Published online Jul 6, 2021. doi: 10.12998/wjcc.v9.i19.5007
Peer-review started: March 16, 2021
First decision: May 1, 2021
Revised: May 3, 2021
Accepted: May 24, 2021
Article in press: May 24, 2021
Published online: July 6, 2021
Processing time: 99 Days and 15.7 Hours
The host immunity is crucial in determining the clinical course and prognosis of coronavirus disease 2019, where some systemic and severe manifestations are associated with excessive or suboptimal responses. Several antigenic epitopes in spike, nucleocapsid and membrane proteins of severe acute respiratory syndrome coronavirus 2 are targeted by the immune system, and a robust response with innate and adaptive components develops in infected individuals. High titer neutralizing antibodies and a balanced T cell response appears to constitute the optimal immune response to severe acute respiratory syndrome coronavirus 2, where innate and mucosal defenses also contribute significantly. Following exposure, immunological memory seems to develop and be maintained for substantial periods. Here, we provide an overview of the main aspects in antiviral immunity involving innate and adaptive responses with insights into virus structure, individual variations pertaining to disease severity as well as long-term protective immunity expected to be attained by vaccination.
Core Tip: Robust cellular and humoral responses are elicited in immunocompetent individuals with severe acute respiratory syndrome coronavirus 2 infection that remain detectable for several months following exposure. A balanced T cell response and neutralizing antibodies in circulation and mucosal surfaces are pivotal in controlling virus infection and for protection. Particular impairments in innate and adaptive immune responses are associated with pathogenesis and severe disease.
- Citation: Polat C, Ergunay K. Insights into the virologic and immunologic features of SARS-COV-2. World J Clin Cases 2021; 9(19): 5007-5018
- URL: https://www.wjgnet.com/2307-8960/full/v9/i19/5007.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i19.5007
An outbreak of pneumonia was reported in Wuhan City, Hubei Province, China in December 2019. In a short time, the World Health Organization declared the epidemic as a public health emergency of international concern[1]. The agent was subsequently named as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the International Committee on Taxonomy of Viruses, and the World Health Organization named the disease caused by SARS-CoV-2 as coronavirus disease-2019 (COVID-19)[2].
SARS-CoV-2 is the third zoonotic human coronavirus that emerged in this century following SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) [3]. Overall, SARS-CoV-2 is less pathogenic than SARS-CoV and MERS-CoV but more transmissible[4].
SARS-CoV-2 is an enveloped, positive-sense, single-stranded RNA virus, classified in the Coronaviridae family, Betacoronavirus genus[5,6]. It is phylogenetically-related to SARS-CoV and bat SARS-like coronavirus strain BatCov RaTG13, with 79.6% and 96.2% identities, respectively[7]. However, the origin of SARS-CoV-2 is yet to be confirmed. Although betacoronaviruses from Malayan pangolins share sequence similarities in the receptor binding domain (RBD) of the spike (S) gene, they are more distantly-related[8,9]. Therefore, pangolins are suggested as the intermediate host for SARS-CoV-2[8].
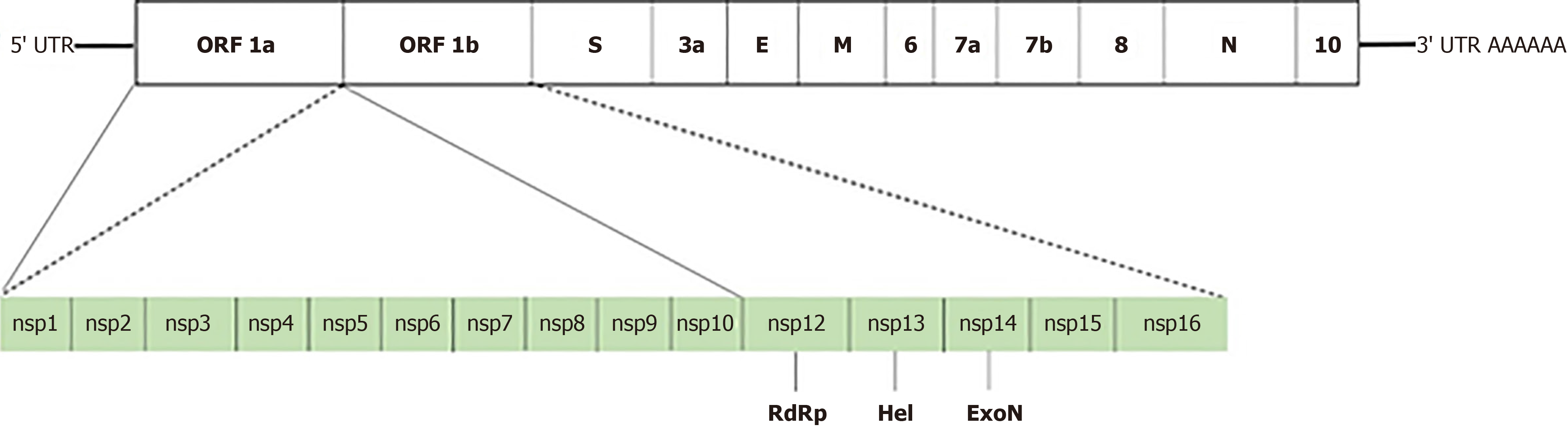
The genome size of SARS-CoV-2 is approximately 29.9 kb with 14 open reading frames encoding 27 proteins[5,10]. These include four structural (nucleocapsid (N), envelope (E), membrane (M) and S), as well as 16 nonstructural (nsp1-16) and seven accessory (ORF3a-ORF8) proteins (Figure 1).
The proteins encoded by the SARS-CoV-2 genome have various functions in virus replication and packaging. The S protein interacts with the host cell receptors and is essential for virus entry into host cells[11]. It is also a major antigen targeted by the immune response. The subunit S1 involves the RBD and binds to the host cell, while S2 fuses the viral and cellular membranes for penetration[12,13]. The RBD of the S protein recognizes the cellular receptor, angiotensin-converting enzyme 2 on the host cell, and SARS-CoV-2 enters into the target cell. The E and M proteins, major components of the virus structure, participate in virion assembly and release[14]. M protein binds to the N and accessory proteins 3a and 7a for the budding of viral particles[15]. Virus capsid formed by the N protein encapsulates the viral genome and contributes to replication and the cell signal pathway[16]. Main features and known functions of the viral nonstructural proteins are provided in Table 1.
| Protein | Function |
| Nsp 1 | RNA processing |
| Fixing of the replication complex to cellular membranes[17] | |
| Nsp 2 | p65 homolog |
| Host cell survival[18] | |
| Nsp 3 | Proteolytic cleavage[19] |
| Nsp 4 | Stabilization of the viral replication-transcription complex[20] |
| Nsp 5 | Polyprotein processing[21] |
| Nsp 6 | Autophagosome expansion inhibition[22] |
| Nsp 7/8 | Primase[23,24] |
| RNA-dependent RNA polymerase (RdRp) cofactor[25] | |
| Nsp 9 | ssRNA-binding protein[26] |
| Nsp 10 | Cap methylation of viral mRNAs[27] |
| Nsp 12 | Catalytic subunit of the RdRp[25,28] |
| Nsp 13 | Helicase (Hel) and NTPase activity[29,30] |
| Interferon antagonist[31] | |
| Nsp 14 | Exoribonuclease (ExoN) and methyltransferase activity[32] |
| Interferon antagonist[31] | |
| Nsp 15 | Nidoviral RNA uridylate-specific endoribonuclease (NendoU)[33] |
| Interferon antagonist[31] | |
| Nsp 16 | 2'-O-ribose methyltransferase |
| mRNA capping[34,35] |
Similar to many other respiratory viruses, a robust immune response with innate and adaptive components develops in individuals infected with SARS-CoV-2[36]. Nearly a year following the declaration of the pandemic, it is now established that SARS-CoV-2 infections produce prolonged immunity with cellular and humoral responses, detectable for several months after exposure. However, the duration and patterns of this response in exposed and vaccinated persons need further elucidation as hallmarks for assessment of protective immunity in individuals and populations. Moreover, the association of variations in individual immune responses and their impact in pathogenesis also need in-depth investigation to fully understand and control severe manifestations of COVID-19[37].
In general, the initial response upon viral infection involves the components of the innate immunity such as induction of type I interferon, the inflammation process, complement, neutrophils and natural killer cells, which are subsequently taken over by adaptive responses involving T and B lymphocytes. Various viral proteins processed by the antigen-presenting cells such as dendritic cells are recognized by T and B cells in lymphoid tissues, resulting in activation of humoral and cellular components of the adaptive response that produce highly-specific defense mechanisms to control ongoing or future infections[36,38]. Here, main findings in antiviral immunity involving particular components of the innate and adaptive immune response are revisited.
As the first line of defense against virus infection, appropriately elicited innate immunity is crucial to control SARS-CoV-2 infections as well as for an optimal adaptive response. Reduced type I interferon responses due to various factors such as mutations in the genome or autoantibodies are associated with severe disease[39-41]. Interestingly, an excessive response has also been observed to exacerbate clinical symptoms, where an overproduction of proinflammatory cytokines, which may be coupled with impaired type I interferon response, is present. Here, the signature cytokines have been identified as interleukin-6 (IL-6), IL-10 and C-reactive protein[42,43]. IL-6 has attracted particular interest due to its involvement as a potential contributing factor in SARS-CoV-2-associated acute respiratory distress syndrome[44]. It regulates dendritic cell differentiation, plasma cell maturation and is associated with ischemic injury. In SARS-CoV-2 infections, excessive macrophage activation and IL-6 production may result in the cytokine storm with subsequent endothelial cell damage, capillary leak and development of acute respiratory distress syndrome. Therefore, the inhibition of IL-6 or receptor binding has been investigated as potential therapeutic options to reduce morbidity and mortality with inconclusive findings so far[38,45-47].
In conjunction with IL-6 and other proinflammatory cytokines, the complement system also contributes to the pathogenesis in severe COVID-19 disease, including thrombotic events. Findings in infected individuals as well as acute lung injury models in mice indicate the involvement of the C5a-C5a receptor axis[48,49]. SARS-CoV-2 N protein is suggested to activate the mannose binding-lectin pathway, which in turn may be a triggering event in acute respiratory distress syndrome development[50]. Hence, complement inhibitors, especially those that can suppress coagulation pathway activation, such as C1 esterase inhibitor, are currently being investigated as novel approaches for treatment of SARS-CoV-2 pneumonia[38,51]. Complement system activation via alternate or classic pathways is also involved in COVID-19 disease, especially following antibody response and may be a key contributor in the immune complex-related injury[38,49].
In addition to the aforementioned contributors, changes in the expression of interferon receptor gene IFNAR2, tyrosine kinase 2, monocyte/macrophage chemotactic receptor CCR2 and particular antiviral restriction enzyme activators (OAS1, OAS2, OAS3) were documented in infected individuals, some associated with disease severity[42,43,52].
Despite aimed to prevent Mycobacterium tuberculosis infections, the Bacillus Calmette-Guérin vaccine has been proposed to affect COVID-19 infections in regions where it has been used in population vaccination[53-54]. Although reduced disease severity and mortality were observed in some reports, a beneficial effect is not universally documented[55-59]. The proposed mechanism of action is the enhanced reactivity of monocytes/macrophages and natural killer cells by epigenetic reprogramming (also termed as “trained immunity”), in the presence of proinflammatory cytokines[60]. Currently, the impact of heterologous protection provided by the Bacillus Calmette-Guérin vaccine in COVID-19 lacks concrete evidence, which is likely to be provided by the ongoing clinical trial[59].
The humoral and cellular components of the adaptive immune response, mainly involving various T and B cell subsets, provides specific defenses for controlling ongoing virus infection as well as neutralizing immunity upon re-exposure. Therefore, information on the mechanisms and dynamics of the adaptive response in COVID-19 is crucial to understand pathogenesis and to develop successful preventive measures[36].
Cellular responses: A robust and enduring CD4+ and CD8+ T cell response is elicited in the majority of SARS-CoV-2-infected individuals, targeting multiple virus epitopes including S, N and M proteins[61,62]. It is detectable from the second week of symptom onset and at the convalescent stage[62,63]. Although the duration and degree of protection is not clear, virus-specific T cells remain detectable 6-8 mo following infection[64,65]. Variations in breadth and magnitude of the T cell response and associations with disease severity is reported in some cohorts[61,63,66,67]. In SARS-CoV-2 infections, virus-specific CD4+ T cells produce tumor necrosis factor, IL-2 and interferon, where CD4+ T cell response is correlated with anti-spike/anti-nucleocapsid IgG/IgA and neutralizing antibody titers[63,64,68-70]. Similarly, CD8+ T cells are commonly present in convalescent plasma of COVID-19 patients in which activated cytotoxic and polyfunctional/stem-like cells prevalent at acute and convalescent stages, respectively[62,63,66,69,71]. Single cell transcriptome investigations provided insights into T cell subsets in different groups of infected individuals, indicating that a coordinated and focused immune response is crucial for successful elimination of the virus[72]. It is also noteworthy that the virus-specific T cell repertoire is maintained following infection for extended periods despite declines in humoral immunity and can swiftly be activated upon recurrent exposure and antigen presentation[63,73]. A polyfunctional and persistent SARS-CoV-2 specific memory develops in recovered patients, which can contribute to a rapid anamnestic response upon re-exposure[74,75]. Interestingly, a prolonged viral RNA positivity in pharyngeal mucosa with reduced risk for transmission was associated with increased SARS-CoV-2-specific CD8+ T cells, suggesting that low-level viral persistence supports immune stimulation and maturation[76].
During acute SARS-CoV-2 infections in adults, a peripheral T cell depletion resulting in lymphopenia might occur[61]. A transient condition that coincides with clinical recovery, it is associated with extensive T cell activation, altered differentiation and diminished function possibly involving proinflammatory cytokines, which in turn may prolong viral clearance and increase morbidity[73,77].
An intriguing observation is the detection of pre-existing CD4+ and CD8+ T cells in persons with no documented virus exposure[73,78,79]. Involving several T-cell epitopes, it suggests that previous exposures to endemic coronaviruses causing seasonal upper respiratory tract infections induce some sort of cross-reactive immunity to SARS-CoV-2[79,80]. The extent and potential impact of the cross-protection is not currently well-defined, but it is probably among the contributing factors to the varying clinical presentations in COVID-19 disease[81].
Humoral responses: In SARS-CoV-2, a polyclonal humoral response with antibodies, mainly targeting virus S and N proteins is mounted[82,83]. Despite the presence of IgM, IgG and IgA antibodies in acute and convalescent COVID-19 patients, virus serology is not practical in diagnosis, as antibodies increase only slightly during early symptomatic disease and are detectable in a portion of the patients[68,82,84]. Subsequently, a gradual increase in virus-specific IgG and IgM levels are observed with IgA dynamics similar to IgM, having attained peak levels before IgG. Most of the patients will have detectable seroconversion within 20 d after the onset of symptoms with a median time of 12 d[82,85,86]. Then, IgM levels begin to decrease in approximately 3 wk after symptom onset, while IgG continues to elevate, peaking at 50-60 d post infection and may last up to 10 mo[87,88]. Despite decreasing antibody levels, memory B cells remain and activate to produce antibodies upon virus re-exposure[89]. In COVID-19 patients, antibody levels are negatively correlated with viral RNA, which indicates their importance in eliminating the viruses in circulation[82,90]. In the respiratory tract, mucosal immunity provided by IgA antibodies and local T cells can effectively abolish SARS-CoV-2 infection and prevent systemic dissemination and further transmission[37].
Neutralizing antibodies are capable of suppressing virus entry into host cells. Therefore, they are pivotal in protection from reinfections. They are produced in human infections as well as animal infection models and observed to persist for 6 mo following infection[87,89,91]. Among antibodies against the S protein, RBD as well as S1 and S2 domains are targeted by neutralizing antibodies[83,89,91,92]. Anti-RBD antibodies have been reported to persist longer, possibly related to the preferential detection of higher-affinity antibodies[91]. Furthermore, despite variations in antibody titers, the number of RBD-specific memory B cells is shown to remain stable over time, producing antibodies with increased potency and resistance to mutations in the virus genome, suggesting ongoing evolution of the humoral response. This is likely to be fueled by persistent B cell exposure to antigens trapped as immune complexes on follicular dendritic cells[89].
Similar to cellular immunity, some form of cross-reactivity between SARS-CoV-2 and other human coronaviruses seem to occur. This is particularly observed in N protein and S1-S2 subunits of the spike protein but not in the RBD region for SARS-CoV and follows genomic similarities in these regions[86,92-94]. The impact of cross-reactive antibodies, especially those induced by endemic respiratory coronaviruses in SARS-CoV-2 immune response remains to be elucidated.
The impact of the virus-specific humoral immune response has led to the practice of administration of convalescent plasma as a therapeutic option, especially in severe disease to benefit from immediate effects of preformed polyclonal antibodies[37,82]. Such therapies have previously been used in particular viral pathogens such as Ebola virus, SARS-CoV and MERS-CoV[82,95]. Currently, sufficient data is lacking to establish the effectiveness as well as indications/limitations of convalescent plasma therapy in COVID-19 disease[82]. Many factors including the timing of administration during clinical course, patient selection criteria and antibody titers in the donor plasma seem to influence the outcome. Due to limited availability of convalescent plasma from donors, monoclonal neutralizing antibodies, antibody cocktails and antibody designs to increase efficiency are currently in clinical trials[95-98].
An optimal immune response to SARS-CoV-2 appears to include high-titer neutralizing antibodies and a balanced T cell response. Factors affecting innate and mucosal immunity also contribute substantially to infection control[37,38]. Currently used vaccines have been approved for population immunization with Emergency Use Authorization in several countries, based on the findings of the interim Phase III clinical trials. Current vaccine prototypes utilize various antigen delivery strategies based on DNA, mRNA or adenovirus-based platforms, recombinant viral subunits/protein and inactivated virus as well as other approaches. The reports on preclinical efficacy, phase I-III clinical trials and the immune responses induced by these vaccines have been extensively reviewed and can be found in detail elsewhere[37]. Protective immunity induced by different vaccine platforms may be based on different immune mechanisms and may require booster administrations to maintain long-term immunity. Therefore, a thorough understanding of immune responses induced by each platform and subsequent outcomes in preventing transmission and controlling infection must be monitored and optimal boosting strategies should be determined.
Robust cellular and humoral responses are elicited in immunocompetent individuals with SARS-CoV-2 infection that remain detectable for several months following exposure. Impaired type I interferon response, imbalances in complement components and production of excessive proinflammatory cytokines such as IL-6 are associated with pathogenesis and severe disease. Several antigenic epitopes in virus S, N and M proteins are recognized by the immune system. A balanced T cell response and neutralizing antibodies in circulation and mucosal surfaces are pivotal in controlling virus infection and for protection. Despite reductions in antibody levels in months following infection, the maintained memory cells can be activated to produce antibodies upon virus re-exposure. Cross-reactive immunity due to previous exposure to other coronaviruses is documented, with currently unknown implications for SARS-CoV-2 infections.
Manuscript source: Invited manuscript
Specialty type: Microbiology
Country/Territory of origin: Turkey
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Barbosa OA S-Editor: Ma YJ L-Editor: Filipodia P-Editor: Xing YX
| 1. | World Health Organization. Statement on the Second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV). 2020. Available from: https://www.scirp.org/reference/referencespapers.aspx?referenceid=2804611. |
| 2. | World Health Organization. WHO Director-General’s remarks at the media briefing on 2019-nCoV on 11 February 2020. 2020. Available from: https://www.capitalethiopia.com/interview/who-director-generals-opening-remarks-at-the-media-briefing-on-covid-19/. |
| 3. | Gralinski LE, Menachery VD. Return of the Coronavirus: 2019-nCoV. Viruses. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 699] [Cited by in RCA: 742] [Article Influence: 148.4] [Reference Citation Analysis (0)] |
| 4. | Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020;382:1199-1207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11224] [Cited by in RCA: 9319] [Article Influence: 1863.8] [Reference Citation Analysis (0)] |
| 5. | Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565-574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8473] [Cited by in RCA: 7602] [Article Influence: 1520.4] [Reference Citation Analysis (0)] |
| 6. | Chan JF, Kok KH, Zhu Z, Chu H, To KK, Yuan S, Yuen KY. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221-236. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1813] [Cited by in RCA: 1953] [Article Influence: 390.6] [Reference Citation Analysis (0)] |
| 7. | Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270-273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15248] [Cited by in RCA: 14131] [Article Influence: 2826.2] [Reference Citation Analysis (1)] |
| 8. | Xiao K, Zhai J, Feng Y, Zhou N, Zhang X, Zou JJ, Li N, Guo Y, Li X, Shen X, Zhang Z, Shu F, Huang W, Li Y, Chen RA, Wu YJ, Peng SM, Huang M, Xie WJ, Cai QH, Hou FH, Chen W, Xiao L, Shen Y. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 414] [Cited by in RCA: 494] [Article Influence: 98.8] [Reference Citation Analysis (0)] |
| 9. | Lam TT, Jia N, Zhang YW, Shum MH, Jiang JF, Zhu HC, Tong YG, Shi YX, Ni XB, Liao YS, Li WJ, Jiang BG, Wei W, Yuan TT, Zheng K, Cui XM, Li J, Pei GQ, Qiang X, Cheung WY, Li LF, Sun FF, Qin S, Huang JC, Leung GM, Holmes EC, Hu YL, Guan Y, Cao WC. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1019] [Cited by in RCA: 1200] [Article Influence: 240.0] [Reference Citation Analysis (0)] |
| 10. | Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, Meng J, Zhu Z, Zhang Z, Wang J, Sheng J, Quan L, Xia Z, Tan W, Cheng G, Jiang T. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe. 2020;27:325-328. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1413] [Cited by in RCA: 1502] [Article Influence: 300.4] [Reference Citation Analysis (0)] |
| 11. | Rastogi M, Pandey N, Shukla A, Singh SK. SARS coronavirus 2: from genome to infectome. Respir Res. 2020;21:318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 12. | Burkard C, Verheije MH, Wicht O, van Kasteren SI, van Kuppeveld FJ, Haagmans BL, Pelkmans L, Rottier PJ, Bosch BJ, de Haan CA. Coronavirus cell entry occurs through the endo-/Lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014;10:e1004502. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 289] [Cited by in RCA: 301] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 13. | Millet JK, Whittaker GR. Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120-134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 622] [Cited by in RCA: 656] [Article Influence: 59.6] [Reference Citation Analysis (0)] |
| 14. | De Maio F, Lo Cascio E, Babini G, Sali M, Della Longa S, Tilocca B, Roncada P, Arcovito A, Sanguinetti M, Scambia G, Urbani A. Improved binding of SARS-CoV-2 Envelope protein to tight junction-associated PALS1 could play a key role in COVID-19 pathogenesis. Microbes Infect. 2020;22:592-597. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 56] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 15. | Tang T, Bidon M, Jaimes JA, Whittaker GR, Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020;178:104792. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 610] [Cited by in RCA: 547] [Article Influence: 109.4] [Reference Citation Analysis (0)] |
| 16. | McBride R, van Zyl M, Fielding BC. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991-3018. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 575] [Cited by in RCA: 679] [Article Influence: 61.7] [Reference Citation Analysis (0)] |
| 17. | Schubert K, Karousis ED, Jomaa A, Scaiola A, Echeverria B, Gurzeler LA, Leibundgut M, Thiel V, Mühlemann O, Ban N. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol. 2020;27:959-966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 298] [Article Influence: 59.6] [Reference Citation Analysis (0)] |
| 18. | Angeletti S, Benvenuto D, Bianchi M, Giovanetti M, Pascarella S, Ciccozzi M. COVID-2019: The role of the nsp2 and nsp3 in its pathogenesis. J Med Virol. 2020;92:584-588. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 231] [Cited by in RCA: 228] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 19. | Lei J, Kusov Y, Hilgenfeld R. Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antiviral Res. 2018;149:58-74. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 392] [Cited by in RCA: 473] [Article Influence: 59.1] [Reference Citation Analysis (0)] |
| 20. | Manolaridis I, Wojdyla JA, Panjikar S, Snijder EJ, Gorbalenya AE, Berglind H, Nordlund P, Coutard B, Tucker PA. Structure of the C-terminal domain of nsp4 from feline coronavirus. Acta Crystallogr D Biol Crystallogr. 2009;65:839-846. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 21. | Roe MK, Junod NA, Young AR, Beachboard DC, Stobart CC. Targeting novel structural and functional features of coronavirus protease nsp5 (3CLpro, Mpro) in the age of COVID-19. J Gen Virol. 2021;102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 62] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 22. | Xia H, Cao Z, Xie X, Zhang X, Chen JY, Wang H, Menachery VD, Rajsbaum R, Shi PY. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020;33:108234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 777] [Cited by in RCA: 718] [Article Influence: 143.6] [Reference Citation Analysis (0)] |
| 23. | Konkolova E, Klima M, Nencka R, Boura E. Structural analysis of the putative SARS-CoV-2 primase complex. J Struct Biol. 2020;211:107548. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 24. | Krichel B, Falke S, Hilgenfeld R, Redecke L, Uetrecht C. Processing of the SARS-CoV pp1a/ab nsp7-10 region. Biochem J. 2020;477:1009-1019. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 88] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 25. | Kirchdoerfer RN, Ward AB. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun. 2019;10:2342. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 643] [Cited by in RCA: 614] [Article Influence: 102.3] [Reference Citation Analysis (0)] |
| 26. | Egloff MP, Ferron F, Campanacci V, Longhi S, Rancurel C, Dutartre H, Snijder EJ, Gorbalenya AE, Cambillau C, Canard B. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc Natl Acad Sci USA. 2004;101:3792-3796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 213] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 27. | Joseph JS, Saikatendu KS, Subramanian V, Neuman BW, Brooun A, Griffith M, Moy K, Yadav MK, Velasquez J, Buchmeier MJ, Stevens RC, Kuhn P. Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs. J Virol. 2006;80:7894-7901. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 98] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 28. | Peng Q, Peng R, Yuan B, Zhao J, Wang M, Wang X, Wang Q, Sun Y, Fan Z, Qi J, Gao GF, Shi Y. Structural and Biochemical Characterization of the nsp12-nsp7-nsp8 Core Polymerase Complex from SARS-CoV-2. Cell Rep. 2020;31:107774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 194] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 29. | Jang KJ, Jeong S, Kang DY, Sp N, Yang YM, Kim DE. A high ATP concentration enhances the cooperative translocation of the SARS coronavirus helicase nsP13 in the unwinding of duplex RNA. Sci Rep. 2020;10:4481. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 77] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 30. | Shu T, Huang M, Wu D, Ren Y, Zhang X, Han Y, Mu J, Wang R, Qiu Y, Zhang DY, Zhou X. SARS-Coronavirus-2 Nsp13 Possesses NTPase and RNA Helicase Activities That Can Be Inhibited by Bismuth Salts. Virol Sin. 2020;35:321-329. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 136] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 31. | Yuen CK, Lam JY, Wong WM, Mak LF, Wang X, Chu H, Cai JP, Jin DY, To KK, Chan JF, Yuen KY, Kok KH. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbes Infect. 2020;9:1418-1428. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 299] [Cited by in RCA: 399] [Article Influence: 79.8] [Reference Citation Analysis (0)] |
| 32. | Ogando NS, Zevenhoven-Dobbe JC, van der Meer Y, Bredenbeek PJ, Posthuma CC, Snijder EJ. The Enzymatic Activity of the nsp14 Exoribonuclease Is Critical for Replication of MERS-CoV and SARS-CoV-2. J Virol. 2020;94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 176] [Cited by in RCA: 184] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 33. | Kim Y, Jedrzejczak R, Maltseva NI, Wilamowski M, Endres M, Godzik A, Michalska K, Joachimiak A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020;29:1596-1605. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 214] [Cited by in RCA: 265] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 34. | Wang Y, Sun Y, Wu A, Xu S, Pan R, Zeng C, Jin X, Ge X, Shi Z, Ahola T, Chen Y, Guo D. Coronavirus nsp10/nsp16 Methyltransferase Can Be Targeted by nsp10-Derived Peptide In Vitro and In Vivo To Reduce Replication and Pathogenesis. J Virol. 2015;89:8416-8427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 119] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 35. | von Grotthuss M, Wyrwicz LS, Rychlewski L. mRNA cap-1 methyltransferase in the SARS genome. Cell. 2003;113:701-702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 95] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 36. | Hope JL, Bradley LM. Lessons in antiviral immunity. Science. 2021;371:464-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 37. | Sui Y, Bekele Y, Berzofsky JA. Potential SARS-CoV-2 Immune Correlates of Protection in Infection and Vaccine Immunization. Pathogens. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 51] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 38. | Jordan SC. Innate and adaptive immune responses to SARS-CoV-2 in humans: relevance to acquired immunity and vaccine responses. Clin Exp Immunol. 2021;204:310-320. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 57] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 39. | Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, Ogishi M, Sabli IKD, Hodeib S, Korol C, Rosain J, Bilguvar K, Ye J, Bolze A, Bigio B, Yang R, Arias AA, Zhou Q, Zhang Y, Onodi F, Korniotis S, Karpf L, Philippot Q, Chbihi M, Bonnet-Madin L, Dorgham K, Smith N, Schneider WM, Razooky BS, Hoffmann HH, Michailidis E, Moens L, Han JE, Lorenzo L, Bizien L, Meade P, Neehus AL, Ugurbil AC, Corneau A, Kerner G, Zhang P, Rapaport F, Seeleuthner Y, Manry J, Masson C, Schmitt Y, Schlüter A, Le Voyer T, Khan T, Li J, Fellay J, Roussel L, Shahrooei M, Alosaimi MF, Mansouri D, Al-Saud H, Al-Mulla F, Almourfi F, Al-Muhsen SZ, Alsohime F, Al Turki S, Hasanato R, van de Beek D, Biondi A, Bettini LR, D'Angio' M, Bonfanti P, Imberti L, Sottini A, Paghera S, Quiros-Roldan E, Rossi C, Oler AJ, Tompkins MF, Alba C, Vandernoot I, Goffard JC, Smits G, Migeotte I, Haerynck F, Soler-Palacin P, Martin-Nalda A, Colobran R, Morange PE, Keles S, Çölkesen F, Ozcelik T, Yasar KK, Senoglu S, Karabela ŞN, Rodríguez-Gallego C, Novelli G, Hraiech S, Tandjaoui-Lambiotte Y, Duval X, Laouénan C; COVID-STORM Clinicians; COVID Clinicians; Imagine COVID Group; French COVID Cohort Study Group; CoV-Contact Cohort; Amsterdam UMC Covid-19 Biobank; COVID Human Genetic Effort; NIAID-USUHS/TAGC COVID Immunity Group; Snow AL, Dalgard CL, Milner JD, Vinh DC, Mogensen TH, Marr N, Spaan AN, Boisson B, Boisson-Dupuis S, Bustamante J, Puel A, Ciancanelli MJ, Meyts I, Maniatis T, Soumelis V, Amara A, Nussenzweig M, García-Sastre A, Krammer F, Pujol A, Duffy D, Lifton RP, Zhang SY, Gorochov G, Béziat V, Jouanguy E, Sancho-Shimizu V, Rice CM, Abel L, Notarangelo LD, Cobat A, Su HC, Casanova JL. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1723] [Cited by in RCA: 1634] [Article Influence: 326.8] [Reference Citation Analysis (0)] |
| 40. | Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann HH, Zhang Y, Dorgham K, Philippot Q, Rosain J, Béziat V, Manry J, Shaw E, Haljasmägi L, Peterson P, Lorenzo L, Bizien L, Trouillet-Assant S, Dobbs K, de Jesus AA, Belot A, Kallaste A, Catherinot E, Tandjaoui-Lambiotte Y, Le Pen J, Kerner G, Bigio B, Seeleuthner Y, Yang R, Bolze A, Spaan AN, Delmonte OM, Abers MS, Aiuti A, Casari G, Lampasona V, Piemonti L, Ciceri F, Bilguvar K, Lifton RP, Vasse M, Smadja DM, Migaud M, Hadjadj J, Terrier B, Duffy D, Quintana-Murci L, van de Beek D, Roussel L, Vinh DC, Tangye SG, Haerynck F, Dalmau D, Martinez-Picado J, Brodin P, Nussenzweig MC, Boisson-Dupuis S, Rodríguez-Gallego C, Vogt G, Mogensen TH, Oler AJ, Gu J, Burbelo PD, Cohen JI, Biondi A, Bettini LR, D'Angio M, Bonfanti P, Rossignol P, Mayaux J, Rieux-Laucat F, Husebye ES, Fusco F, Ursini MV, Imberti L, Sottini A, Paghera S, Quiros-Roldan E, Rossi C, Castagnoli R, Montagna D, Licari A, Marseglia GL, Duval X, Ghosn J; HGID Lab; NIAID-USUHS Immune Response to COVID Group; COVID Clinicians; COVID-STORM Clinicians; Imagine COVID Group; French COVID Cohort Study Group; Milieu Intérieur Consortium; CoV-Contact Cohort; Amsterdam UMC Covid-19 Biobank; COVID Human Genetic Effort; Tsang JS, Goldbach-Mansky R, Kisand K, Lionakis MS, Puel A, Zhang SY, Holland SM, Gorochov G, Jouanguy E, Rice CM, Cobat A, Notarangelo LD, Abel L, Su HC, Casanova JL. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1932] [Cited by in RCA: 1919] [Article Influence: 383.8] [Reference Citation Analysis (0)] |
| 41. | Wadman M. Flawed interferon response spurs severe illness. Science. 2020;369:1550-1551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 42. | Laing AG, Lorenc A, Del Molino Del Barrio I, Das A, Fish M, Monin L, Muñoz-Ruiz M, McKenzie DR, Hayday TS, Francos-Quijorna I, Kamdar S, Joseph M, Davies D, Davis R, Jennings A, Zlatareva I, Vantourout P, Wu Y, Sofra V, Cano F, Greco M, Theodoridis E, Freedman JD, Gee S, Chan JNE, Ryan S, Bugallo-Blanco E, Peterson P, Kisand K, Haljasmägi L, Chadli L, Moingeon P, Martinez L, Merrick B, Bisnauthsing K, Brooks K, Ibrahim MAA, Mason J, Lopez Gomez F, Babalola K, Abdul-Jawad S, Cason J, Mant C, Seow J, Graham C, Doores KJ, Di Rosa F, Edgeworth J, Shankar-Hari M, Hayday AC. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat Med. 2020;26:1623-1635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 696] [Cited by in RCA: 682] [Article Influence: 136.4] [Reference Citation Analysis (0)] |
| 43. | Lavillegrand JR, Garnier M, Spaeth A, Mario N, Hariri G, Pilon A, Berti E, Fieux F, Thietart S, Urbina T, Turpin M, Darrivière L, Fartoukh M, Verdonk F, Dumas G, Tedgui A, Guidet B, Maury E, Chantran Y, Voiriot G, Ait-Oufella H. Elevated plasma IL-6 and CRP levels are associated with adverse clinical outcomes and death in critically ill SARS-CoV-2 patients: inflammatory response of SARS-CoV-2 patients. Ann Intensive Care. 2021;11:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 59] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 44. | Jordan SC, Zakowski P, Tran HP, Smith EA, Gaultier C, Marks G, Zabner R, Lowenstein H, Oft J, Bluen B, Le C, Shane R, Ammerman N, Vo A, Chen P, Kumar S, Toyoda M, Ge S, Huang E. Compassionate Use of Tocilizumab for Treatment of SARS-CoV-2 Pneumonia. Clin Infect Dis. 2020;71:3168-3173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 62] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 45. | Somers EC, Eschenauer GA, Troost JP, Golob JL, Gandhi TN, Wang L, Zhou N, Petty LA, Baang JH, Dillman NO, Frame D, Gregg KS, Kaul DR, Nagel J, Patel TS, Zhou S, Lauring AS, Hanauer DA, Martin E, Sharma P, Fung CM, Pogue JM. Tocilizumab for treatment of mechanically ventilated patients with COVID-19. Clin Infect Dis. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 202] [Cited by in RCA: 310] [Article Influence: 77.5] [Reference Citation Analysis (0)] |
| 46. | Roschewski M, Lionakis MS, Sharman JP, Roswarski J, Goy A, Monticelli MA, Roshon M, Wrzesinski SH, Desai JV, Zarakas MA, Collen J, Rose K, Hamdy A, Izumi R, Wright GW, Chung KK, Baselga J, Staudt LM, Wilson WH. Inhibition of Bruton tyrosine kinase in patients with severe COVID-19. Sci Immunol. 2020;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 287] [Cited by in RCA: 278] [Article Influence: 55.6] [Reference Citation Analysis (0)] |
| 47. | Guaraldi G, Meschiari M, Cozzi-Lepri A, Milic J, Tonelli R, Menozzi M, Franceschini E, Cuomo G, Orlando G, Borghi V, Santoro A, Di Gaetano M, Puzzolante C, Carli F, Bedini A, Corradi L, Fantini R, Castaniere I, Tabbì L, Girardis M, Tedeschi S, Giannella M, Bartoletti M, Pascale R, Dolci G, Brugioni L, Pietrangelo A, Cossarizza A, Pea F, Clini E, Salvarani C, Massari M, Viale PL, Mussini C. Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lancet Rheumatol. 2020;2:e474-e484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 604] [Cited by in RCA: 688] [Article Influence: 137.6] [Reference Citation Analysis (0)] |
| 48. | Carvelli J, Demaria O, Vély F, Batista L, Chouaki Benmansour N, Fares J, Carpentier S, Thibult ML, Morel A, Remark R, André P, Represa A, Piperoglou C; Explore COVID-19 IPH group; Explore COVID-19 Marseille Immunopole group; Cordier PY, Le Dault E, Guervilly C, Simeone P, Gainnier M, Morel Y, Ebbo M, Schleinitz N, Vivier E. Association of COVID-19 inflammation with activation of the C5a-C5aR1 axis. Nature. 2020;588:146-150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 352] [Cited by in RCA: 387] [Article Influence: 77.4] [Reference Citation Analysis (0)] |
| 49. | Cao X. COVID-19: immunopathology and its implications for therapy. Nat Rev Immunol. 2020;20:269-270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 972] [Cited by in RCA: 1118] [Article Influence: 223.6] [Reference Citation Analysis (0)] |
| 50. | Gao T, Hu M, Zhang X, Li H, Zhu L, Liu H, Dong Q, Zhang Z, Wang Z, Hu Y, Fu Y, Jin Y, Li K, Zhao S, Xiao Y, Luo S, Li L, Zhao L, Liu J, Zhao H, Liu Y, Yang W, Peng J, Chen X, Li P, Xie Y, Song J, Zhang L, Ma Q, Bian X, Chen W, Liu X, Mao Q, Cao C. Highly pathogenic coronavirus N protein aggravates lung injury by MASP-2-mediated complement over-activation. medRxiv2020. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 174] [Reference Citation Analysis (0)] |
| 51. | Urwyler P, Moser S, Charitos P, Heijnen IAFM, Rudin M, Sommer G, Giannetti BM, Bassetti S, Sendi P, Trendelenburg M, Osthoff M. Treatment of COVID-19 With Conestat Alfa, a Regulator of the Complement, Contact Activation and Kallikrein-Kinin System. Front Immunol. 2020;11:2072. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 52. | Pairo-Castineira E, Clohisey S, Klaric L, Bretherick AD, Rawlik K, Pasko D, Walker S, Parkinson N, Fourman MH, Russell CD, Furniss J, Richmond A, Gountouna E, Wrobel N, Harrison D, Wang B, Wu Y, Meynert A, Griffiths F, Oosthuyzen W, Kousathanas A, Moutsianas L, Yang Z, Zhai R, Zheng C, Grimes G, Beale R, Millar J, Shih B, Keating S, Zechner M, Haley C, Porteous DJ, Hayward C, Yang J, Knight J, Summers C, Shankar-Hari M, Klenerman P, Turtle L, Ho A, Moore SC, Hinds C, Horby P, Nichol A, Maslove D, Ling L, McAuley D, Montgomery H, Walsh T, Pereira AC, Renieri A; GenOMICC Investigators; ISARIC4C Investigators; COVID-19 Human Genetics Initiative; 23andMe Investigators; BRACOVID Investigators; Gen-COVID Investigators; Shen X, Ponting CP, Fawkes A, Tenesa A, Caulfield M, Scott R, Rowan K, Murphy L, Openshaw PJM, Semple MG, Law A, Vitart V, Wilson JF, Baillie JK. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591:92-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1045] [Cited by in RCA: 934] [Article Influence: 233.5] [Reference Citation Analysis (0)] |
| 53. | O'Neill LAJ, Netea MG. BCG-induced trained immunity: can it offer protection against COVID-19? Nat Rev Immunol. 2020;20:335-337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 291] [Cited by in RCA: 347] [Article Influence: 69.4] [Reference Citation Analysis (0)] |
| 54. | Covián C, Retamal-Díaz A, Bueno SM, Kalergis AM. Could BCG Vaccination Induce Protective Trained Immunity for SARS-CoV-2? Front Immunol. 2020;11:970. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 74] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 55. | Lindestam Arlehamn CS, Sette A, Peters B. Lack of evidence for BCG vaccine protection from severe COVID-19. Proc Natl Acad Sci USA. 2020;117:25203-25204. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 56. | Hensel J, McAndrews KM, McGrail DJ, Dowlatshahi DP, LeBleu VS, Kalluri R. Protection against SARS-CoV-2 by BCG vaccination is not supported by epidemiological analyses. Sci Rep. 2020;10:18377. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 57. | Escobar LE, Molina-Cruz A, Barillas-Mury C. BCG vaccine protection from severe coronavirus disease 2019 (COVID-19). Proc Natl Acad Sci USA. 2020;117:17720-17726. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 331] [Cited by in RCA: 300] [Article Influence: 60.0] [Reference Citation Analysis (0)] |
| 58. | Marín-Hernández D, Nixon DF, Hupert N. Anticipated reduction in COVID-19 mortality due to population-wide BCG vaccination: evidence from Germany. Hum Vaccin Immunother. 2021;1-3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 59. | Madsen AMR, Schaltz-Buchholzer F, Benfield T, Bjerregaard-Andersen M, Dalgaard LS, Dam C, Ditlev SB, Faizi G, Johansen IS, Kofoed PE, Kristensen GS, Loekkegaard ECL, Mogensen CB, Mohamed L, Ostenfeld A, Oedegaard ES, Soerensen MK, Wejse C, Jensen AKG, Nielsen S, Krause TG, Netea MG, Aaby P, Benn CS. Using BCG vaccine to enhance non-specific protection of health care workers during the COVID-19 pandemic: A structured summary of a study protocol for a randomised controlled trial in Denmark. Trials. 2020;21:799. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 60. | Netea MG, Joosten LA, Latz E, Mills KH, Natoli G, Stunnenberg HG, O'Neill LA, Xavier RJ. Trained immunity: A program of innate immune memory in health and disease. Science. 2016;352:aaf1098. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1360] [Cited by in RCA: 1771] [Article Influence: 196.8] [Reference Citation Analysis (0)] |
| 61. | Shrotri M, van Schalkwyk MCI, Post N, Eddy D, Huntley C, Leeman D, Rigby S, Williams SV, Bermingham WH, Kellam P, Maher J, Shields AM, Amirthalingam G, Peacock SJ, Ismail SA. T cell response to SARS-CoV-2 infection in humans: A systematic review. PLoS One. 2021;16:e0245532. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 93] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 62. | Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, Marrama D, de Silva AM, Frazier A, Carlin AF, Greenbaum JA, Peters B, Krammer F, Smith DM, Crotty S, Sette A. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020; 181: 1489-1501. e15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2147] [Cited by in RCA: 2781] [Article Influence: 556.2] [Reference Citation Analysis (0)] |
| 63. | Peng Y, Mentzer AJ, Liu G, Yao X, Yin Z, Dong D, Dejnirattisai W, Rostron T, Supasa P, Liu C, López-Camacho C, Slon-Campos J, Zhao Y, Stuart DI, Paesen GC, Grimes JM, Antson AA, Bayfield OW, Hawkins DEDP, Ker DS, Wang B, Turtle L, Subramaniam K, Thomson P, Zhang P, Dold C, Ratcliff J, Simmonds P, de Silva T, Sopp P, Wellington D, Rajapaksa U, Chen YL, Salio M, Napolitani G, Paes W, Borrow P, Kessler BM, Fry JW, Schwabe NF, Semple MG, Baillie JK, Moore SC, Openshaw PJM, Ansari MA, Dunachie S, Barnes E, Frater J, Kerr G, Goulder P, Lockett T, Levin R, Zhang Y, Jing R, Ho LP; Oxford Immunology Network Covid-19 Response T cell Consortium; ISARIC4C Investigators; Cornall RJ, Conlon CP, Klenerman P, Screaton GR, Mongkolsapaya J, McMichael A, Knight JC, Ogg G, Dong T. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol. 2020;21:1336-1345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1024] [Cited by in RCA: 938] [Article Influence: 187.6] [Reference Citation Analysis (0)] |
| 64. | Zuo J, Dowell AC, Pearce H, Verma K, Long HM, Begum J, Aiano F, Amin-Chowdhury Z, Hallis B, Stapley L, Borrow R, Linley E, Ahmad S, Parker B, Horsley A, Amirthalingam G, Brown K, Ramsay ME, Ladhani S, Moss P. Robust SARS-CoV-2-specific T cell immunity is maintained at 6 mo following primary infection. Nat Immunol. 2021;22:620-626. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 262] [Cited by in RCA: 272] [Article Influence: 68.0] [Reference Citation Analysis (0)] |
| 65. | Sherina N, Piralla A, Du L, Wan H, Kumagai-Braesch M, Andréll J, Braesch-Andersen S, Cassaniti I, Percivalle E, Sarasini A, Bergami F, Di Martino R, Colaneri M, Vecchia M, Sambo M, Zuccaro V, Bruno R, Sachs M, Oggionni T, Meloni F, Abolhassani H, Bertoglio F, Schubert M, Byrne-Steele M, Han J, Hust M, Xue Y, Hammarström L, Baldanti F, Marcotte H, Pan-Hammarström Q. Persistence of SARS-CoV-2-specific B and T cell responses in convalescent COVID-19 patients 6-8 months after the infection. Med (N Y) 2021; 2: 281-295. e4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 126] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 66. | Sekine T, Perez-Potti A, Rivera-Ballesteros O, Strålin K, Gorin JB, Olsson A, Llewellyn-Lacey S, Kamal H, Bogdanovic G, Muschiol S, Wullimann DJ, Kammann T, Emgård J, Parrot T, Folkesson E; Karolinska COVID-19 Study Group; Rooyackers O, Eriksson LI, Henter JI, Sönnerborg A, Allander T, Albert J, Nielsen M, Klingström J, Gredmark-Russ S, Björkström NK, Sandberg JK, Price DA, Ljunggren HG, Aleman S, Buggert M. Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell 2020; 183: 158-168. e14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1425] [Cited by in RCA: 1388] [Article Influence: 277.6] [Reference Citation Analysis (0)] |
| 67. | Schulien I, Kemming J, Oberhardt V, Wild K, Seidel LM, Killmer S, Sagar, Daul F, Salvat Lago M, Decker A, Luxenburger H, Binder B, Bettinger D, Sogukpinar O, Rieg S, Panning M, Huzly D, Schwemmle M, Kochs G, Waller CF, Nieters A, Duerschmied D, Emmerich F, Mei HE, Schulz AR, Llewellyn-Lacey S, Price DA, Boettler T, Bengsch B, Thimme R, Hofmann M, Neumann-Haefelin C. Characterization of pre-existing and induced SARS-CoV-2-specific CD8+ T cells. Nat Med. 2021;27:78-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 277] [Article Influence: 55.4] [Reference Citation Analysis (0)] |
| 68. | Ni L, Ye F, Cheng ML, Feng Y, Deng YQ, Zhao H, Wei P, Ge J, Gou M, Li X, Sun L, Cao T, Wang P, Zhou C, Zhang R, Liang P, Guo H, Wang X, Qin CF, Chen F, Dong C. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity 2020; 52: 971-977. e3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 774] [Cited by in RCA: 814] [Article Influence: 162.8] [Reference Citation Analysis (0)] |
| 69. | Neidleman J, Luo X, Frouard J, Xie G, Gill G, Stein ES, McGregor M, Ma T, George AF, Kosters A, Greene WC, Vasquez J, Ghosn E, Lee S, Roan NR. SARS-CoV-2-Specific T Cells Exhibit Phenotypic Features of Helper Function, Lack of Terminal Differentiation, and High Proliferation Potential. Cell Rep Med. 2020;1:100081. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 128] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 70. | Wang Y, Zhang L, Sang L, Ye F, Ruan S, Zhong B, Song T, Alshukairi AN, Chen R, Zhang Z, Gan M, Zhu A, Huang Y, Luo L, Mok CKP, Al Gethamy MM, Tan H, Li Z, Huang X, Li F, Sun J, Zhang Y, Wen L, Li Y, Chen Z, Zhuang Z, Zhuo J, Chen C, Kuang L, Wang J, Lv H, Jiang Y, Li M, Lin Y, Deng Y, Tang L, Liang J, Huang J, Perlman S, Zhong N, Zhao J, Malik Peiris JS. Kinetics of viral load and antibody response in relation to COVID-19 severity. J Clin Invest. 2020;130:5235-5244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 364] [Cited by in RCA: 419] [Article Influence: 83.8] [Reference Citation Analysis (0)] |
| 71. | Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, Chng MHY, Lin M, Tan N, Linster M, Chia WN, Chen MI, Wang LF, Ooi EE, Kalimuddin S, Tambyah PA, Low JG, Tan YJ, Bertoletti A. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1275] [Cited by in RCA: 1455] [Article Influence: 291.0] [Reference Citation Analysis (0)] |
| 72. | Zhang JY, Wang XM, Xing X, Xu Z, Zhang C, Song JW, Fan X, Xia P, Fu JL, Wang SY, Xu RN, Dai XP, Shi L, Huang L, Jiang TJ, Shi M, Zhang Y, Zumla A, Maeurer M, Bai F, Wang FS. Single-cell landscape of immunological responses in patients with COVID-19. Nat Immunol. 2020;21:1107-1118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 475] [Article Influence: 95.0] [Reference Citation Analysis (0)] |
| 73. | Bonifacius A, Tischer-Zimmermann S, Dragon AC, Gussarow D, Vogel A, Krettek U, Gödecke N, Yilmaz M, Kraft ARM, Hoeper MM, Pink I, Schmidt JJ, Li Y, Welte T, Maecker-Kolhoff B, Martens J, Berger MM, Lobenwein C, Stankov MV, Cornberg M, David S, Behrens GMN, Witzke O, Blasczyk R, Eiz-Vesper B. COVID-19 immune signatures reveal stable antiviral T cell function despite declining humoral responses. Immunity 2021; 54: 340-354. e6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 164] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 74. | Breton G, Mendoza P, Hägglöf T, Oliveira TY, Schaefer-Babajew D, Gaebler C, Turroja M, Hurley A, Caskey M, Nussenzweig MC. Persistent cellular immunity to SARS-CoV-2 infection. J Exp Med. 2021;218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 98] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 75. | Quast I, Tarlinton D. B cell memory: understanding COVID-19. Immunity. 2021;54:205-210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 100] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 76. | Vibholm LK, Nielsen SSF, Pahus MH, Frattari GS, Olesen R, Andersen R, Monrad I, Andersen AHF, Thomsen MM, Konrad CV, Andersen SD, Højen JF, Gunst JD, Østergaard L, Søgaard OS, Schleimann MH, Tolstrup M. SARS-CoV-2 persistence is associated with antigen-specific CD8 T-cell responses. EBioMedicine. 2021;64:103230. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 107] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 77. | Chen Z, John Wherry E. T cell responses in patients with COVID-19. Nat Rev Immunol. 2020;20:529-536. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 692] [Cited by in RCA: 598] [Article Influence: 119.6] [Reference Citation Analysis (0)] |
| 78. | Braun J, Loyal L, Frentsch M, Wendisch D, Georg P, Kurth F, Hippenstiel S, Dingeldey M, Kruse B, Fauchere F, Baysal E, Mangold M, Henze L, Lauster R, Mall MA, Beyer K, Röhmel J, Voigt S, Schmitz J, Miltenyi S, Demuth I, Müller MA, Hocke A, Witzenrath M, Suttorp N, Kern F, Reimer U, Wenschuh H, Drosten C, Corman VM, Giesecke-Thiel C, Sander LE, Thiel A. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 803] [Cited by in RCA: 933] [Article Influence: 186.6] [Reference Citation Analysis (0)] |
| 79. | Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JM, Burger ZC, Rawlings SA, Smith DM, Phillips E, Mallal S, Lammers M, Rubiro P, Quiambao L, Sutherland A, Yu ED, da Silva Antunes R, Greenbaum J, Frazier A, Markmann AJ, Premkumar L, de Silva A, Peters B, Crotty S, Sette A, Weiskopf D. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89-94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1024] [Cited by in RCA: 879] [Article Influence: 175.8] [Reference Citation Analysis (0)] |
| 80. | Nelde A, Bilich T, Heitmann JS, Maringer Y, Salih HR, Roerden M, Lübke M, Bauer J, Rieth J, Wacker M, Peter A, Hörber S, Traenkle B, Kaiser PD, Rothbauer U, Becker M, Junker D, Krause G, Strengert M, Schneiderhan-Marra N, Templin MF, Joos TO, Kowalewski DJ, Stos-Zweifel V, Fehr M, Rabsteyn A, Mirakaj V, Karbach J, Jäger E, Graf M, Gruber LC, Rachfalski D, Preuß B, Hagelstein I, Märklin M, Bakchoul T, Gouttefangeas C, Kohlbacher O, Klein R, Stevanoviæ S, Rammensee HG, Walz JS. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat Immunol. 2021;22:74-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 285] [Cited by in RCA: 438] [Article Influence: 87.6] [Reference Citation Analysis (0)] |
| 81. | Lipsitch M, Grad YH, Sette A, Crotty S. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat Rev Immunol. 2020;20:709-713. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 210] [Cited by in RCA: 194] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 82. | Lagunas-Rangel FA, Chávez-Valencia V. What do we know about the antibody responses to SARS-CoV-2? Immunobiology. 2021;226:152054. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 83. | Ladner JT, Henson SN, Boyle AS, Engelbrektson AL, Fink ZW, Rahee F, D'ambrozio J, Schaecher KE, Stone M, Dong W, Dadwal S, Yu J, Caligiuri MA, Cieplak P, Bjørås M, Fenstad MH, Nordbø SA, Kainov DE, Muranaka N, Chee MS, Shiryaev SA, Altin JA. Epitope-resolved profiling of the SARS-CoV-2 antibody response identifies cross-reactivity with endemic human coronaviruses. Cell Rep Med. 2021;2:100189. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 130] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 84. | Padoan A, Sciacovelli L, Basso D, Negrini D, Zuin S, Cosma C, Faggian D, Matricardi P, Plebani M. IgA-Ab response to spike glycoprotein of SARS-CoV-2 in patients with COVID-19: A longitudinal study. Clin Chim Acta. 2020;507:164-166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 258] [Cited by in RCA: 235] [Article Influence: 47.0] [Reference Citation Analysis (0)] |
| 85. | Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, Wang X, Yuan J, Li T, Li J, Qian S, Hong C, Wang F, Liu Y, Wang Z, He Q, Li Z, He B, Zhang T, Fu Y, Ge S, Liu L, Zhang J, Xia N, Zhang Z. Antibody Responses to SARS-CoV-2 in Patients With Novel Coronavirus Disease 2019. Clin Infect Dis. 2020;71:2027-2034. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1590] [Cited by in RCA: 1840] [Article Influence: 368.0] [Reference Citation Analysis (0)] |
| 86. | Long QX, Liu BZ, Deng HJ, Wu GC, Deng K, Chen YK, Liao P, Qiu JF, Lin Y, Cai XF, Wang DQ, Hu Y, Ren JH, Tang N, Xu YY, Yu LH, Mo Z, Gong F, Zhang XL, Tian WG, Hu L, Zhang XX, Xiang JL, Du HX, Liu HW, Lang CH, Luo XH, Wu SB, Cui XP, Zhou Z, Zhu MM, Wang J, Xue CJ, Li XF, Wang L, Li ZJ, Wang K, Niu CC, Yang QJ, Tang XJ, Zhang Y, Liu XM, Li JJ, Zhang DC, Zhang F, Liu P, Yuan J, Li Q, Hu JL, Chen J, Huang AL. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26:845-848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2411] [Cited by in RCA: 2047] [Article Influence: 409.4] [Reference Citation Analysis (0)] |
| 87. | Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CE, Grifoni A, Ramirez SI, Haupt S, Frazier A, Nakao C, Rayaprolu V, Rawlings SA, Peters B, Krammer F, Simon V, Saphire EO, Smith DM, Weiskopf D, Sette A, Crotty S. Immunological memory to SARS-CoV-2 assessed for up to 8 mo after infection. Science. 2021;371. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2187] [Cited by in RCA: 1934] [Article Influence: 483.5] [Reference Citation Analysis (0)] |
| 88. | Barnes CO, Jette CA, Abernathy ME, Dam KA, Esswein SR, Gristick HB, Malyutin AG, Sharaf NG, Huey-Tubman KE, Lee YE, Robbiani DF, Nussenzweig MC, West AP Jr, Bjorkman PJ. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1404] [Cited by in RCA: 1230] [Article Influence: 246.0] [Reference Citation Analysis (0)] |
| 89. | Gaebler C, Wang Z, Lorenzi JCC, Muecksch F, Finkin S, Tokuyama M, Cho A, Jankovic M, Schaefer-Babajew D, Oliveira TY, Cipolla M, Viant C, Barnes CO, Bram Y, Breton G, Hägglöf T, Mendoza P, Hurley A, Turroja M, Gordon K, Millard KG, Ramos V, Schmidt F, Weisblum Y, Jha D, Tankelevich M, Martinez-Delgado G, Yee J, Patel R, Dizon J, Unson-O'Brien C, Shimeliovich I, Robbiani DF, Zhao Z, Gazumyan A, Schwartz RE, Hatziioannou T, Bjorkman PJ, Mehandru S, Bieniasz PD, Caskey M, Nussenzweig MC. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 972] [Cited by in RCA: 1249] [Article Influence: 312.3] [Reference Citation Analysis (0)] |
| 90. | Du Z, Zhu F, Guo F, Yang B, Wang T. Detection of antibodies against SARS-CoV-2 in patients with COVID-19. J Med Virol. 2020;92:1735-1738. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 84] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 91. | L'Huillier AG, Meyer B, Andrey DO, Arm-Vernez I, Baggio S, Didierlaurent A, Eberhardt CS, Eckerle I, Grasset-Salomon C, Huttner A, Posfay-Barbe KM, Royo IS, Pralong JA, Vuilleumier N, Yerly S, Siegrist CA, Kaiser L; Geneva Centre for Emerging Viral Diseases. Antibody persistence in the first 6 months following SARS-CoV-2 infection among hospital workers: a prospective longitudinal study. Clin Microbiol Infect. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 97] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 92. | Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, Yu J, Shan S, Zhou B, Song S, Tang X, Lan J, Yuan J, Wang H, Zhao J, Zhang S, Wang Y, Shi X, Liu L, Wang X, Zhang Z, Zhang L. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1001] [Cited by in RCA: 1285] [Article Influence: 257.0] [Reference Citation Analysis (0)] |
| 93. | Ng KW, Faulkner N, Cornish GH, Rosa A, Harvey R, Hussain S, Ulferts R, Earl C, Wrobel AG, Benton DJ, Roustan C, Bolland W, Thompson R, Agua-Doce A, Hobson P, Heaney J, Rickman H, Paraskevopoulou S, Houlihan CF, Thomson K, Sanchez E, Shin GY, Spyer MJ, Joshi D, O'Reilly N, Walker PA, Kjaer S, Riddell A, Moore C, Jebson BR, Wilkinson M, Marshall LR, Rosser EC, Radziszewska A, Peckham H, Ciurtin C, Wedderburn LR, Beale R, Swanton C, Gandhi S, Stockinger B, McCauley J, Gamblin SJ, McCoy LE, Cherepanov P, Nastouli E, Kassiotis G. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339-1343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 665] [Cited by in RCA: 634] [Article Influence: 126.8] [Reference Citation Analysis (0)] |
| 94. | Okba NMA, Müller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, Lamers MM, Sikkema RS, de Bruin E, Chandler FD, Yazdanpanah Y, Le Hingrat Q, Descamps D, Houhou-Fidouh N, Reusken CBEM, Bosch BJ, Drosten C, Koopmans MPG, Haagmans BL. Severe Acute Respiratory Syndrome Coronavirus 2-Specific Antibody Responses in Coronavirus Disease Patients. Emerg Infect Dis. 2020;26:1478-1488. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1123] [Cited by in RCA: 1120] [Article Influence: 224.0] [Reference Citation Analysis (0)] |
| 95. | Tiberghien P, de Lamballerie X, Morel P, Gallian P, Lacombe K, Yazdanpanah Y. Collecting and evaluating convalescent plasma for COVID-19 treatment: why and how? Vox Sang. 2020;115:488-494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 106] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 96. | Baum A, Ajithdoss D, Copin R, Zhou A, Lanza K, Negron N, Ni M, Wei Y, Mohammadi K, Musser B, Atwal GS, Oyejide A, Goez-Gazi Y, Dutton J, Clemmons E, Staples HM, Bartley C, Klaffke B, Alfson K, Gazi M, Gonzalez O, Dick E Jr, Carrion R Jr, Pessaint L, Porto M, Cook A, Brown R, Ali V, Greenhouse J, Taylor T, Andersen H, Lewis MG, Stahl N, Murphy AJ, Yancopoulos GD, Kyratsous CA. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110-1115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 431] [Cited by in RCA: 421] [Article Influence: 84.2] [Reference Citation Analysis (0)] |
| 97. | Yang L, Liu W, Yu X, Wu M, Reichert JM, Ho M. COVID-19 antibody therapeutics tracker: a global online database of antibody therapeutics for the prevention and treatment of COVID-19. Antib Ther. 2020;3:205-212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 98. | Hussen J, Kandeel M, Hemida MG, Al-Mubarak AIA. Antibody-Based Immunotherapeutic Strategies for COVID-19. Pathogens. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |