Published online Sep 16, 2021. doi: 10.12998/wjcc.v9.i26.7693
Peer-review started: February 18, 2021
First decision: April 13, 2021
Revised: April 15, 2021
Accepted: July 2, 2021
Article in press: July 2, 2021
Published online: September 16, 2021
Processing time: 197 Days and 4.8 Hours
Chlamydia psittaci (C. psittaci) is a gram-negative intracellular parasitic pathogenic bacterium that can infect avian and mammalian hosts, including humans. The detection of C. psittaci infections typically relies on traditional antigen-based immunoassays or serological testing that often lack sensitivity and/or specificity. Metagenomic next generation sequencing (mNGS) is an emerging tool for diag
To demonstrate that mNGS represents a valuable tool for rapid, sensitive, and accurate pathogen detection including C. psittaci infections.
Four cases of psittacosis pneumonia and one case of pediatric psittacosis meningitis were diagnosed between December 2019 and May 2020 using mNGS at Changzhou Second People’s Hospital affiliated to Nanjing Medical University. Patients’ clinical characteristics, manifestations, and treatment histories were retrospectively evaluated.
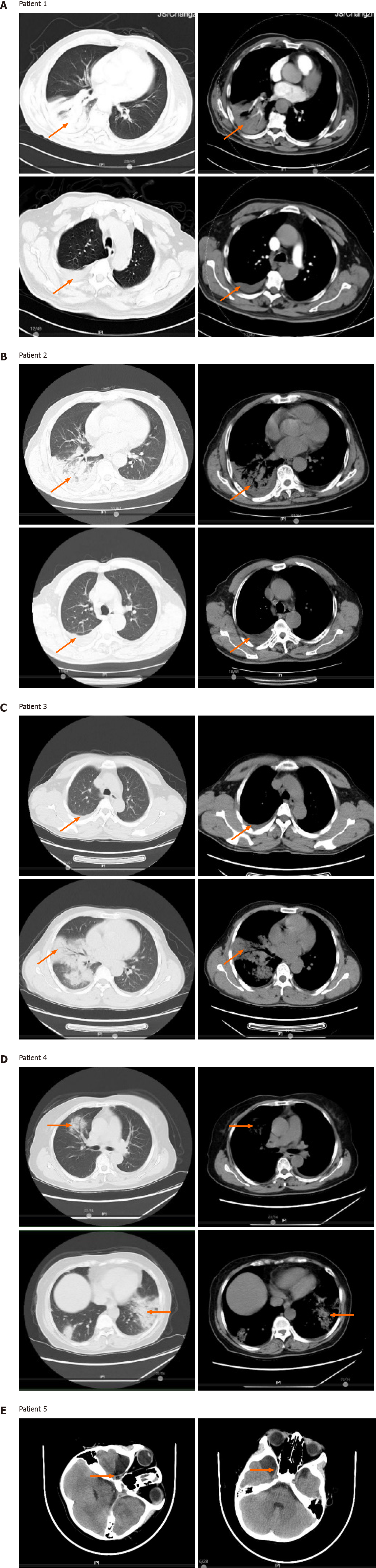
All five patients had a history of exposure to wild (psittacine or other birds) or domesticated birds (chickens). All patients had a high fever (> 39℃) and three of them (60%) experienced organ insufficiency during the disease. The laboratory data showed normal to slightly increased leucocyte and neutrophil counts, and elevated procalcitonin levels in all five cases, and very high C-reactive protein levels in psittacosis pneumonia patients. mNGS identified a potential pathogen, C. psittaci, in patients’ bronchoalveolar lavage fluid or cerebrospinal fluid. Computed tomography revealed lung air-space consolidation, pleural thickening, and effu
This study not only demonstrated that mNGS represents a valuable tool for rapid, sensitive, and accurate pathogen detection, but also raised public health concerns over C. psittaci infections.
Core Tip: Detection of Chlamydia psittaci (C. psittaci) infections relies on traditional antigen-based immunoassays or serological testing that often lack sensitivity and/or specificity. Our data not only reinforce that metagenomic next generation sequencing represents a valuable tool for rapid, sensitive, and accurate pathogen detection, in
- Citation: Yin XW, Mao ZD, Zhang Q, Ou QX, Liu J, Shao Y, Liu ZG. Clinical metagenomic sequencing for rapid diagnosis of pneumonia and meningitis caused by Chlamydia psittaci. World J Clin Cases 2021; 9(26): 7693-7703
- URL: https://www.wjgnet.com/2307-8960/full/v9/i26/7693.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i26.7693
Chlamydia are gram-negative intracellular parasitic pathogenic bacteria that can infect avian and mammalian hosts, including humans. Chlamydia can be parasitic in bird tissues, blood and feces, and is also a zoonotic disease with bird breeders, in particular, at increased risk of developing chlamydia infections.
