the membranes were incubated with secondary antibody for 1 h at 37°C. Protein expression was detected with a chemiluminescent detection system.
SPSS 20.0 software (SPSS, Chicago, IL, United States) was used for statistical analysis. Data are presented as the mean ± SD. Differences among groups were analyzed by one-way ANOVA or a Student's t-test. Each experiment was conducted three times. A value of P < 0.05 was considered to be significant.
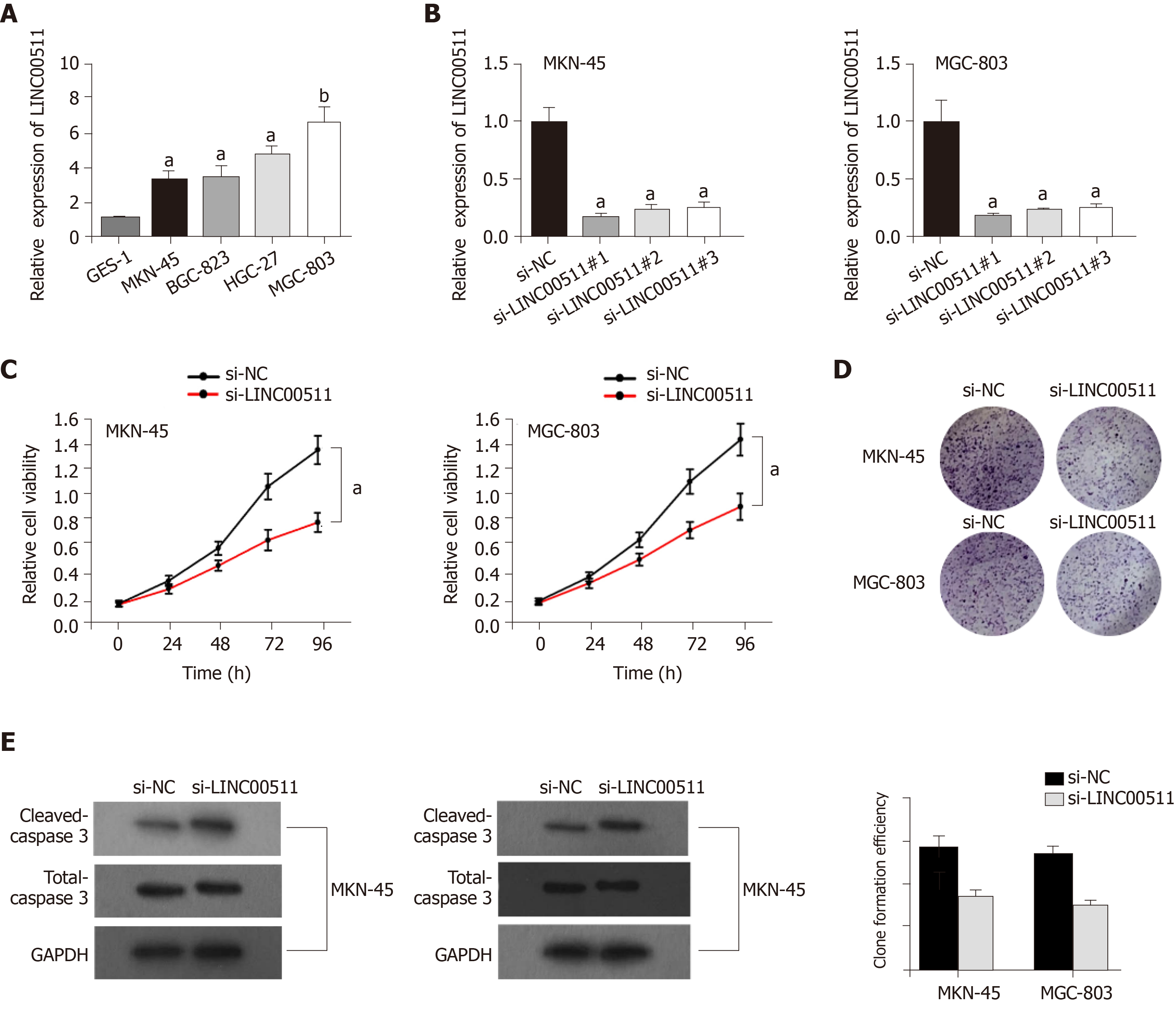
Multiple reports have demonstrated that LINC00511 is high expression in various cancers and that LINC00511 can accelerate tumorigenesis[10,12,13]. Nevertheless, whether LINC00511 can also promote tumorigenesis in GC remains unknown. Based on a qRT-PCR assay, we found that the expression of LINC00511 in MKN-45, BGC-823, HGC-27, and MGC-803 cells was clearly higher than that in GES-1 (Figure 1A). Moreover, a significant decrease in LINC00511 expression was observed in MKN-45, and MGC-803 cells that had been transfected with si-LINC00511#1, si-LINC00511#2 and si-LINC00511#3 (Figure 1B) compared to that of the controls. Additionally, cell proliferation was clearly decreased in MKN-45 and MGC-803 cells the si-LINC00511 group compared with that in the cells that had been transfected with the scrambled control (Figure 1C). A colony formation assay was performed. We found that the number of countable colonies in the si-LINC00511 group was lower than that in the scramble control group for the MKN-45 and MGC-803 cells (Figure 1D). Based on a Western blot assay, the expression of Cleaved-Caspase 3 was increased by knocking down LINC00511 compared to that in the controls, which meant that the LINC00511 knockdown promoted apoptosis in the MKN-45 and MGC-803 cell lines (Figure 1E). In summary, LINC00511 is highly expressed in GC cells, promoting cell proliferation and limiting cell apoptosis.
