Published online Jul 16, 2021. doi: 10.12998/wjcc.v9.i20.5621
Peer-review started: February 17, 2021
First decision: March 11, 2021
Revised: March 31, 2021
Accepted: April 20, 2021
Article in press: April 20, 2021
Published online: July 16, 2021
Processing time: 139 Days and 23.5 Hours
Mycobacterium mucogenicum (M. mucogenicum) belongs to the group of rapidly growing Nontuberculous mycobacteria. This microorganism is associated with a wide spectrum of infectious diseases. Due to a low detection rate or the time required for conventional culture methodology, a rapid and broad-spectrum method is necessary to identify rare pathogens.
A 12-year-old immunocompetent girl presented with painful masses for five months. The first mass was found in the right upper quadrant of the abdomen, and was about 1 cm × 1.5 cm in size, tough but pliable in texture, with an irregular margin and tenderness. An abscess gradually formed and ulcerated with suppuration of the mass. Three new masses appeared on the back one by one. Chest computed tomography showed patchy and streaky cloudy opacities in both lungs. Needle aspiration of the abscess was performed, but the smear and conventional culture were negative, and the pathological examination showed no pathogens. We then performed next-generation sequencing using a formalin-fixed, paraffin-embedded specimen to identify the pathogen. A significantly high abundance of M. mucogenicum was detected. The patient’s abscesses gradually decreased in size, while inflammation in both lungs improved following 12-wk of treatment. No recurrence was observed four months after the end of the one-year treatment period.
Next-generation sequencing is a promising tool for the rapid and accurate diagno
Core Tip: Mycobacterium mucogenicum (M. mucogenicum) belongs to the group of Nontuberculous mycobacteria, and is associated with a wide spectrum of clinical diseases, including osteomyelitis, respiratory tract, bloodstream, and disseminated infections in both immunocompetent and immunosuppressed individuals. However, time-consuming techniques and a low detection rate for identifying this pathogen usually lead to delayed or missed diagnosis. We present a case of disseminated infection with M. mucogenicum diagnosed in a formalin-fixed, paraffin-embedded (FFPE) specimen using next-generation sequencing (NGS). NGS is a promising tool for the rapid and accurate diagnosis of rare pathogens, even when using a FFPE speci
- Citation: Liu J, Lei ZY, Pang YH, Huang YX, Xu LJ, Zhu JY, Zheng JX, Yang XH, Lin BL, Gao ZL, Zhuo C. Rapid diagnosis of disseminated Mycobacterium mucogenicum infection in formalin-fixed, paraffin-embedded specimen using next-generation sequencing: A case report. World J Clin Cases 2021; 9(20): 5621-5630
- URL: https://www.wjgnet.com/2307-8960/full/v9/i20/5621.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i20.5621
Nontuberculous mycobacteria (NTM) were initially recognized as pathogens in the mid 1950’s[1]. More and more species were discovered due to recognition by medical workers and improvements in examination techniques. Some species may cause diseases in humans[1]. NTM has been reported to include more than 170 species worldwide, including two types: rapidly growing mycobacteria (RGM) and slow growing mycobacteria[2]. Mycobacterium mucogenicum (M. mucogenicum) belongs to the group of RGM, and is considered an environmental organism with a worldwide distribution. It is associated with a wide spectrum of clinical diseases, including osteomyelitis, respiratory tract, bloodstream, and disseminated infections in both immunocompetent and immunosuppressed individuals[3]. However, the time required and low detection rate to identify this pathogen usually leads to delayed or missed diagnosis. Thus, a rapid and broad-spectrum method is necessary to identify rare pathogens. Next-generation sequencing (NGS) is an unbiased approach whose RefSeq contains 4152 whole genome sequences of viral taxa, 3446 bacterial genomes or scaffolds, 206 fungi related to human infection, and 140 parasites associated with human diseases. It identifies the nucleotides in the target samples and compares the detected nucleotides against the catalogue library of pathogens, thereby listing the possible causative agents in the clinical samples[4]. The specimens examined are usually fresh ones, such as blood, pus, bronchoalveolar lavage fluid, cerebrospinal fluid, lymph nodes, tissue, and so on[5-7]. However, there are few reports on formalin-fixed, paraffin-embedded (FFPE) specimens, and it is unclear whether the clinical significance of the detection results using FFPE is useful.
Here, we report a case with disseminated infection of M. mucogenicum detected by NGS using a FFPE specimen.
A 12-year-old Han Chinese girl (30 kg), who was a middle school student, presented to our hospital with several masses on the abdomen and back with low-grade fever for five months.