Poultry infections are typically asymptomatic; however, once humans are infected with Chlamydia, it may cause multiple diseases, including those of the respiratory tract, gastrointestinal tract, cerebrospinal fluid (CSF), eyes, and serous cavity. Such infec
Unlike traditional antigen-based immunoassays or serological testing, metagenomic next generation sequencing (mNGS) is an emerging technique to support pathogen detection[4-6]. mNGS can not only overcome the limitations of routine clinical mi
Patients were admitted to the Department of Respiratory and Critical Care Medicine and the Department of Pediatrics at Changzhou Second People’s Hospital affiliated to Nanjing Medical University between December 2019 and May 2020. An etiologic diagnosis was not known at the time of admission. Patients’ clinical characteristics, manifestations, and treatment history were retrospectively evaluated. The treating physicians were informed about the research-based mNGS results through a reporting mechanism approved by the Institutional Review Board of Changzhou Second People’s Hospital. Written informed consent was obtained from each patient prior to sample collection.
mNGS was performed in a Clinical Laboratory Improvement Amendments-certified and College of American Pathologists-accredited laboratory (Nanjing Geneseeq Technology, Jiangsu Province, China). BALF and CSF samples were collected from patients as previously described[7]. Genomic DNA was extracted using the TIANamp Magnetic DNA Kit (Tiangen) according to the manufacturer’s protocols. The quantity and quality of DNA were assessed using Qubit (Thermo Fisher Scientific) and Nano
An in-house bioinformatics pipeline was developed for pathogen detection. Briefly, Trimmomatic v.0.36 software[8] was used to generate high-quality sequencing data by removing low quality reads, adapter contamination, duplicated and short (length < 36 bp) reads. Human host sequences were filtered by mapping to the human reference genome (hs37d5) using Bowtie2 software[9]. Kraken 2 (v2.0.7) software[10] was used to assign nonhuman sequences to the microbial genome database, which contains more than 20,000 genome sequences of bacteria, fungi, viruses, and parasites for sequence alignment (http://ftp://ftp.ncbi.nlm.nih.gov/genomes/genbank/). To distinguish putative pathogens from contaminating microbial sequences derived from skin, collection tubes, laboratory agents, or the environment, negative ‘no-template’ control (NTC) samples were analyzed in parallel. Unique nonhuman sequences were iden