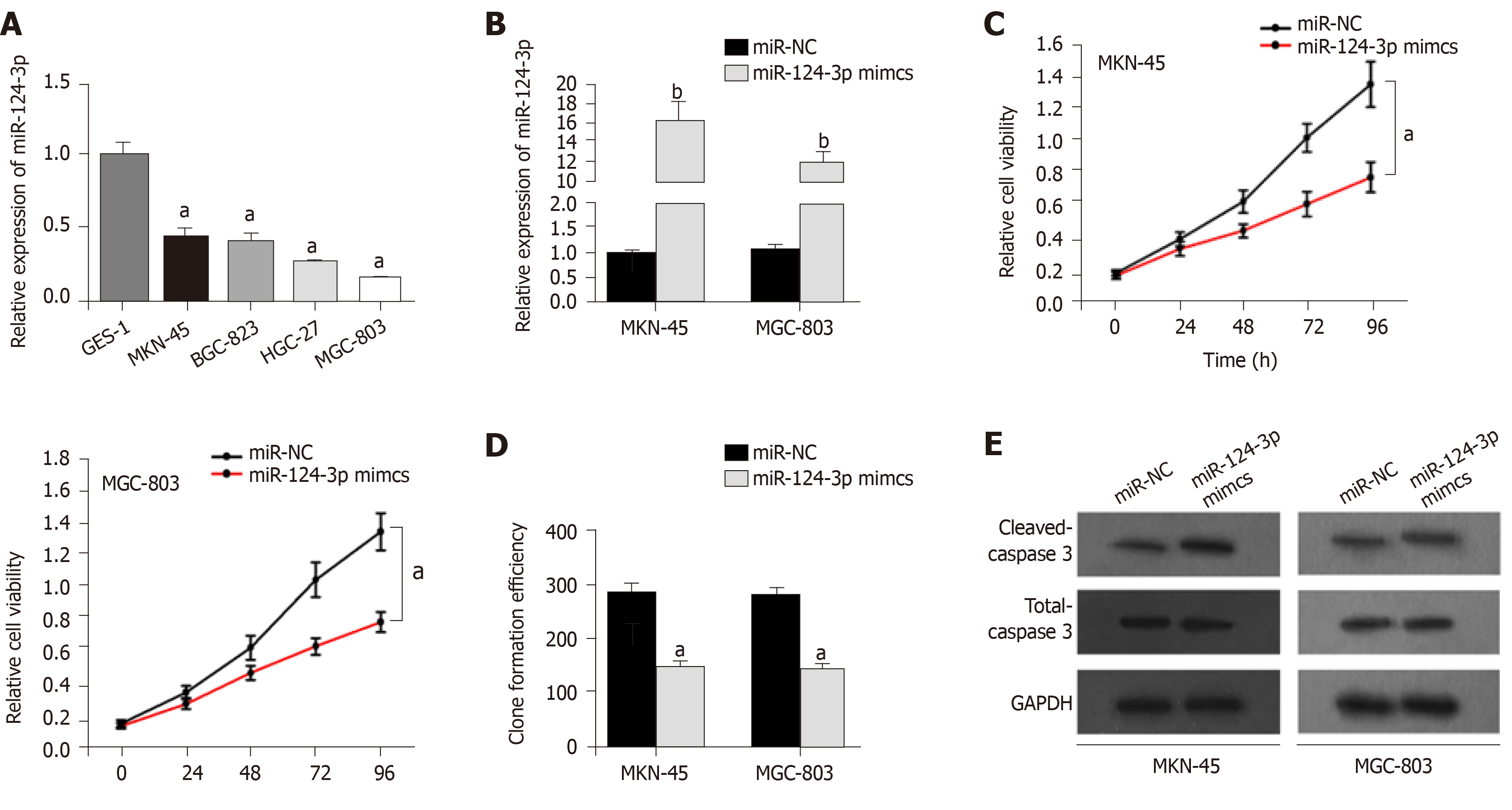
Although miR-124-3p has been reported to suppress tumorigenesis in colorectal cancer[14,15], the biological function of miR-124-3p in GC has not yet been reported. First, the qRT-PCR results indicated that miR-124-3p expression was downregulated in GC cell lines (MKN-45, BGC-823, HGC-27, and MGC-803) (Figure 2A) compared to that in other cell types. Then, miR-124-3p mimics were transfected into MKN-45 and MGC-803 cells, and miR-NC was used as a scrambled control. We found that miR-124-3p expression was remarkably increased by miR-124-3p mimics in MKN-45 and MGC-803 cells (Figure 2B) compared to that in the controls. Finally, we noticed that miR-124-3p overexpression limited proliferation but accelerated apoptosis in MKN-45 and MGC-803 cells (Figure 2C-E) compared to those in the controls. In conclusion, miR-124-3p is downregulated in GC, which prevents cell proliferation and accelerates cell apoptosis.
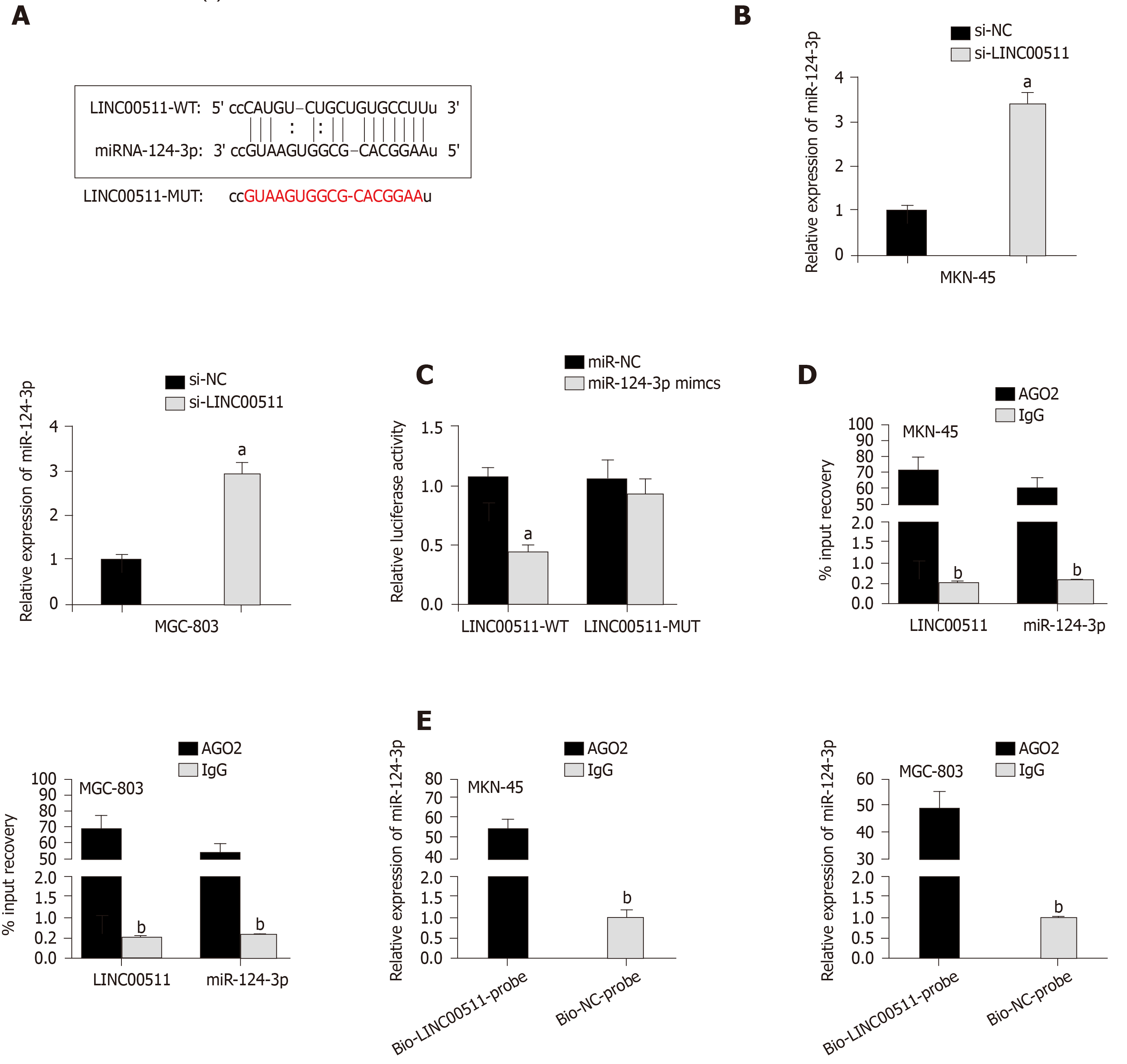
According to the starBase database, miR-124-3p contains binding sites for LINC00511 (Figure 3A), and miR-124-3p was reported to function as a tumor suppressor in non-small cell lung cancer[16]. Hence, miR-124-3p was chosen for further exploration. To analyze the relationship between LINC00511 and miR-124-3p, the following assays were performed. Initially, the qRT-PCR assay was used to demonstrate that the expression of miR-124-3p was higher in si-LINC00511-transfected cells than in the scrambled control cells (Figure 3B). Then, a luciferase assay showed that the luciferase activity of pGL3-LINC00511-Wt was prominently reduced in MKN-45 and MGC-803 cells transfected with miR-124-3p mimics compared to that in the controls. However, no apparent change in luciferase activity was observed in the pGL3-LINC00511-Mut group, indicating that LINC00511 could bind miR-124-3p (Figure 3C). Additionally, the RIP assay revealed that LINC00511 and miR-124-3p could be coimmuno-precipitated by anti-Ago2 but not with anti-IgG (Figure 3D). Last but not least, compared to Bio-NC-Probe, higher expression of miR-124-3p in the Bio-LINC00511 Probe group was observed in MKN-45 and MGC-803 cells (Figure 3E). These experimental results confirmed that LINC00511 could bind miR-124-3p, negatively modulating the expression of miR-124-3p in GC.
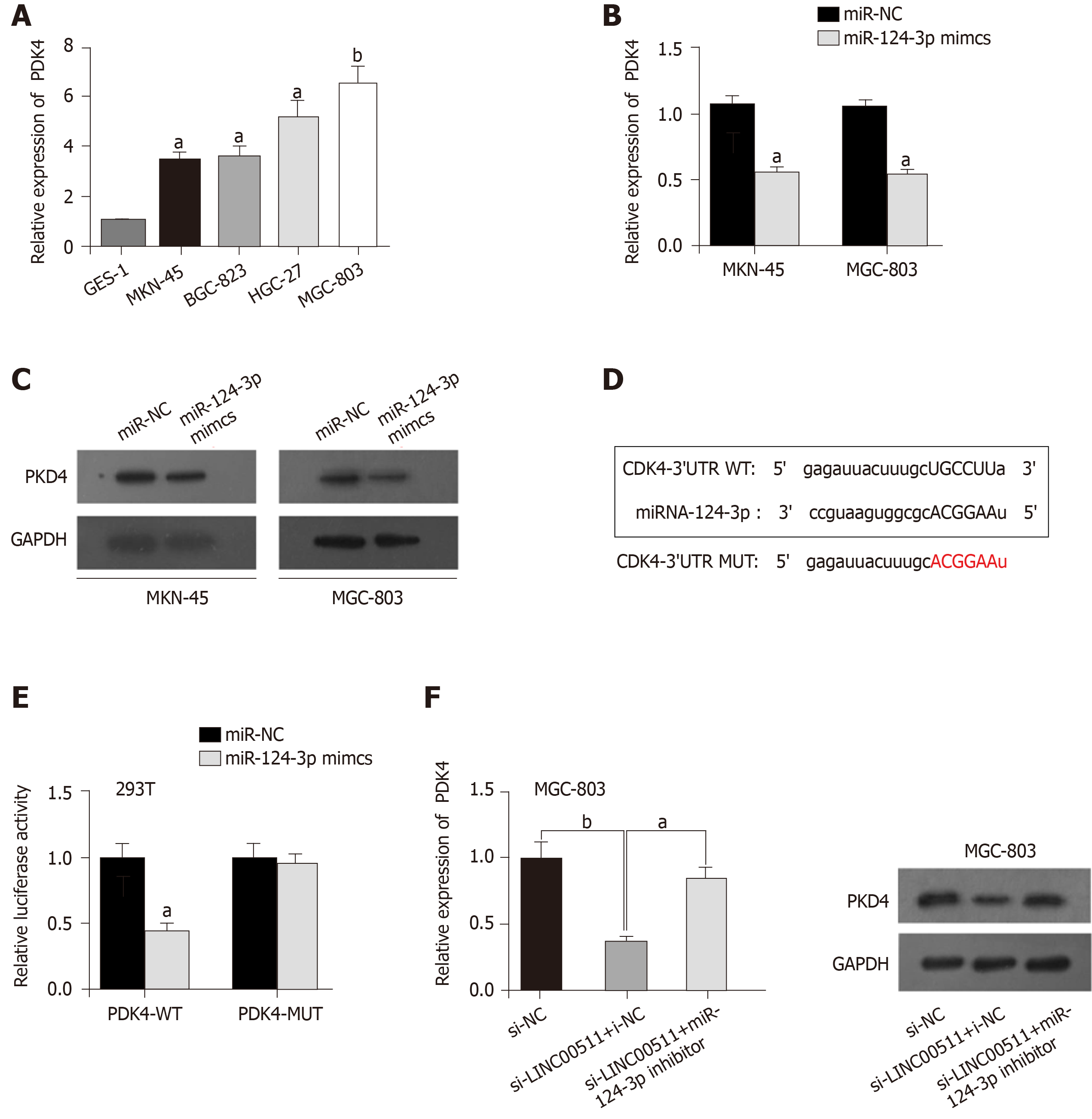
An increasing number of reports have shown that PDK4 functions as an oncogene in a variety of cancers[17-19], but its expression status in GC remains unclear. Obviously, higher expression of PDK4 was observed in MKN-45, BGC-823, HGC-27, and MGC-803 cell lines (Figure 4A) compared to that in other types of cells. As shown in Figure 4B, the overexpression of miR-124-3p clearly downregulated the expression of PDK4 in MKN-45 and MGC-803 cells compared to that in the controls. Western blot assays verified that miR-124-3p overexpression prevented the protein expression of PDK4 in MKN-45 and MGC-803 cells (Figure 4C). The StarBase database was utilized to identify potential target genes, and we found that PDK4 could bind miR-124-3p (Figure 4D). Figure 4E shows that the luciferase activity of the LINC00511-Wt vectors was prominently reduced in 293T cells that were transfected with miR-124-3p mimics compared to that in the controls. However, no obvious change in luciferase activity was observed in the group of LINC00511-Mut vectors, which suggested that LINC00511 could bind miR-124-3p (Figure 4E). Finally, Western blot analysis showed that miR-124-3p inhibitors reversed the inhibition of PDK4 that was induced by LINC00511 knockdown (Figure 4F) in MGC-803 cells. Taken together, LINC00511 competes with binding with miR-124-3p to regulate PDK4 expression.
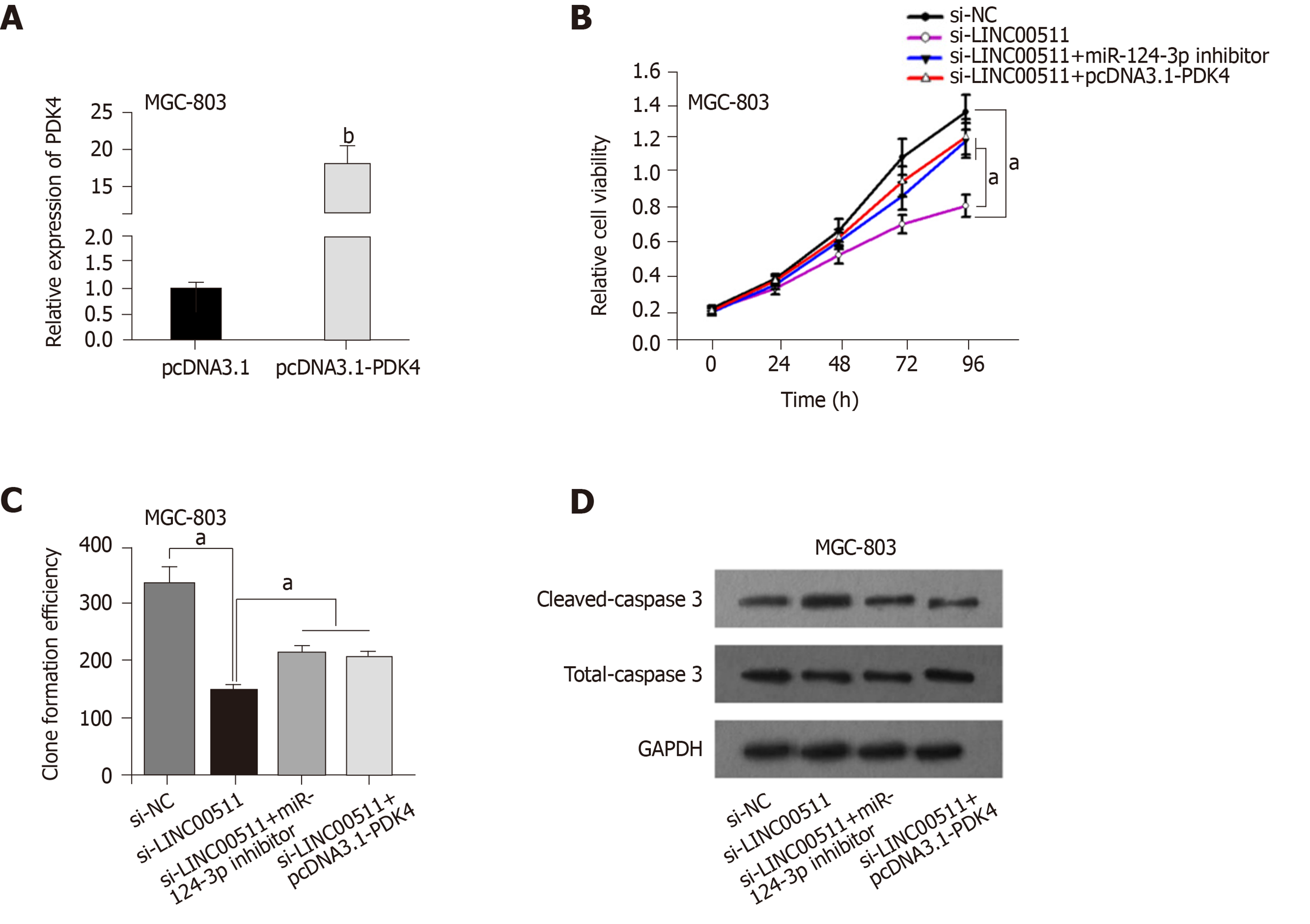
To confirm whether LINC00511 acts as a ceRNA in GC, rescue assays were performed. PDK4 expression was conspicuously increased in MGC-803 cells transfected with pcDNA3.1-PDK4 compared that in cells that had been transfected with the scrambled control (Figure 5A). The CCK-8 assay and colony formation assay demonstrated that PDK4 overexpression or miR-124-3p downregulation partially reversed the inhibitory effects of LINC00511 knockdown on cell proliferation (Figure 5B and C). Finally, the promotion of apoptosis that was caused by LINC00511 knockdown was partially counteracted by the overexpression of PDK4 or the downregulation of miR-124-3p (Figure 5D). Collectively, these results revealed that LINC00511 facilitates GC cell growth by regulating the miR-124-3p/PDK4 axis.
Numerous studies have revealed that lncRNAs influence the progression of different kinds of cancers, such as bladder cancer, lung cancer, hepatocellular carcinoma, and esophageal squamous cell carcinoma[7,20-23]. It has been reported that the lncRNA LINC00511 contributes to tumorigenesis and tumor progression in breast cancer by enhancing the miR-185-3p/E2F1/Nanog axis[10]. The long noncoding RNA LINC00511 promotes proliferation and suppresses apoptosis of bladder cancer cells via the Wnt/β-catenin signaling pathway[11]. LINC00511 modulates tongue squamous cell carcinoma progression by interacting with miR-765 to regulate LAMC2 expression[12]. Nevertheless, the biological function and molecular regulatory mechanisms of LINC00511 in GC remain unclear. In our study, we found that LINC00511 was more highly expressed in GC cell lines than in corresponding normal cell lines. Furthermore, the knockdown of LINC00511 inhibited cell proliferation while promoting cell apoptosis. All of these data indicated that LINC00511 promoted the progression of GC.
MircoRNAs (miRNAs) are a class of short noncoding RNAs containing[20-24] nucleotides that play significant roles in the post-transcriptional regulation of the expression of genes[24,25]. Recently, it has been reported that lncRNAs modulate some diseases by acting as sponges to combine with miRNAs. For example, the long noncoding RNA LINC00202 promotes tumor progression by sponging miR-3619-5p in retinoblastoma[26]. AC016405.3, a novel lncRNA, acts as a tumor suppressor by modulating TET2 through miR-19a-5p sponging in glioblastoma[27]. LINC00511 promotes the progression of tongue squamous cell carcinoma by interacting with miR-765[12]. However, the interaction of LINC00511 with its downstream miRNA in GC cells remains to be examined. In this study, we identified miR-124-3p as a potential target miRNA that could bind with LINC00511 with starBase. According to previous studies, miR-124-3p has antitumor effects in various cancers[28-30]. In our study, we further confirmed that LINC00511 could bind miR-124-3p and negatively modulate the expression of miR-124-3p in GC. All of these data indicated that LINC00511 sponged miR-124-3p to regulate GC progression.
PDK4 is reported to have an oncogenic effect in many human cancers[18,31]. The overexpression of PDK4 is associated with cell proliferation, drug resistance, and poor prognosis in ovarian cancer[18]. The loss of PDK4 switches the hepatic NF-κB/TNF pathway from having a prosurvival effect to having a pro-apoptosis impact[31]. It was predicted that PDK4 could bind with miR-124-3p based upon bioinformatics analysis. We found that miR-124-3p could bind PDK4 to regulate PDK4 expression negatively. Lastly, rescue assays demonstrated that the overexpression of PDK4 or the inhibition of miR-124-3p could partially rescue the effects of LINC00511 knockdown on cell proliferation and apoptosis.
In summary, our study showed that LINC00511 promotes the tumorigenesis and development of GC by targeting miR-124-3p/PDK4, indicating the potential of the LINC00511/miR-124-3p/PDK4 axis as a new biomarker for GC prognosis.
It has been reported that LINC00511 contributes to tumorigenesis in various diseases. However, the role of LINC00511 in gastric cancer (GC) cell growth remains mostly unknown.
The specific biological function and regulatory mechanism of LINC00511 in GC have not been extensively explored.
In this study, the authors aimed to determine whether the long noncoding RNAs LINC00511 exerted its carcinogenic function in GC via the miR-124-3p/PDK4 axis.
The cell culture and transfection, RNA extraction and quantitative real-time PCR, CCK-8 assay, Colony formation assay, Luciferase reporter assay, RIP assay, RNA pull-down assay, and Western blot analysis were used to show expression and mechanisms of LINC00511 in GC progression and apoptosis. Rescue assays were performed to verify the relationships among LINC00511, miR-124-3p and PDK4 further.
The expression of LINC00511 was remarkably upregulated in GC cells compared to that in corresponding normal cell lines. Compared to the controls, cell proliferation was inhibited, and cell apoptosis was increased upon LINC00511 knockdown, demonstrating that LINC00511 influenced GC cell growth. An exploration of the molecular mechanism revealed that LINC00511 functioned as a molecular sponge of miR-124-3p and that PDK4 was a downstream target of miR-124-3p in GC.
The results of the study demonstrate that LINC00511 promotes gastric cancer cell growth by acting as a ceRNA to regulate the miR-124-3p/PDK4 axis, which may be a promising therapeutic target for GC.
Manuscript source: Unsolicited manuscript
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Klaiber U, LaRue S, Martelotto S, S-Editor: Wang JL L-Editor: A E-Editor: Xing YX
| 1. | Li X, Yan X, Wang F, Yang Q, Luo X, Kong J, Ju S. Down-regulated lncRNA SLC25A5-AS1 facilitates cell growth and inhibits apoptosis via miR-19a-3p/PTEN/PI3K/AKT signalling pathway in gastric cancer. J Cell Mol Med. 2019;23:2920-2932. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 58] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 2. | Zeng H, Zheng R, Guo Y, Zhang S, Zou X, Wang N, Zhang L, Tang J, Chen J, Wei K, Huang S, Wang J, Yu L, Zhao D, Song G, Chen J, Shen Y, Yang X, Gu X, Jin F, Li Q, Li Y, Ge H, Zhu F, Dong J, Guo G, Wu M, Du L, Sun X, He Y, Coleman MP, Baade P, Chen W, Yu XQ. Cancer survival in China, 2003-2005: a population-based study. Int J Cancer. 2015;136:1921-1930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 415] [Cited by in RCA: 499] [Article Influence: 49.9] [Reference Citation Analysis (0)] |
| 3. | Sgambato D, Miranda A, Romano L, Romano M. Gut microbiota and gastric disease. Minerva Gastroenterol Dietol. 2017;63:345-354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 4. | Wang Q, Gao S, Li H, Lv M, Lu C. Long noncoding RNAs (lncRNAs) in triple negative breast cancer. J Cell Physiol. 2017;232:3226-3233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 5. | Mattick JS. The State of Long Non-Coding RNA Biology. Noncoding RNA. 2018;4:17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 6. | Silva A, Bullock M, Calin G. The Clinical Relevance of Long Non-Coding RNAs in Cancer. Cancers (Basel). 2015;7:2169-2182. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 118] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 7. | Zhang Y, Li R, Ding X, Zhang K, Qin W. Upregulation of long non-coding RNA SNHG6 promote esophageal squamous cell carcinoma cell malignancy and its diagnostic value. Am J Transl Res. 2019;11:1084-1091. [PubMed] |
| 8. | Hu X, Duan L, Liu H, Zhang L. Long noncoding RNA LINC01296 induces non-small cell lung cancer growth and progression through sponging miR-5095. Am J Transl Res. 2019;11:895-903. [PubMed] |
| 9. | Shin VY, Chen J, Cheuk IW, Siu MT, Ho CW, Wang X, Jin H, Kwong A. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis. 2019;10:270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 181] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 10. | Lu G, Li Y, Ma Y, Lu J, Chen Y, Jiang Q, Qin Q, Zhao L, Huang Q, Luo Z, Huang S, Wei Z. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J Exp Clin Cancer Res. 2018;37:289. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 217] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 11. | Li J, Li Y, Meng F, Fu L, Kong C. Knockdown of long non-coding RNA linc00511 suppresses proliferation and promotes apoptosis of bladder cancer cells via suppressing Wnt/β-catenin signaling pathway. Biosci Rep. 2018;38:BSR20171701. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 12. | Ding J, Yang C, Yang S. LINC00511 interacts with miR-765 and modulates tongue squamous cell carcinoma progression by targeting LAMC2. J Oral Pathol Med. 2018;47:468-476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 67] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 13. | Zhao X, Liu Y, Li Z, Zheng S, Wang Z, Li W, Bi Z, Li L, Jiang Y, Luo Y, Lin Q, Fu Z, Rufu C. Linc00511 acts as a competing endogenous RNA to regulate VEGFA expression through sponging hsa-miR-29b-3p in pancreatic ductal adenocarcinoma. J Cell Mol Med. 2018;22:655-667. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 115] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 14. | Wang Y, Chen L, Wu Z, Wang M, Jin F, Wang N, Hu X, Liu Z, Zhang CY, Zen K, Chen J, Liang H, Zhang Y, Chen X. miR-124-3p functions as a tumor suppressor in breast cancer by targeting CBL. BMC Cancer. 2016;16:826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 89] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 15. | Yang JX, Liu B, Yang BY, Meng Q. Long non-coding RNA homeobox (HOX) A11-AS promotes malignant progression of glioma by targeting miR-124-3p. Neoplasma. 2018;65:505-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 16. | Tang LX, Chen GH, Li H, He P, Zhang Y, Xu XW. Long non-coding RNA OGFRP1 regulates LYPD3 expression by sponging miR-124-3p and promotes non-small cell lung cancer progression. Biochem Biophys Res Commun. 2018;505:578-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 17. | Guda MR, Asuthkar S, Labak CM, Tsung AJ, Alexandrov I, Mackenzie MJ, Prasad DV, Velpula KK. Targeting PDK4 inhibits breast cancer metabolism. Am J Cancer Res. 2018;8:1725-1738. [PubMed] |
| 18. | Wang J, Qian Y, Gao M. Overexpression of PDK4 is associated with cell proliferation, drug resistance and poor prognosis in ovarian cancer. Cancer Manag Res. 2019;11:251-262. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 19. | Woolbright BL, Choudhary D, Mikhalyuk A, Trammel C, Shanmugam S, Abbott E, Pilbeam CC, Taylor JA. The Role of Pyruvate Dehydrogenase Kinase-4 (PDK4) in Bladder Cancer and Chemoresistance. Mol Cancer Ther. 2018;17:2004-2012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 84] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 20. | Zhu Y, Dai B, Zhang H, Shi G, Shen Y, Ye D. Long non-coding RNA LOC572558 inhibits bladder cancer cell proliferation and tumor growth by regulating the AKT-MDM2-p53 signaling axis. Cancer Lett. 2016;380:369-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 21. | Park SM, Choi EY, Bae DH, Sohn HA, Kim SY, Kim YJ. The LncRNA EPEL Promotes Lung Cancer Cell Proliferation Through E2F Target Activation. Cell Physiol Biochem. 2018;45:1270-1283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 22. | Lin Q, Zheng H, Xu J, Zhang F, Pan H. LncRNA SNHG16 aggravates tumorigenesis and development of hepatocellular carcinoma by sponging miR-4500 and targeting STAT3. J Cell Biochem. 2019;120:11604–11615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 23. | Yang F, Wen S, Zhang Y, Xu Y, Lv H, Zhu Y, Wang M, Su P, Huang C, Tian Z. Identifying potential metastasis-related long non-coding RNAs, microRNAs, and message RNAs in the esophageal squamous cell carcinoma. J Cell Biochem. 2019;120:13202-13215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 24. | Srivastava SK, Bhardwaj A, Leavesley SJ, Grizzle WE, Singh S, Singh AP. MicroRNAs as potential clinical biomarkers: emerging approaches for their detection. Biotech Histochem. 2013;88:373-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 25. | D'Angelo B, Benedetti E, Cimini A, Giordano A. MicroRNAs: A Puzzling Tool in Cancer Diagnostics and Therapy. Anticancer Res. 2016;36:5571-5575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 82] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 26. | Yan G, Su Y, Ma Z, Yu L, Chen N. Long Noncoding RNA LINC00202 Promotes Tumor Progression by Sponging miR-3619-5p in Retinoblastoma. Cell Struct Funct. 2019;44:51-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 27. | Ren S, Xu Y. AC016405.3, a novel long noncoding RNA, acts as a tumor suppressor through modulation of TET2 by microRNA-19a-5p sponging in glioblastoma. Cancer Sci. 2019;110:1621-1632. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 28. | Wang JR, Liu B, Zhou L, Huang YX. MicroRNA-124-3p suppresses cell migration and invasion by targeting ITGA3 signaling in bladder cancer. Cancer Biomark. 2019;24:159-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 29. | Luo L, Chi H, Ling J. MiR-124-3p suppresses glioma aggressiveness via targeting of Fra-2. Pathol Res Pract. 2018;214:1825-1834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 30. | Liu H, Deng H, Zhao Y, Li C, Liang Y. LncRNA XIST/miR-34a axis modulates the cell proliferation and tumor growth of thyroid cancer through MET-PI3K-AKT signaling. J Exp Clin Cancer Res. 2018;37:279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 193] [Cited by in RCA: 242] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 31. | Wu J, Zhao Y, Park YK, Lee JY, Gao L, Zhao J, Wang L. Loss of PDK4 switches the hepatic NF-κB/TNF pathway from pro-survival to pro-apoptosis. Hepatology. 2018;68:1111-1124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 61] [Article Influence: 8.7] [Reference Citation Analysis (0)] |