Published online Aug 16, 2021. doi: 10.12998/wjcc.v9.i23.6867
Peer-review started: March 17, 2021
First decision: April 4, 2021
Revised: April 13, 2021
Accepted: June 28, 2021
Article in press: June 28, 2021
Published online: August 16, 2021
Processing time: 141 Days and 5.2 Hours
The FGFR signaling pathway is activated in multiple tumor types through gene amplifications, single base substitutions, or gene fusions. Novel FGFR gene fusions may represent candidate targets for the development of tyrosine kinase inhibitors.
Herein, we report a patient with colorectal cancer (CRC) harboring a novel FGFR2 fusion gene. A 59-year-old man felt discomfort in his right upper abdomen with loss of appetite for 6 mo. An abdominal computed tomography scan revealed the existence of a space-occupying lesion in the ascending colon. The pathological diagnosis was a poorly differentiated adenocarcinoma. Subsequent biopsy specimen was subjected to next-generation sequencing analysis, and a novel FGFR2-TSC22D1 fusion with complete kinase structure of FGFR2 protein was identified.
We report the first case of CRC harboring FGFR2-TSC22D1, which enriches the FGFR2 fusion spectrum. FGFR2 inhibitors might be effective in the later treatment for this patient.
Core Tip: A colorectal cancer patient had a novel FGFR2-TSC22D1 fusion which included exons 1-17 of FGFR2 and exons 3 of TSC22D1. This fusion contains FGFR2 kinase domain and coil coiled domains encoded by TSC22D1 exon 3, which might induce oncogenesis. Our case enriches the FGFR2 fusion spectrum. We believe that these novel findings have important implications in the strategy development of therapy for colorectal cancer.
- Citation: Kao XM, Zhu X, Zhang JL, Chen SQ, Fan CG. FGFR2-TSC22D1, a novel FGFR2 fusion gene identified in a patient with colorectal cancer: A case report. World J Clin Cases 2021; 9(23): 6867-6871
- URL: https://www.wjgnet.com/2307-8960/full/v9/i23/6867.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i23.6867
Colorectal cancer (CRC) is one of the most common causes of cancer-related death worldwide and the 3rd most common cancer in males[1]. In recent years, the burden of CRC is increasing rapidly in China[2]. With the advent of targeted drugs such as epidermal growth factor or vascular endothelial growth factor tyrosine kinase inhibitors (TKIs), the overall survival of CRC increased from the past 8-12 mo to 30 mo nowadays[3-5].
FGFR fusions in solid tumors are caused by chromosomal rearrangements. FGFR families includes FGFR1-4, four highly conserved receptor tyrosine kinases, among which FGFR2 has been proven to be a potential target of FGFR TKI inhibitors. At the same time, in many solid tumors, such as urothelial carcinomas and intrahepatic cholangiogarcinomas, FGFR activating molecular alterations (including FGFR3 mutations and FGFR2 fusions) have been found, expanding the options for patients who may benefit from FGFR inhibitors[6]. With the development of next generation sequencing (NGS), some uncommon genomic mutations are detected, such as FGFR2-PPHLN1[7], FGFR2-BICC1[8], and FGFR2-CCDC6 fusion mutations[9]. In this case, with the help of NGS detection technology, we found a CRC patient carrying a rare FGFR2-TSC22D1 fusion gene, which further expanded the FGFR2 fusion variant spectrum.
A 59-year-old male patient with right upper quadrant pain and discomfort, and no appetite for 6 mo, was referred to our hospital for further treatment.
Six months ago, the patient developed right upper abdominal distension, pain, and discomfort without obvious inducement, accompanied by anorexia. No nausea, vomiting, chills, high fever, palpitation, shortness of breath, or yellow staining of skin and mucous membrane was noted. At a local hospital, colorectal examination revealed an ulcerative mass in the liver flexure of the ascending colon. There was a 0.6 cm × 0.6 cm polyp in the middle of the transverse colon. Pathological report suggested that the ascending colon mass was poorly differentiated adenocarcinoma.
Physical examination revealed that the patient’s body temperature was 36.8 °C, heart rate 78 bpm/min, respiratory 16 bpm/min, and blood pressure 122/78 mmHg.
The white cell count was 9.46 × 109/L, hemoglobin was 62 g/mL , and platelet count was 451 × 109/L. Serum tumor markers carcinoembryonic antigen (7.58 ng/mL), carbohydrate antigen (CA)19-9 (21.63 U/mL), CA125 (44.85 U/mL), CA242 (21.57 IU/mL), CA724 (8.38 IU/mL), and CA153 (4.59 IU/mL) were detectable.
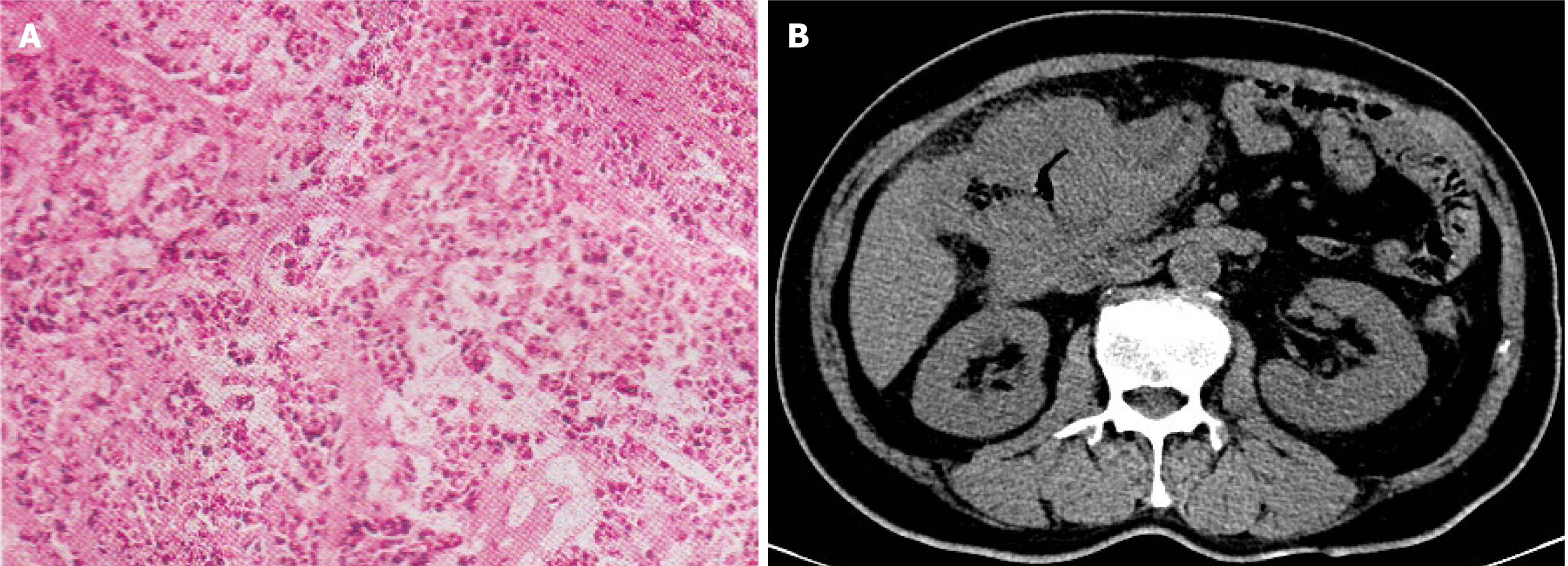
The pathological diagnosis under ascending colonoscopy was poorly differentiated adenocarcinoma (cT4N+M0, Figure 1).
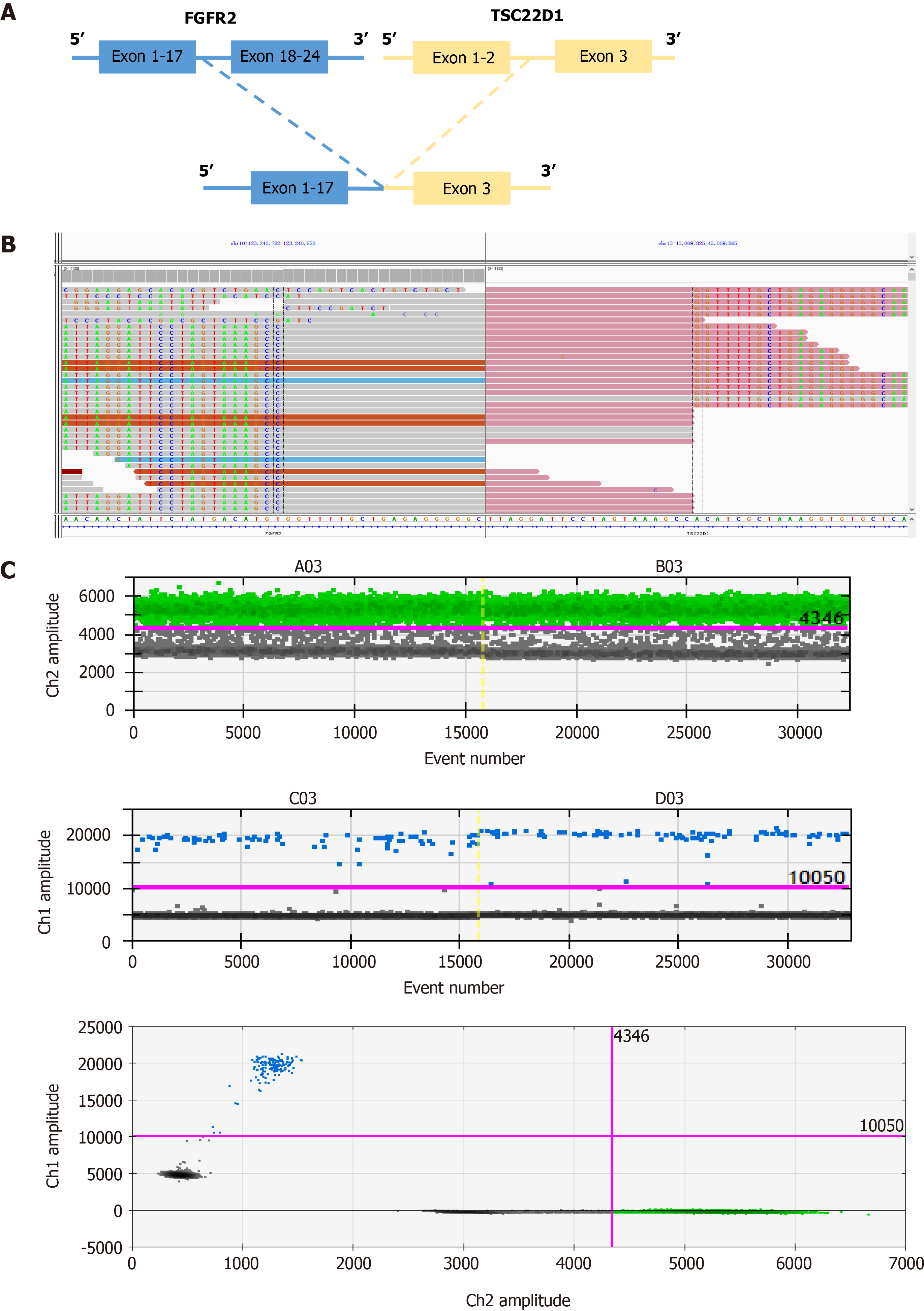
The tissue biopsy specimens were then analyzed by NGS and a rare intergenic region between FGFR2 and TSC22D1 fusion variation was detected (Figure 2A and B). TSC22D1 (TSC22 domain family protein 1) gene is a transcription factor belonging to a large family of early response. Dimers of TSC22D1 act as a transcription factor and have tumor suppressor function. The FGFR2-TSC22D1 fusion gene includes exons 1-17 of FGFR2 and exons 3 of TSC22D1, retaining the complete kinase structure of FGFR2. The ratio of FAM-positivity (FGFR2 gene) to VIC (internal reference gene) was 0.0048 in droplet digital polymerase chain reaction (ddPCR), indicating weakly FGFR2 expression (Figure 2C) and proving that FGFR2 was positive.
A computed tomography scan of the abdomen showed the presence of a space-occupying lesion in the liver flexure of the ascending colon.
The patient was finally diagnosed as having stage cT4N+M0 CRC and carrying the novel FGFR2-TSC22D1 fusion gene.
Based on pathological staging, the patient was given preoperative chemotherapy for tumor down-staging. The regimen is oxaliplatin (200 mg, D1, intravenous drips) plus capecitabine (1500 mg, D1-14, oral). After six cycles of preoperative chemotherapy, the patient underwent radical resection for CRC.
Postoperative pathology showed that no residual cells were found in the duodenal wall, which suggested that a pathological complete response has been achieved.
FGFR2 fusions have been identified as a novel oncogenic target for drug development in a number of cancers including breast cancer[10] and intrahepatic cholangiocarcinoma[7-9]. Interestingly, a novel FGFR2 gene fusion was identified in the CRC patient reported in this paper, suggesting that this event may represent a new candidate therapeutic target for which similar strategies could be used in the clinical management.
A comprehensive understanding of FGFR2 fusion information seems to be necessary. NGS could be used as a supplementary method for FGFR2 variation for high-throughput molecular analysis, while detecting gene copy number alterations, fusions, insertions, and deletions simultaneously.
In our case, a rare FGFR2 fusion gene, confirmed by ddPCR, was found, which enriched the FGFR2 fusion spectrum. FGFR2 inhibitors might be effective in the later treatment for this patient.
Manuscript source: Unsolicited manuscript
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A, A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kraja F, Valiveti CK S-Editor: Fan JR L-Editor: Wang TQ P-Editor: Li JH
| 1. | Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467-1480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1570] [Cited by in RCA: 3003] [Article Influence: 500.5] [Reference Citation Analysis (3)] |
| 2. | Feng RM, Zong YN, Cao SM, Xu RH. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Commun (Lond). 2019;39:22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 733] [Cited by in RCA: 1121] [Article Influence: 186.8] [Reference Citation Analysis (1)] |
| 3. | Heinemann V, von Weikersthal LF, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran SE, Heintges T, Lerchenmüller C, Kahl C, Seipelt G, Kullmann F, Stauch M, Scheithauer W, Hielscher J, Scholz M, Müller S, Link H, Niederle N, Rost A, Höffkes HG, Moehler M, Lindig RU, Modest DP, Rossius L, Kirchner T, Jung A, Stintzing S. FOLFIRI plus cetuximab vs FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): a randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15:1065-1075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1117] [Cited by in RCA: 1328] [Article Influence: 120.7] [Reference Citation Analysis (0)] |
| 4. | Van Cutsem E, Köhne CH, Láng I, Folprecht G, Nowacki MP, Cascinu S, Shchepotin I, Maurel J, Cunningham D, Tejpar S, Schlichting M, Zubel A, Celik I, Rougier P, Ciardiello F. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29:2011-2019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1314] [Cited by in RCA: 1452] [Article Influence: 103.7] [Reference Citation Analysis (0)] |
| 5. | Loupakis F, Cremolini C, Masi G, Lonardi S, Zagonel V, Salvatore L, Cortesi E, Tomasello G, Ronzoni M, Spadi R, Zaniboni A, Tonini G, Buonadonna A, Amoroso D, Chiara S, Carlomagno C, Boni C, Allegrini G, Boni L, Falcone A. Initial therapy with FOLFOXIRI and bevacizumab for metastatic colorectal cancer. N Engl J Med. 2014;371:1609-1618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 689] [Cited by in RCA: 794] [Article Influence: 72.2] [Reference Citation Analysis (0)] |
| 6. | Facchinetti F, Hollebecque A, Bahleda R, Loriot Y, Olaussen KA, Massard C, Friboulet L. Facts and New Hopes on Selective FGFR Inhibitors in Solid Tumors. Clin Cancer Res. 2020;26:764-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 92] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 7. | Sia D, Losic B, Moeini A, Cabellos L, Hao K, Revill K, Bonal D, Miltiadous O, Zhang Z, Hoshida Y, Cornella H, Castillo-Martin M, Pinyol R, Kasai Y, Roayaie S, Thung SN, Fuster J, Schwartz ME, Waxman S, Cordon-Cardo C, Schadt E, Mazzaferro V, Llovet JM. Massive parallel sequencing uncovers actionable FGFR2-PPHLN1 fusion and ARAF mutations in intrahepatic cholangiocarcinoma. Nat Commun. 2015;6:6087. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 233] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 8. | Ying X, Tu J, Wang W, Li X, Xu C, Ji J. FGFR2-BICC1: A Subtype Of FGFR2 Oncogenic Fusion Variant In Cholangiocarcinoma And The Response To Sorafenib. Onco Targets Ther. 2019;12:9303-9307. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 9. | Wang Y, Ding X, Wang S, Moser CD, Shaleh HM, Mohamed EA, Chaiteerakij R, Allotey LK, Chen G, Miyabe K, McNulty MS, Ndzengue A, Barr Fritcher EG, Knudson RA, Greipp PT, Clark KJ, Torbenson MS, Kipp BR, Zhou J, Barrett MT, Gustafson MP, Alberts SR, Borad MJ, Roberts LR. Antitumor effect of FGFR inhibitors on a novel cholangiocarcinoma patient derived xenograft mouse model endogenously expressing an FGFR2-CCDC6 fusion protein. Cancer Lett. 2016;380:163-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 71] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 10. | Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, Lonigro RJ, Vats P, Wang R, Lin SF, Cheng AJ, Kunju LP, Siddiqui J, Tomlins SA, Wyngaard P, Sadis S, Roychowdhury S, Hussain MH, Feng FY, Zalupski MM, Talpaz M, Pienta KJ, Rhodes DR, Robinson DR, Chinnaiyan AM. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636-647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 504] [Cited by in RCA: 578] [Article Influence: 48.2] [Reference Citation Analysis (0)] |