A final sequencing list of suspected pathogenic microorganisms was obtained after removal of common background microorganisms compared to the NTCs. The top-ranked taxa were considered with respect to the clinical features of the patient. Mi
Four adult patients (three males and one female) and one male pediatric patient were retrospectively studied. The characteristics of all patients are summarized in Table 1. All patients had avian exposure history and were admitted to the hospital due to remittent fever of higher than 39°C. Other symptoms were also observed, including coughing in three adult patients and dizziness, vomiting, and convulsions in the pediatric patient (patient 5). Four patients developed chills, chest tightness, and asthma, and three patients developed dry cough and chest pain. The clinical symp
| Patient No. | 1 | 2 | 3 | 4 | 5 |
| Sex | M | M | M | F | M |
| Age (yr) | 75 | 73 | 44 | 66 | 5 |
| Symptoms | Fever, cough | Fever | Fever, cough | Fever, cough | Fever, dizziness, vomiting, convulsions |
| Chronic disease | Hypertension, diabetes, coronary heart disease, lacunar infarction, prostatic hyperplasia, amoebic dysentery | Hepatitis B | No history | No history | No history |
| Exposure history | Birds | Chickens | Birds | Psittacine birds | Birds |
| Fever | 39.7℃ | 40.9℃ | 40.1℃ | 40.0℃ | 39.9℃ |
| Leucocytes | 8.46 × 109/L | 2.98 × 109/L | 14.51 × 109/L | 2.93 × 109/L | 24.01 × 109/L |
| Neutrophils | 83.20% | 83.20% | 85.20% | 49.90% | 88.20% |
| Platelet count | 106.00 × 109/L | 106.00 × 109/L | 191.00 × 109/L | 286.00 × 109/L | 301.00 × 109/L |
| Procalcitonin | 1.65 ng/mL | 0.49 ng/mL | 0.01 ng/mL | 0.19 ng/mL | 4.28 ng/mL |
| C-reactive protein | 241.70 mg/L | 169.00 mg/L | 225.40 mg/L | 96.30 mg/L | 6.30 mg/L |
| Aspartate aminotransferase | 53.5 U/L | Normal | Normal | 65.1 U/L | Normal |
| Glutamate aminotransferase | Normal | Normal | 124.2 U/L | 172.3 U/L | Normal |
| Urea nitrogen | 14.5 mmol/L | Normal | Normal | Normal | Normal |
| Creatinine | 353.0 μmol/L | Normal | Normal | Normal | Normal |
| Mycoplasma pneumoniae IgG titer | 1:40 | Negative | 1:40 | Negative | 1:160 |
| CSF culture | N/A | N/A | N/A | N/A | Streptococcus pneumoniae |
| Diagnosis | Psittacosis pneumonia | Psittacosis pneumonia | Psittacosis pneumonia | Psittacosis pneumonia | Purulent meningitis, Psittacosis meningitis |
| Antibiotics | Piperacillin, tazobactam, etimicin, moxifloxacin | Ganciclovir, tigecycline | Piperacillin, tazobactam, moxifloxacin, tigecycline | Piperacillin, tazobactam, moxifloxacin | Ceftazidime, vancomycin, ceftriaxone sodium, azithromycin, rifampicin |
| Hospitalization (d) | 13 | 11 | 11 | 10 | 23 |
All patients underwent physical examinations and blood tests upon hospital admi
Computed tomography (CT) imaging showed varying degrees of lung inflamma
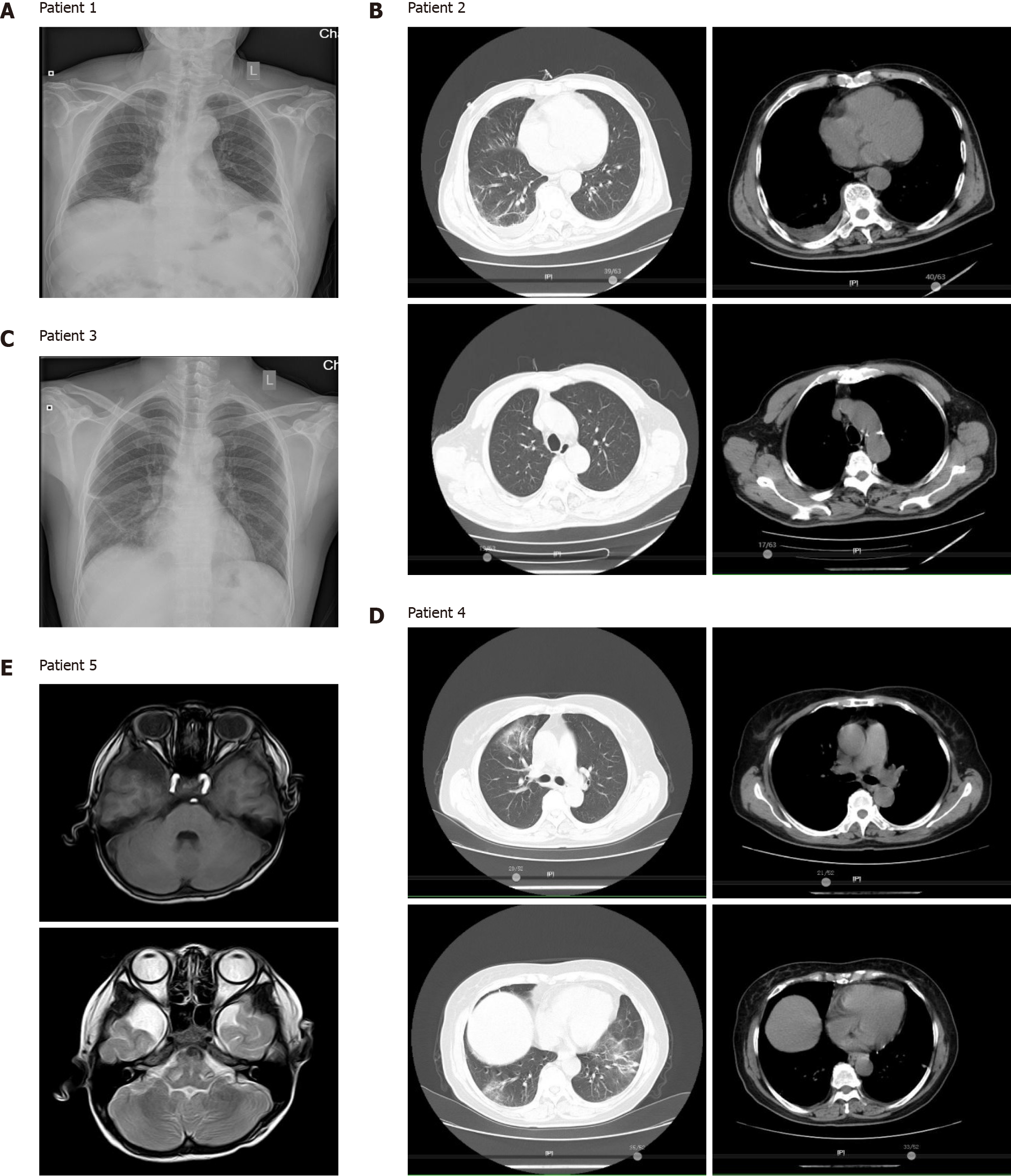
Patient 1 was initially treated with piperacillin, tazobactam, and etimicin, but no benefit was observed. A fiberoptic bronchoscopy was then performed and BALF was immediately subjected to mNGS testing. The mNGS results revealed the presence of Chlamydia psittaci (C. psittaci) (Table 2). The patient then received moxifloxacin and recovered rapidly, as shown by chest radiographs (Figure 2A).
| Patient No. | Total reads | Chlamydia psittaci | Other microbes detected | ||
| Reads | Genome coverage rate | Reads | Genome coverage rate | ||
| 1 | 22128603 | 231 | 1.45% | - | - |
| 2 | 33738995 | 305 | 1.81% | 207 | 0.11% (Chlamydia albicans) |
| 3 | 50359059 | 2 | 0.01% | - | - |
| 4 | 41903841 | 6 | 0.02% | - | - |
| 5 | 18745727 | 2137 | 7.76% | 388 | 0.73% (Streptococcus pneumonia) |
Patient 2 had self-administered acetaminophen prior to hospital admission and was initially treated with ganciclovir plus tigecycline. The patient’s BALF was subjected to mNGS and was diagnosed with a co-infection of C. psittaci and Candida albicans (Table 2). After receiving ganciclovir plus tigecycline for 10 d, CT scans revealed that the patient’s lung lesions were significantly absorbed (Figure 2B).
Patient 3 had taken acetaminophen prior to hospital admission and was initially treated with piperacillin, tazobactam, and moxifloxacin. However, no benefits from these therapies were observed. mNGS testing of the patient’s BALF was performed and revealed two C. psittaci-specific reads, but no other potential pathogenic microbe sequences (Table 2). After receiving tigecycline, the patient’s body temperature de
Patient 4 had self-administered acetaminophen, and was initially administered piperacillin, tazobactam, and moxifloxacin following hospital admission. The patient’s BALF underwent mNGS after her body temperature decreased, and she was diag
The pediatric patient (patient 5) had no history of medication. He was initially treated with ceftazidime after hospital admission, with very little benefit. He was then switched to vancomycin and ceftriaxone; however, no benefit was observed. The patient’s CSF was then collected for mNGS, which revealed the presence of C. psittaci and Streptococcus pneumoniae (S. pneumoniae) (Table 2). The patient received azithro
None of the patients had any symptoms such as fever or cough after hospital discharge.
In this study, we found that all five patients had a history of exposure to birds (psi
Another interesting finding of this study was that Chlamydia was not only detected in BALF but also in CSF, a finding that has not been widely reported and requires further investigation. Furthermore, we demonstrated the clinical utility of mNGS to sensitively detect microbial infections, including those containing different bacteria, viruses, and fungi. mNGS revealed that two patients in our cohort were also positive for C. albicans (patient 2) and S. pneumoniae (patient 5), in addition to C. psittaci. For instance, mNGS results revealed the co-occurrence of C. psittaci and S. pneumoniae in the CSF specimen of patient 5, while the genome coverage rate of C. psittaci (7.76%) was much greater than that of S. pneumoniae (0.73%) indicative of a higher pathogen load of C. psittaci. The patient fully recovered following anti-Chlamydia treatment; however, it did not rule out possible co-infection as S. pneumonia can also cause meningitis and induce inflammation resulting in blood-brain barrier disruption[11,12]. Those findings emphasize the complexity of diseases related to microbial in
This study also raised public health concerns over C. psittaci infections. C. psittaci research began in the 1930s when psittacosis infections were spreading globally. Sir Frank Macfarlane Burnet, a famous Australian virologist, immunologist, and Nobel laureate, was the first person to study this bacterium[13]. Infections with C. psittaci, a zoonotic pathogen, are rarely reported in humans, and approximately 1% of commu
Despite the complexity of different infectious diseases, mNGS technology can provide rapid and comprehensive diagnoses. mNGS detects all microbial nucleic acids in a sample and identifies suspected pathogenic microorganisms. Current mNGS technologies can detect 20000 types of bacteria (including 400 types of Mycobacteria and 400 types of Mycoplasma/Chlamydia/Rickettsia), 4600 types of viruses, 1800 types of fungi, and 100 types of parasites[18]. Moreover, when coupled with machine learning analyses, mNGS can also identify suspected drug resistance genes, which broadens the application of mNGS in disease diagnosis. Overall, our findings demonstrated that mNGS represents a valuable tool for rapid, sensitive, and accurate pathogen detection.
Our study highlights the risk of bird/poultry exposure and C. psittaci infections, as they may underlie a potential public health problem. mNGS represents a valuable tool for rapid, sensitive, and accurate pathogen detection, including the detection of C. psittaci; thus, enabling clinicians to adopt personalized and appropriate treatments.
Chlamydia psittaci (C. psittaci) is a gram-negative intracellular parasitic pathogenic bacterium that can infect avian and mammalian hosts, including humans. The detec
This study provides evidence to illustrate metagenomic next generation sequencing (mNGS) as a promising clinical-microbiology tool for pathogen detection.
This study aimed to demonstrate that mNGS represents a valuable tool for rapid, sensitive, and accurate pathogen detection including C. psittaci infections.
Four cases of psittacosis pneumonia and one case of pediatric psittacosis meningitis were diagnosed using mNGS. Patients’ clinical characteristics, manifestations, and treatment histories were retrospectively evaluated.
All five patients had a history of exposure to wild (psittacine or other birds) or domesticated birds (chickens). All patients had a high fever and three of them experienced organ insufficiency during the disease. The laboratory data showed normal to slightly increased leucocyte and neutrophil counts, and elevated procalcitonin levels in all five cases, and very high C-reactive protein levels in psittacosis pneumonia patients. mNGS identified a potential pathogen, C. psittaci, in patients’ bronchoalveolar lavage fluid or cerebrospinal fluid. Computed tomography revealed lung air-space consolidation, pleural thickening, and effusion fluid buildup in psittacosis pneumonia cases, and an arachnoid cyst in the right temporal lobe of the pediatric psittacosis meningitis patient. All patients experienced complete recovery following the administration of targeted anti-Chlamydia therapy.
Our data not only reinforce that mNGS represents a valuable tool for rapid, sensitive, and accurate pathogen detection, including C. psittaci, but also raise public health concerns over C. psittaci infections.
Despite the complexity of different infectious diseases, mNGS technology can provide rapid and comprehensive diagnoses. The application of mNGS in disease diagnosis can be further broadened by coupling with machine learning algorithms in various aspects including the identification of suspected drug resistance genes.
We would like to thank the patients and their families for providing consent for publication. We also thank the research staff and co-investigators involved in this study.
Manuscript source: Unsolicited manuscript
Specialty type: Infectious diseases
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Wan X S-Editor: Yan JP L-Editor: Webster JR P-Editor: Yuan YY
| 1. | Hogerwerf L, DE Gier B, Baan B, VAN DER Hoek W. Chlamydia psittaci (psittacosis) as a cause of community-acquired pneumonia: a systematic review and meta-analysis. Epidemiol Infect. 2017;145:3096-3105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 144] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 2. | Hogerwerf L, Holstege MMC, Benincà E, Dijkstra F, van der Hoek W. Temporal and spatial analysis of psittacosis in association with poultry farming in the Netherlands, 2000-2015. BMC Infect Dis. 2017;17:519. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 3. | Nieuwenhuizen AA, Dijkstra F, Notermans DW, van der Hoek W. Laboratory methods for case finding in human psittacosis outbreaks: a systematic review. BMC Infect Dis. 2018;18:442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 72] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 4. | Hoffmann B, Tappe D, Höper D, Herden C, Boldt A, Mawrin C, Niederstraßer O, Müller T, Jenckel M, van der Grinten E, Lutter C, Abendroth B, Teifke JP, Cadar D, Schmidt-Chanasit J, Ulrich RG, Beer M. A Variegated Squirrel Bornavirus Associated with Fatal Human Encephalitis. N Engl J Med. 2015;373:154-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 183] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 5. | Wilson MR, O'Donovan BD, Gelfand JM, Sample HA, Chow FC, Betjemann JP, Shah MP, Richie MB, Gorman MP, Hajj-Ali RA, Calabrese LH, Zorn KC, Chow ED, Greenlee JE, Blum JH, Green G, Khan LM, Banerji D, Langelier C, Bryson-Cahn C, Harrington W, Lingappa JR, Shanbhag NM, Green AJ, Brew BJ, Soldatos A, Strnad L, Doernberg SB, Jay CA, Douglas V, Josephson SA, DeRisi JL. Chronic Meningitis Investigated via Metagenomic Next-Generation Sequencing. JAMA Neurol. 2018;75:947-955. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 213] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 6. | Gu W, Miller S, Chiu CY. Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection. Annu Rev Pathol. 2019;14:319-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 389] [Cited by in RCA: 830] [Article Influence: 118.6] [Reference Citation Analysis (0)] |
| 7. | Chang B, Wei X, Wang X, Tang Y, Zhu J, Zheng X, Zhang C, Li S. Metagenomic next-generation sequencing of viruses, bacteria, and fungi in the epineurium of the facial nerve with Bell's palsy patients. J Neurovirol. 2020;26:727-733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 8. | Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114-2120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30322] [Cited by in RCA: 40989] [Article Influence: 3726.3] [Reference Citation Analysis (1)] |
| 9. | Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39162] [Cited by in RCA: 36968] [Article Influence: 2843.7] [Reference Citation Analysis (0)] |
| 10. | Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1317] [Cited by in RCA: 3403] [Article Influence: 567.2] [Reference Citation Analysis (1)] |
| 11. | Iovino F, Orihuela CJ, Moorlag HE, Molema G, Bijlsma JJ. Interactions between blood-borne Streptococcus pneumoniae and the blood-brain barrier preceding meningitis. PLoS One. 2013;8:e68408. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 72] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 12. | van Sorge NM, Doran KS. Defense at the border: the blood-brain barrier versus bacterial foreigners. Future Microbiol. 2012;7:383-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 87] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 13. | Meyer KF, Eddie B, Stevens IM. Recent Studies on Psittacosis. Am J Public Health Nations Health. 1935;25:571-579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 14. | Greco G, Corrente M, Martella V. Detection of Chlamydophila psittaci in asymptomatic animals. J Clin Microbiol. 2005;43:5410-1; author reply 5410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 15. | Rane V, Khailin K, Williams J, Francis M, Kotsanas D, Korman TM, Graham M. Underdiagnosis of Chlamydia trachomatis and Chlamydia psittaci revealed by introduction of respiratory multiplex PCR assay with Chlamydiaceae family primers. Diagn Microbiol Infect Dis. 2018;90:163-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 16. | Charles PG, Whitby M, Fuller AJ, Stirling R, Wright AA, Korman TM, Holmes PW, Christiansen KJ, Waterer GW, Pierce RJ, Mayall BC, Armstrong JG, Catton MG, Nimmo GR, Johnson B, Hooy M, Grayson ML; Australian CAP Study Collaboration. The etiology of community-acquired pneumonia in Australia: why penicillin plus doxycycline or a macrolide is the most appropriate therapy. Clin Infect Dis. 2008;46:1513-1521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 158] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 17. | de Gier B, Hogerwerf L, Dijkstra F, van der Hoek W. Disease burden of psittacosis in the Netherlands. Epidemiol Infect. 2018;146:303-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 18. | Armstrong GL, MacCannell DR, Taylor J, Carleton HA, Neuhaus EB, Bradbury RS, Posey JE, Gwinn M. Pathogen Genomics in Public Health. N Engl J Med. 2019;381:2569-2580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 163] [Article Influence: 27.2] [Reference Citation Analysis (0)] |