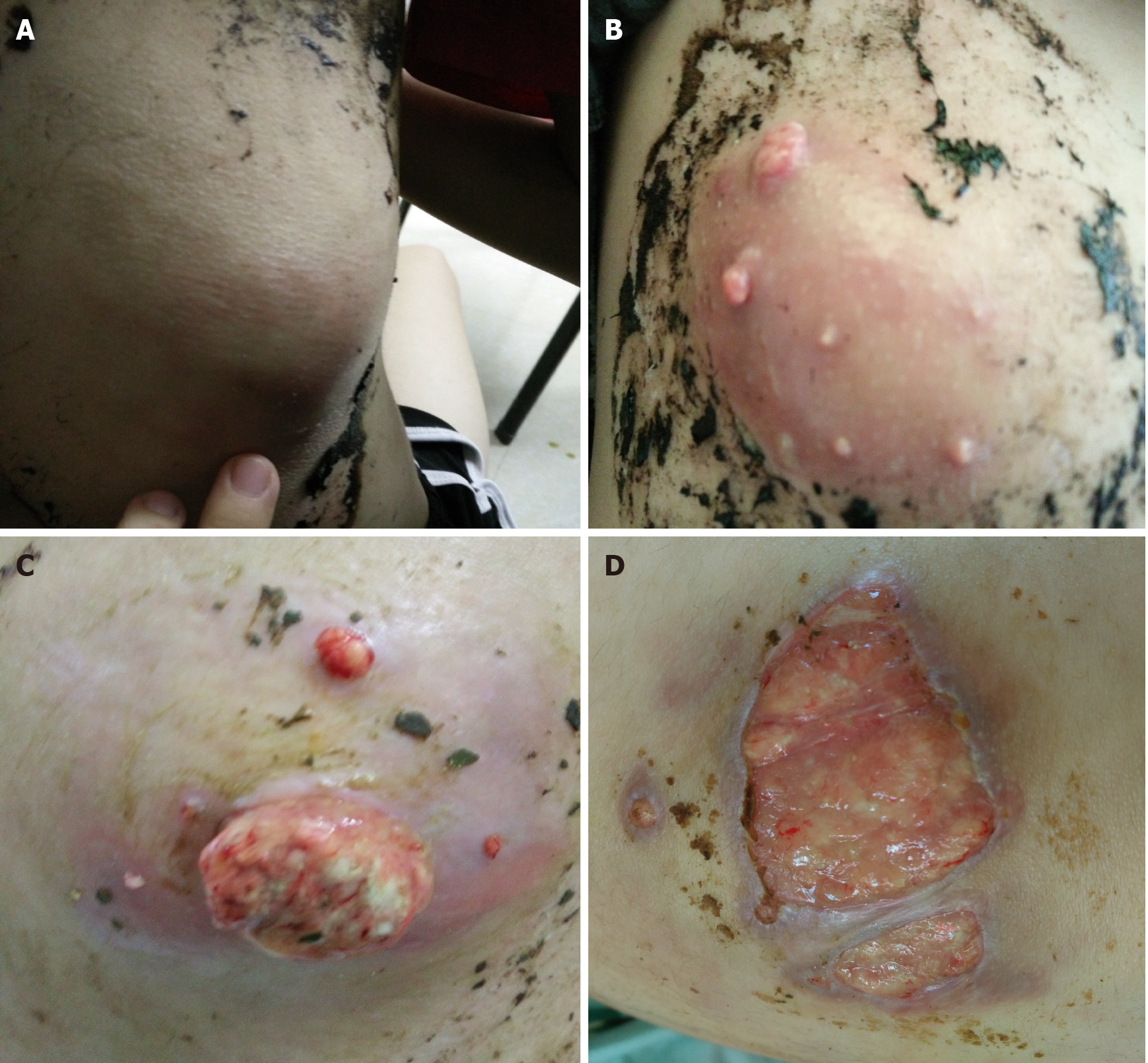
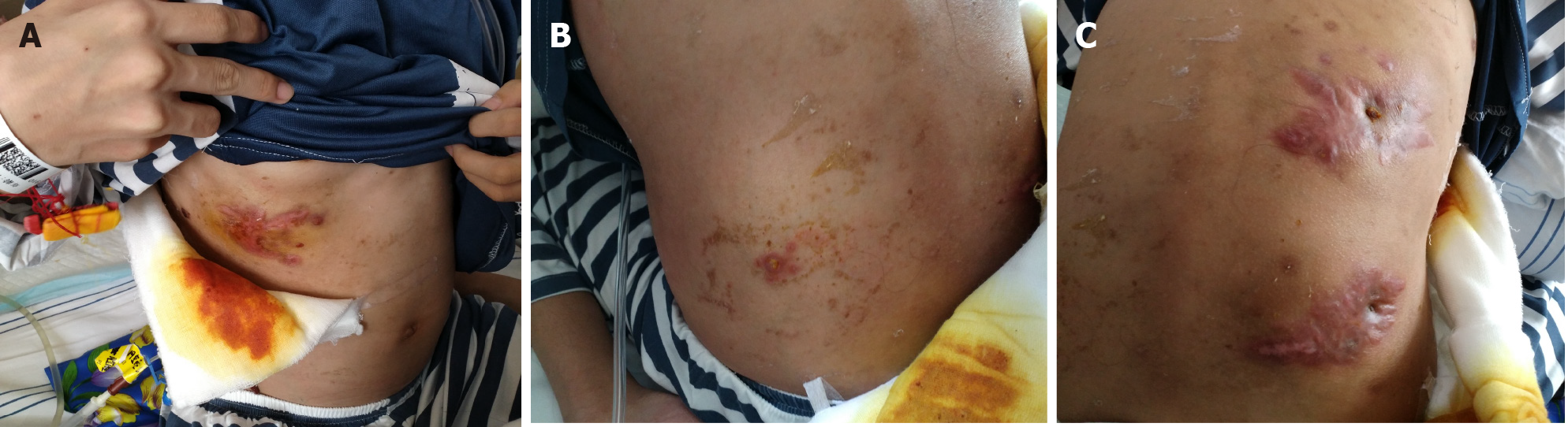
Five months previously, a painful tender mass was found in the right upper quadrant of the girl’s abdomen. The patient reported low-grade fever with a temperature of 37.5℃ and fatigue. Chinese herbal medicines for external application were used for the mass; however, the mass enlarged and an abscess gradually formed, and then ulcerated with suppuration. Subsequently, three other masses appeared, one by one, with one on the left back and two on the right back, which were similar to the first mass (Figure 1). She also had a mild cough without sputum. There was no night sweating, diarrhea, or nausea.
The patient did not recall any previous back or abdominal injury.
The patient and her family had no previous medical history.
The patient’s temperature was 37.5℃, heart rate was 105 bpm, respiratory rate was 20 breaths/min, blood pressure was 105/62 mmHg and oxygen saturation in room air was 99%. Four painful subcutaneous masses were palpable, one on the right upper abdomen, one on the left back, and two on the right back. The masses ranged from 3.0 to 4.0 cm in diameter, were tough but pliable in texture, with tenderness, an irregular margin and discharging pus. Moist rales were found at the bottom of both lungs.
Complete blood cell counts showed that the white blood cell count (WBC) was 25.24 × 109/L with 40% neutrophils, 8.20% lymphocytes, red blood cell count (RBC) was 2.48 × 1012/L, and hemoglobin (HGB) was 54 G/L. The erythrocyte sedimentation rate (ESR) was > 140 mm/h and procalcitonin (PCT) was 2.39 ng/mL.
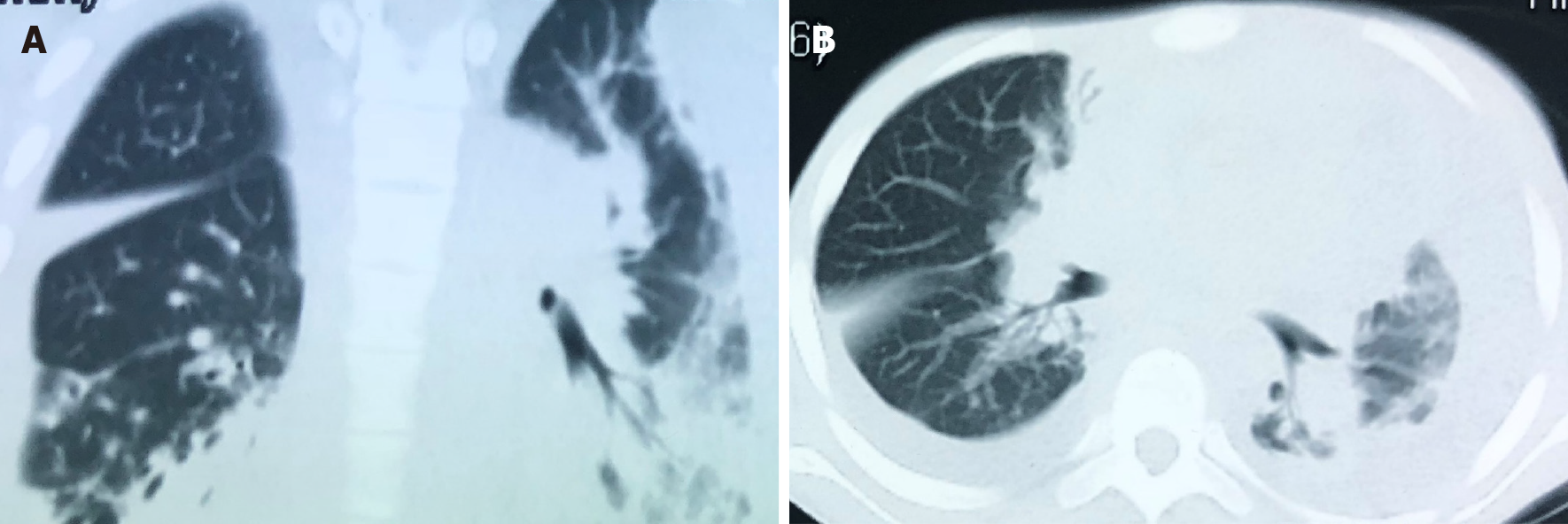
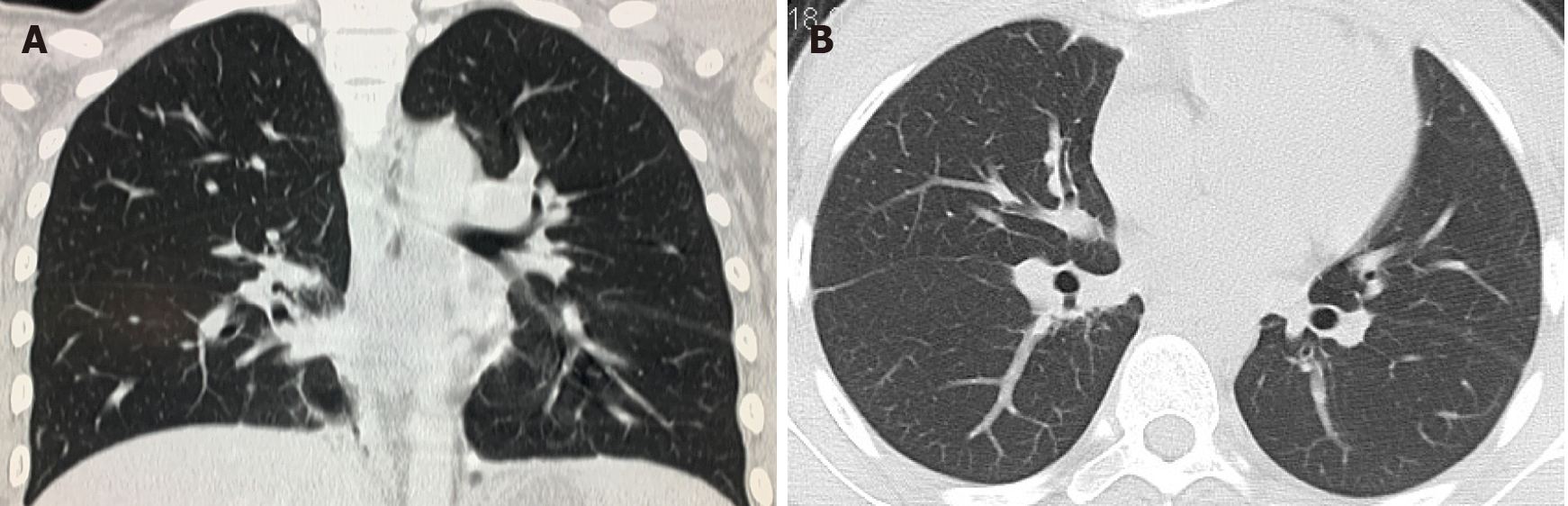
Whole abdomen computed tomography (CT) on September 13th showed obvious swelling of both bilateral psoas, iliopsoas and back muscles, suggesting infection and multiple abscesses, peritonitis and a moderate amount of ascites. The chest CT on September 21st showed massive pleural effusion in the left chest, a small amount of pleural effusion on the right, patchy and streaky cloudy opacities in both lungs, suggesting inflammation (Figure 2). Color Doppler echocardiography on September 28th showed a markedly enlarged right atrium and left atrium, aortic insufficiency, tricuspid insufficiency, pulmonary hypertension, mild to moderate pericardial effusion, left ventricular contractile and diastolic dysfunction.
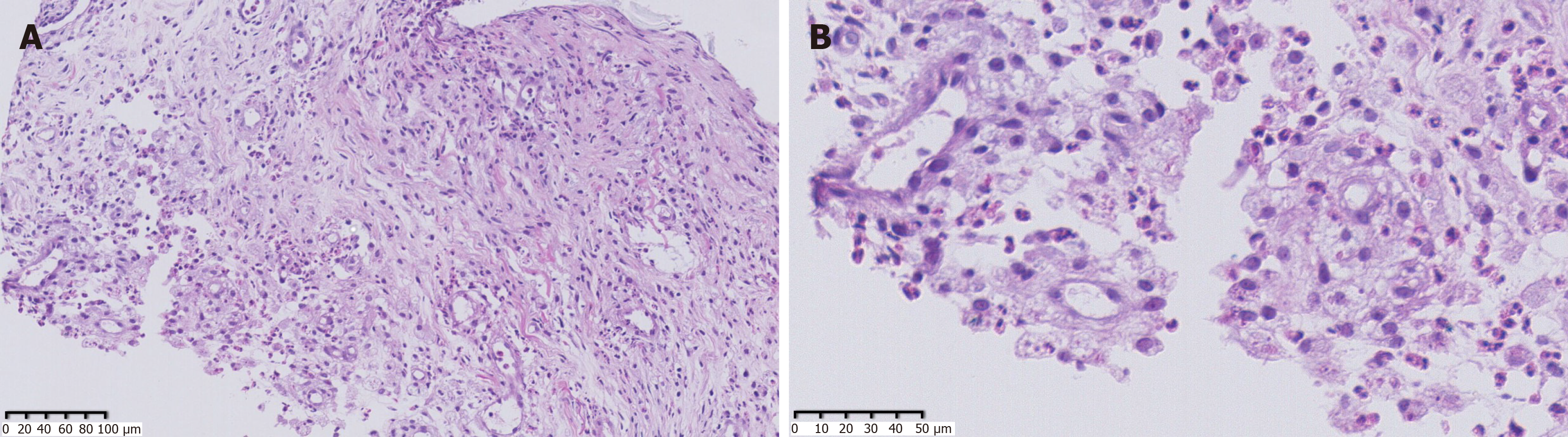
As the patient had signs of heart failure and multiple abscesses, she was transferred to the intensive care unit of the local hospital. A series of assays were performed as follows: C-reactive protein (CRP) 74.2 mg/L, serum amyloid A (SAA) 421 mg/L, PCT 1.69 ng/mL, creatine kinase isoenzyme MB (CKMB) 0.97 ng/mL, and N-terminal prohormone of brain natriuretic peptide (NT-proBNP): 18136 pg/mL. Her hepatic and renal function were normal. HIV screen was negative, and CD4 and CD8 counts were normal. Tuberculosis-antibody and T-SPOT were negative. All autoantibodies were negative except for anticardiolipin antibodies. She was treated with intravenous piperacillin-tazobactam for 9 d, plus linezolid for 3 d for anti-infection, and deslanoside and digoxin for anti-heart failure. The child’s chest congestion and polyserositis improved after these treatments, but the abscesses and lesions in both lungs did not improve. Abscess aspiration and drainage were then performed. No pathogens were found in pus from the abscess, ascites or sputum smear including acid-fast smear. Multiple culture of blood, aspirated pus, ascites and sputum, including common bacteria and fungi culture, as well as incubation in Loewenstein-Jensen medium at 37℃ for 42 d, were all negative. Adenosine deaminase (ADA) in abscess drainage fluid was 2 U/L. The pathological manifestations in abscess tissue from the right psoas showed numerous lymphocytes and neutrophils infiltration. A small number of multinuclear giant cells, and granulation tissues were found, indicating suppurative and granulomatous inflammation (Figure 3). Due to her cardiac insufficiency, fiber bronchoscopy was not feasible at the time.
Therefore, NGS of the patient’s FFPE specimen was performed to identify the causative pathogen.
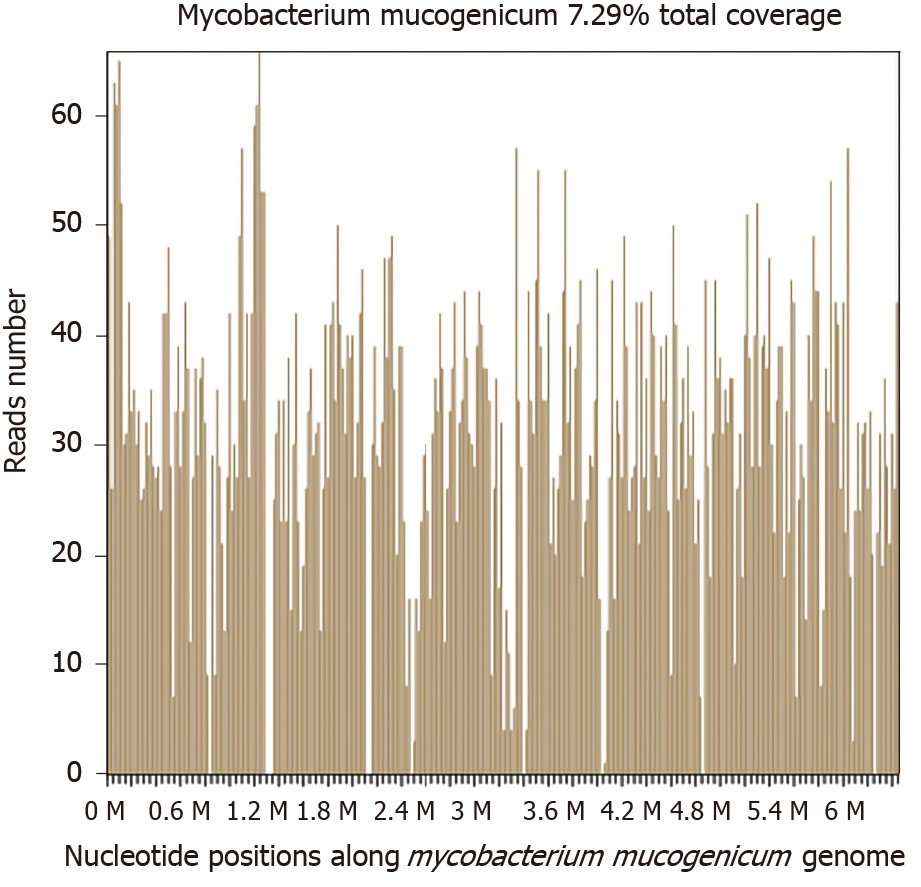
Two days later, a significantly high abundance of Mycobacterium was detected with 9261 reads, 6225 of which matched M. mucogenicum. Two other pathogens were also found: Aggregatibacter (7246 reads, including Aggregatibacter actinomycetemcomitans 6912 reads and Aggregatibacter segnis 4 reads) and Meiothermus (6938 reads, including Meiothermus silvanus 6461 reads and Meiothermus ruber 287 reads) (Figure 4). The former is normally found in the human oropharynx, which seldom causes disease, but occasionally causes infective endocarditis, pneumonia, brain abscess, etc. There are no reports on skin infections caused by Aggregatibacter. The latter is a Gram-negative thermophilic bacterium. To the best of our knowledge, there are no reports of human diseases caused by Meiothermus.
In addition, the patient and her parents underwent analysis of genes related to immunologic function, but no clinically significant findings were observed. No evidence of interferon-γ receptor deficiency or hyper-IgE syndrome was found.
M. mucogenicum was treated as the causative pathogen of the abscesses. M. mucogenicum is closely related to the M. fortuitum group[8], which is usually sensitive to clarithromycin, trimethoprim–sulfamethoxazole (TMP-SMX), doxycycline, quinolones, aminoglycosides and so on as suggested by the SANFORD GUIDE[9]. Due to the limitation of quinolones and aminoglycoside antibiotics use in children in China, she was given clarithromycin 250 mg bid plus TMP-SMX 2# bid plus doxycycline 50 mg bid.
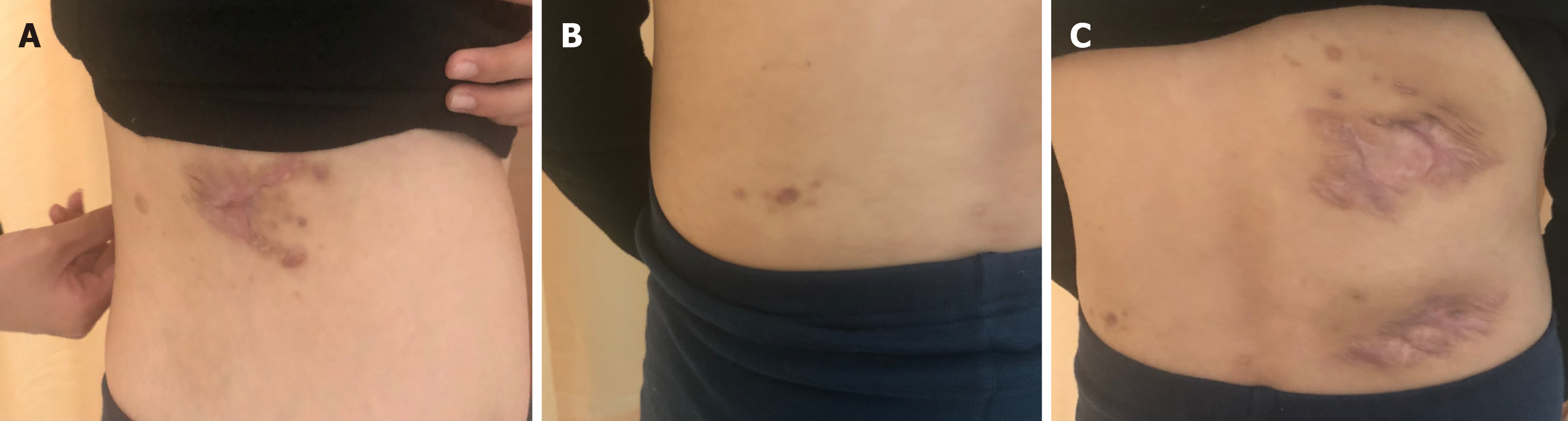
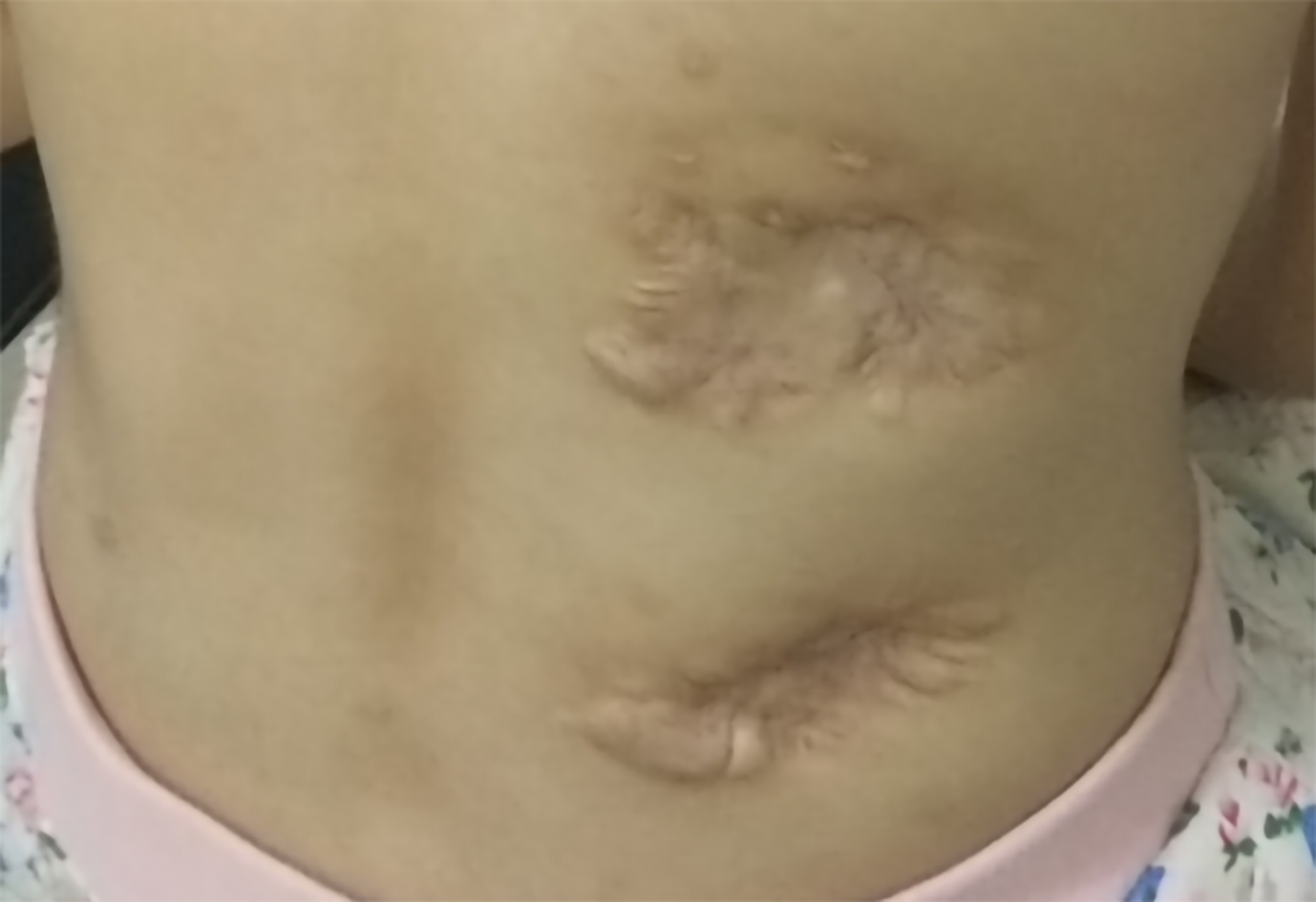
All abscesses decreased in size (Figure 5), her cough disappeared, anemia recovered and the child gained weight 12 d later. The WBC count, neutrophils%, ESR, CRP, PCT, NT-proBNP, and CKMB gradually returned to normal. The wounds healed gradually with scars within 12 wk (Figure 6), while inflammation in both lungs significantly improved but not all was absorbed and a little fibrosis was present, suggesting chronic infection (Figure 7). There were no notable adverse reactions in this patient during the one-year treatment period. The patient had a normal biochemical index with no recurrence four months after the end of the one-year treatment period (Figure 8).
M. mucogenicum, a ubiquitous RGM, has been recognized as a species since 1995. The organism was first called M. chelonae-like organism and caused diseases in humans in 1982, and during two outbreaks of peritonitis associated with peritoneal dialysis in the United States in 1976 and 1978[10]. M. mucogenicum is more closely related to the M. fortuitum group by 16S rDNA sequencing than other Mycobacterium groups[8]. The mucoid surface provides the ability to form a protective biofilm, which contributes to infection of central venous catheters and skin soft-tissue usually post-trauma, which are clinically the most common sites of M. mucogenicum infections[11]. Moreover, it can cause respiratory infections, bloodstream infections, osteomyelitis, and disseminated infections, which are common in immunocompromised hosts[3,12]. Equipment exposed to M. mucogenicum contaminated water can lead to nosocomial infections, including central venous catheter-related and hemodialysis-related infections[1,13].
There was no evidence of immunodeficiency in our patient, even after the detection of genes related to immunologic function in the patient and her parents. Undoubtedly, it was necessary to determine the source of infection, but we did not have evidence of pneumonia with NTM infection. We could not perform fiber bronchoscopy in this patient as she had cardiac insufficiency. However, her pneumonia rapidly improved after anti-NTM treatment. It is unlikely that the infection was nosocomial. Due to the pulmonary lesions and multiple skin lesions and the good therapeutic effect, it is reasonable to presume that the source of infection with this microorganism was via the upper or lower respiratory tract, then a bloodstream infection, followed by a skin soft-tissue infection.
For identification of this microorganism, there are several major methods. The first is conventional culture methodology. This is still difficult and delays in culture lead to missed diagnosis[14]. The second is DNA sequencing with 16S rRNA gene, rpoB, and hsp65 being recognized as useful targets[15-17]. However, due to its targeting, limitations are unavoidable in the identification of uncertain pathogens. The third is matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). Several investigators have demonstrated that MALDI-TOF MS can accurately identify mycobacteria[18,19]. However, these methods are not available in many laboratories.
In the present study, smears and cultures for bacteria, fungi and tuberculosis culture were all negative. Moreover, the results of the biopsy on the right psoas were not satisfactory. Therefore, we performed NGS using a FFPE specimen. Metagenomic NGS has emerged as a promising technological platform for directly identifying and classifying pathogens in complex clinical specimens from patients in a short time at reasonable cost[4]. It works well with short nucleic acid sequences, thus, it can be used to analyze fragmented DNA and RNA extracted from standard FFPE clinical specimens in theory. FFPE specimens have gained interest as an alternative to fresh/frozen tissue for storage, due to FFPE tissue validation in archival samples of biomarkers of exposure, prognosis and disease[20]. FFPE tissue processing remains the most economical approach for longitudinal tissue specimen storage. The ability to apply high throughput genomic applications to FFPE specimens can expand clinical study[21]. We have performed the mNGS technique to analyze the presence of pathogens in FFPE tissues which is one of the most widely available techniques for preserving clinical specimens. The macromolecules extracted from blocks stored for over 11-12 years, 5-7 years, or 1-2 years were not significantly different to recently prepared blocks[22]. FFPE tissue specimens are collected following IRB-approved protocols and stored on site in our climate-controlled biorepository. The FFPE tissue can match both clinical and molecular research endpoints. FFPE specimens collected in a routine clinical environment are a good source of DNA that can be applied in mNGS analysis for clinical pathogen detection[23]. In previous research, FFPE specimens were used for whole genome sequencing, mainly in cancer patients[23,24]. Using FFPE tissue for pathogen identification was also reported in recent years, such as 16S PCR of brain tissue[25], RNA sequencing of 1918 influenza samples[26] and corneal infection diagnosis[27].
To our knowledge, this is the first report on M. mucogenicum confirmed by NGS using a FFPE specimen. In this study, it was feasible to use metagenomic NGS of DNA extracted from a FFPE clinical specimen to identify the causative microorganism. This method holds great promise for the relatively rapid detection of microorganisms, including rare pathogens and cases in which conventional cultures were not attempted or failed to yield positive results.
Following combination therapy with clarithromycin, doxycycline and TMP-SMX, the patient was treated for over 1 yr, and improvement was observed. However, the optimal duration of antimicrobial therapy for disseminated infection with M. mucogenicum is unknown. It is certain that prolonged therapy for one to two years even more contributes to the eradication of infection and reduces the chance of recurrence[1].
Next-generation sequencing is a promising tool for the rapid and accurate diagnosis of rare pathogens based on negative results with the conventional culture, even when using a formalin-fixed, paraffin-embedded specimen.
We thank the patient and her parents for their support and cooperation in publishing this work. We also thank the doctors of the local hospital and another hospital in Guangzhou for the patients’ initial diagnosis and treatment, and the doctor of the First Affiliated Hospital of Sun Yat-Sen University for the patient’s pathological diagnosis. We also thank BGI Genomics Co., Ltd for identification of M. mucogenicum.
Manuscript source: Unsolicited manuscript
Specialty type: Infectious diseases
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ozair A S-Editor: Gong ZM L-Editor: Webster JR P-Editor: Li JH
| 1. | Fleming GA, Frangoul H, Dermody TS, Halasa N. A cord blood transplant recipient with Mycobacterium mucogenicum central venous catheter infection after infusion of tap water. Pediatr Infect Dis J. 2006;25:567-569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 2. | Decostere A, Hermans K, Haesebrouck F. Piscine mycobacteriosis: a literature review covering the agent and the disease it causes in fish and humans. Vet Microbiol. 2004;99:159-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 115] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 3. | El Helou G, Hachem R, Viola GM, El Zakhem A, Chaftari AM, Jiang Y, Tarrand J, Raad II. Management of rapidly growing mycobacterial bacteremia in cancer patients. Clin Infect Dis. 2013;56:843-846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 4. | Gu W, Miller S, Chiu CY. Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection. Annu Rev Pathol. 2019;14:319-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 389] [Cited by in RCA: 810] [Article Influence: 115.7] [Reference Citation Analysis (0)] |
| 5. | Miao Q, Ma Y, Wang Q, Pan J, Zhang Y, Jin W, Yao Y, Su Y, Huang Y, Wang M, Li B, Li H, Zhou C, Li C, Ye M, Xu X, Li Y, Hu B. Microbiological Diagnostic Performance of Metagenomic Next-generation Sequencing When Applied to Clinical Practice. Clin Infect Dis. 2018;67:S231-S240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 579] [Article Influence: 96.5] [Reference Citation Analysis (0)] |
| 6. | Koehler JW, Douglas CE, Minogue TD. A highly multiplexed broad pathogen detection assay for infectious disease diagnostics. PLoS Negl Trop Dis. 2018;12:e0006889. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 7. | Wilson MR, Naccache SN, Samayoa E, Biagtan M, Bashir H, Yu G, Salamat SM, Somasekar S, Federman S, Miller S, Sokolic R, Garabedian E, Candotti F, Buckley RH, Reed KD, Meyer TL, Seroogy CM, Galloway R, Henderson SL, Gern JE, DeRisi JL, Chiu CY. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014;370:2408-2417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 609] [Cited by in RCA: 672] [Article Influence: 61.1] [Reference Citation Analysis (0)] |
| 8. | Springer B, Böttger EC, Kirschner P, Wallace RJ Jr. Phylogeny of the Mycobacterium chelonae-like organism based on partial sequencing of the 16S rRNA gene and proposal of Mycobacterium mucogenicum sp. nov. Int J Syst Bacteriol. 1995;45:262-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 104] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 9. | Gilbert DN, Eliopoulos GM, Chambers HF, Saag MS, Pavia AT. The Sanford Guide to Antimicrobial Therapy 2016 (46th Edition). Antimicrobial Therapy, Incorporated, 2016: 145. |
| 10. | Band JD, Ward JI, Fraser DW, Peterson NJ, Silcox VA, Good RC, Ostroy PR, Kennedy J. Peritonitis due to a mycobacterium chelonei-like organism associated with intermittent chronic peritoneal dialysis. J Infect Dis. 1982;145:9-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 137] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 11. | Armbruster CR, Forster TS, Donlan RM, O'Connell HA, Shams AM, Williams MM. A biofilm model developed to investigate survival and disinfection of Mycobacterium mucogenicum in potable water. Biofouling. 2012;28:1129-1139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 12. | Abidi MZ, Ledeboer N, Banerjee A, Hari P. Mycobacterium mucogenicum bacteremia in immune-compromised patients, 2008-2013. Diagn Microbiol Infect Dis. 2016;85:182-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 13. | Maybrook RJ, Campsen J, Wachs ME, Levi ME. A case of Mycobacterium mucogenicum infection in a liver transplant recipient and a review of the literature. Transpl Infect Dis. 2013;15:E260-E263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 14. | Oriani AS, Marfil MJ, Zumárraga MJ, Baldini MD. Prevalence and species diversity of nontuberculous mycobacteria in drinking water supply system of Bahía Blanca City, Argentina. Int J Mycobacteriol. 2019;8:138-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 15. | Set R, Shastri J. Laboratory aspects of clinically significant rapidly growing mycobacteria. Indian J Med Microbiol. 2011;29:343-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 16. | Hall L, Doerr KA, Wohlfiel SL, Roberts GD. Evaluation of the MicroSeq system for identification of mycobacteria by 16S ribosomal DNA sequencing and its integration into a routine clinical mycobacteriology laboratory. J Clin Microbiol. 2003;41:1447-1453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 170] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 17. | Adékambi T, Berger P, Raoult D, Drancourt M. rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int J Syst Evol Microbiol. 2006;56:133-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 254] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 18. | Kodana M, Tarumoto N, Kawamura T, Saito T, Ohno H, Maesaki S, Ikebuchi K. Utility of the MALDI-TOF MS method to identify nontuberculous mycobacteria. J Infect Chemother. 2016;22:32-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 19. | Buckwalter SP, Olson SL, Connelly BJ, Lucas BC, Rodning AA, Walchak RC, Deml SM, Wohlfiel SL, Wengenack NL. Evaluation of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for Identification of Mycobacterium species, Nocardia species, and Other Aerobic Actinomycetes. J Clin Microbiol. 2016;54:376-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 93] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 20. | Azimzadeh O, Barjaktarovic Z, Aubele M, Calzada-Wack J, Sarioglu H, Atkinson MJ, Tapio S. Formalin-fixed paraffin-embedded (FFPE) proteome analysis using gel-free and gel-based proteomics. J Proteome Res. 2010;9:4710-4720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 74] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 21. | Zhang P, Lehmann BD, Shyr Y, Guo Y. The Utilization of Formalin Fixed-Paraffin-Embedded Specimens in High Throughput Genomic Studies. Int J Genomics. 2017;2017:1926304. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 22. | Kokkat TJ, Patel MS, McGarvey D, LiVolsi VA, Baloch ZW. Archived formalin-fixed paraffin-embedded (FFPE) blocks: A valuable underexploited resource for extraction of DNA, RNA, and protein. Biopreserv Biobank. 2013;11:101-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 156] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 23. | Robbe P, Popitsch N, Knight SJL, Antoniou P, Becq J, He M, Kanapin A, Samsonova A, Vavoulis DV, Ross MT, Kingsbury Z, Cabes M, Ramos SDC, Page S, Dreau H, Ridout K, Jones LJ, Tuff-Lacey A, Henderson S, Mason J, Buffa FM, Verrill C, Maldonado-Perez D, Roxanis I, Collantes E, Browning L, Dhar S, Damato S, Davies S, Caulfield M, Bentley DR, Taylor JC, Turnbull C, Schuh A; 100; 000 Genomes Project. Clinical whole-genome sequencing from routine formalin-fixed, paraffin-embedded specimens: pilot study for the 100,000 Genomes Project. Genet Med. 2018;20:1196-1205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 109] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 24. | Menon R, Deng M, Boehm D, Braun M, Fend F, Biskup S, Perner S. Exome enrichment and SOLiD sequencing of formalin fixed paraffin embedded (FFPE) prostate cancer tissue. Int J Mol Sci. 2012;13:8933-8942. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 25. | Emery DC, Shoemark DK, Batstone TE, Waterfall CM, Coghill JA, Cerajewska TL, Davies M, West NX, Allen SJ. 16S rRNA Next Generation Sequencing Analysis Shows Bacteria in Alzheimer's Post-Mortem Brain. Front Aging Neurosci. 2017;9:195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 210] [Cited by in RCA: 230] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 26. | Xiao YL, Kash JC, Beres SB, Sheng ZM, Musser JM, Taubenberger JK. High-throughput RNA sequencing of a formalin-fixed, paraffin-embedded autopsy lung tissue sample from the 1918 influenza pandemic. J Pathol. 2013;229:535-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 27. | Li Z, Breitwieser FP, Lu J, Jun AS, Asnaghi L, Salzberg SL, Eberhart CG. Identifying Corneal Infections in Formalin-Fixed Specimens Using Next Generation Sequencing. Invest Ophthalmol Vis Sci. 2018;59:280-288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |