Published online Dec 26, 2020. doi: 10.12998/wjcc.v8.i24.6465
Peer-review started: August 28, 2020
First decision: September 24, 2020
Revised: October 8, 2020
Accepted: October 26, 2020
Article in press: October 26, 2020
Published online: December 26, 2020
Processing time: 113 Days and 13.5 Hours
Bainbridge-Ropers syndrome (BRPS) is a severe disorder characterized by failure to thrive, facial dysmorphism, and severe developmental delay. BRPS is caused by a heterozygous loss-of-function mutation in the ASXL3 gene. Due to limited knowledge of the disease and lack of specific features, clinical diagnosis of this syndrome is challenging. With the use of trio-based whole exome sequencing, we identified a novel ASXL3 mutation in a Chinese boy with BRPS and performed a literature review.
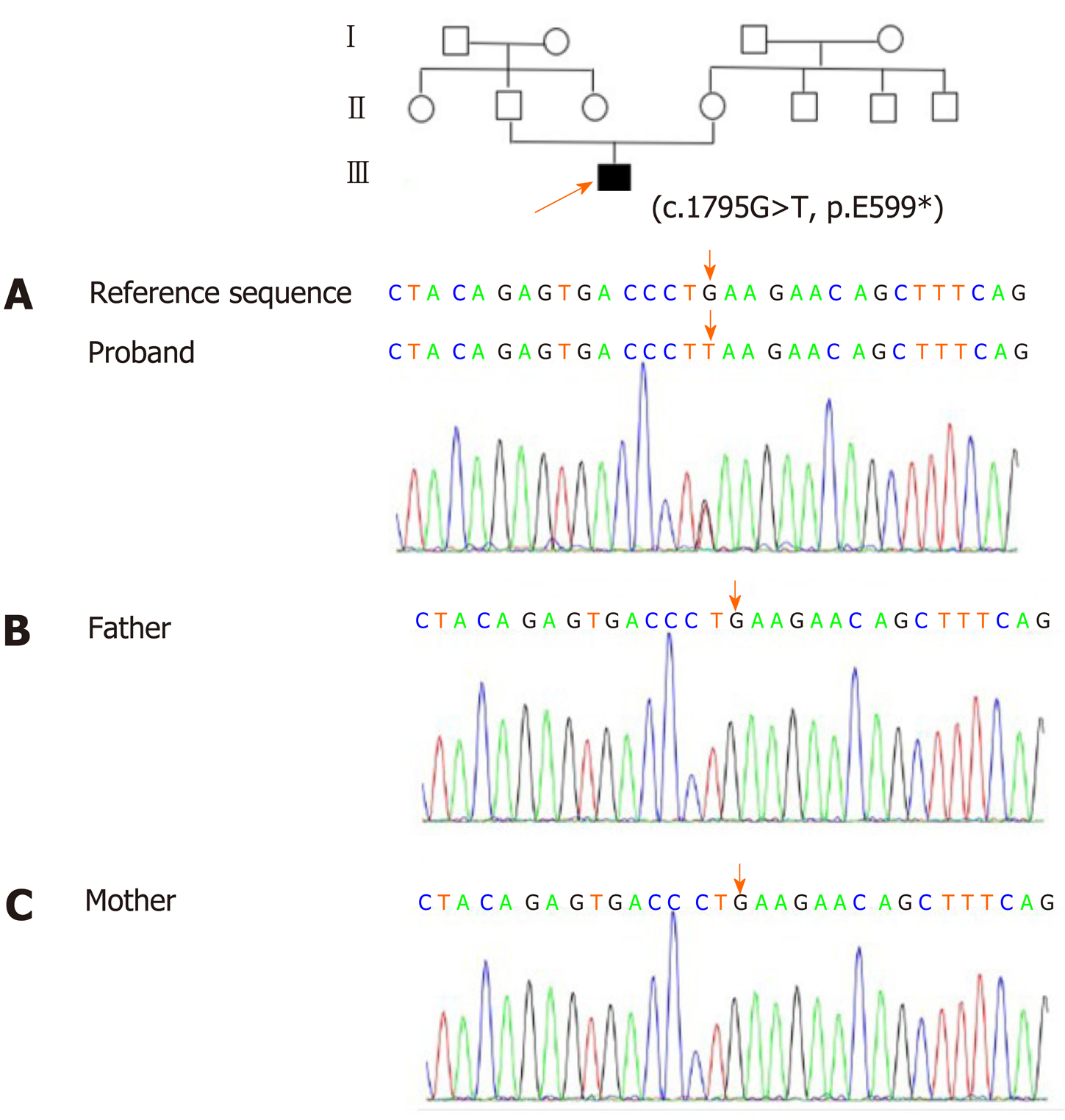
A 3-year-old Chinese boy was referred to our hospital due to progressive postnatal microcephaly and intellectual disability with severe speech impairment for 2 years. His other remarkable clinical features were shown as follows: Facial dysmorphism, feeding difficulties, poor growth, motor delay, and abnormal behavior. For the proband, regular laboratory tests, blood tandem mass spectrometry, urine gas chromatographic mass spectrometry, karyotype, hearing screening, and brain magnetic resonance imaging were performed, with negative results. Therefore, for the proband and his unaffected parents, trio-based whole exome sequencing and subsequent validation by Sanger sequencing were performed. A novel nonsense variant in exon 11 of the ASXL3 gene (c.1795G>T; p.E599*) was detected, present in the patient but absent from his parents. Taking into account the concordant phenotypic features of our patient with reported BRPS patients and the detected truncated variant located in the known mutational cluster region, we confirmed a diagnosis of BRPS for this proband. The rehabilitation treatment seemed to have a mild effect.
In this case, a novel nonsense mutation (c.1795G>T, p.E599*) in ASXL3 gene was identified in a Chinese boy with BRPS. This finding not only contributed to better genetic counseling and prenatal diagnosis for this family but also expanded the pathogenic mutation spectrum of ASXL3 gene and provided key information for clinical diagnosis of BRPS.
Core Tip: This report introduces a Chinese boy referred to our hospital mainly due to progressive postnatal microcephaly and intellectual disability with severe speech impairment. A novel pathogenic mutation (c.1795G>T; p.E599*) was detected in the patient by trio-based whole exome sequencing. The proband’s clinical features largely conformed to reported Bainbridge-Ropers syndrome patients in the literature. These clinical and genetic findings improved our understanding of Bainbridge-Ropers syndrome and also aided in the definitive diagnosis and genetic counseling for this family.
- Citation: Li JR, Huang Z, Lu Y, Ji QY, Jiang MY, Yang F. Novel mutation in the ASXL3 gene in a Chinese boy with microcephaly and speech impairment: A case report. World J Clin Cases 2020; 8(24): 6465-6472
- URL: https://www.wjgnet.com/2307-8960/full/v8/i24/6465.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v8.i24.6465
Bainbridge-Ropers syndrome (BRPS), first described by Bainbridge and colleagues in 2013, is a severe disorder with clinical phenotype that includes early feeding difficulties, profound speech impairment, intellectual disability, autistic features, hypotonia, and dysmorphic craniofacial features[1,2]. So far, about 25 cases have been reported, with a total of 49 patients including seven Chinese patients in the literature. BRPS is caused by a heterozygous loss-of-function mutation in the ASXL3 gene on chromosome 18q12.1, a gene belonging to the ASXL gene family involved in transcriptional regulation of many genes through direct actions or epigenetically via histone modifications[3]. BRPS is not an easily recognizable syndrome due to the limited knowledge of reported cases and the absence of specific clinical features, especially since its phenotype overlaps with that of Bohring-Opitz syndrome, which is caused by a heterogeneous pathogenic mutation in the ASXL1 gene, and Shashi-Pena syndrome, which is caused by a heterogeneous pathogenic mutation in the ASXL2 gene[4]. To our knowledge, all reported BRPS cases were diagnosed through next generation sequencing. Herein, we report a case of BRPS in a Chinese boy with a novel heterozygous nonsense variant in the ASXL3 gene identified by next generation sequencing. In addition, the genetic and phenotypic spectrum of reported BRPS cases are reviewed and summarized.
A 3-year-old Chinese boy was referred to our hospital due to progressive postnatal microcephaly and intellectual disability with severe speech impairment for about 2 years.
The patient’s chief clinical features were noticed by his parents about 2 years ago with gradually prominent facial dysmorphism, poor growth, motor delay, and some behavioral problems such as periodic agitation, self-injurious behavior, and autistic features, including sudden shrieking and disabilities of language and eye contact.
The proband was the first born of a non-consanguineous Chinese couple (mother and father both born in 1990), and there was no family history of neurodevelopmental disorders. The pregnancy and delivery at 38 wk gestation were uneventful. At birth, the weight was 3000 g [-1 SD to the median (World Health Organization (WHO)], the length was 48 cm [-1 SD to the median (WHO)], and the occipitofrontal circumference (OFC) was 32.5 cm (3rd- 15th percentile (WHO)] (Supplementary Figure 1).
Due to weak suction, he could not be breastfed, and he required tube feeding. He showed a muscular hypotonia and growth retardation (his growth charts are shown in Supplementary Figure 1). At 2 mo of age, the child presented with vomiting, bloody stools, and diarrhea, which was diagnosed as a cow's milk protein allergy, and he received a deep hydrolysis formula for 6 mo. After 1 year of age, the child could tolerate a normal diet and was not fed with a tube.
His developmental milestones were delayed. He could raise his head at 5 mo of age, rollover at 6 mo of age, sit unaided at 1 year of age, walk dependently at 30 mo of age, and walk and jump independently at 3 years of age. Notably, he could only speak a few meaningful words, such as yes, no, papa, and mama, which were not used for communication. However, his parents failed to pay attention to these abnormalities and did not pursue medical consultation of professionals in the early years.
The patient previously consulted a dentist and ophthalmologist for the crowded teeth and strabismus, respectively. The patient had not received the related treatment yet.
Nonspecific.
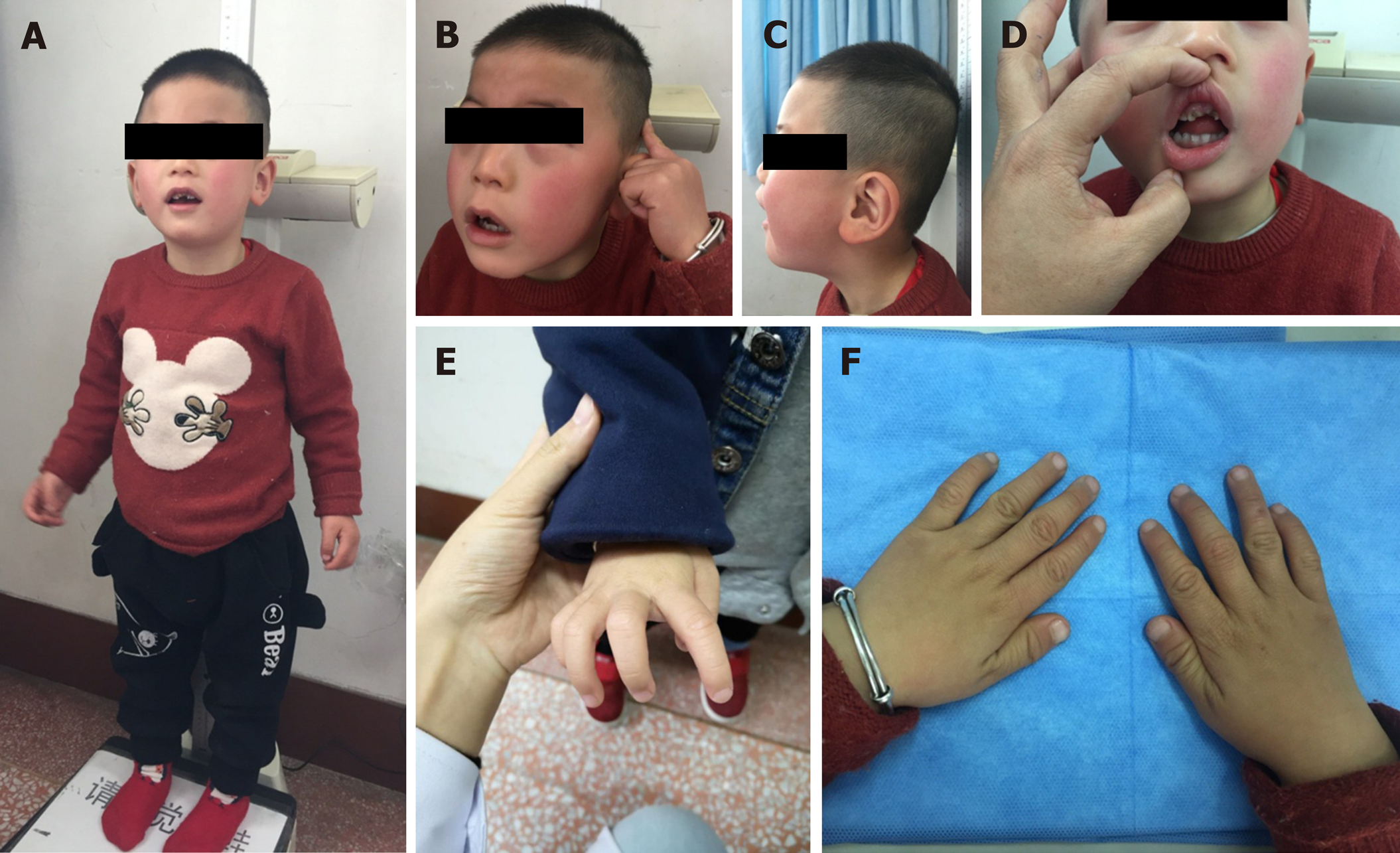
At 3-year-old, his weight was 11.5 kg [-2 SD to -1 SD (WHO)], length was 90 cm [-2 SD to -1 SD (WHO)], and the OFC was 46 cm [< 3rd percentile (WHO)], fitting the diagnosis of microcephaly (Supplementary Figure 1). He had a prominent forehead, arched eyebrows, edematous periorbital region, strabismus, hypertelorism, downslanting palpebral fissures, low columella, low-set ears, thin upper lip, lower lip valgus, crowed teeth, enamel hypoplasia, and slightly claw-shaped hands (Figure 1).
Routine test results for complete blood count, liver and kidney function, electrolytes, myocardial enzymes, and thyroid function were normal. Levels of blood lactate and ammonia, blood tandem mass spectrometry, and urine gas chromatographic mass spectrometry were unremarkable. The result of karyotype analysis was 46, XY.
There was no abnormality in his skeletal X-ray imaging, brain magnetic resonance imaging, or hearing screening. His childhood autism rating scale score for autism was 35, indicating moderate autism. The developmental screen test showed abnormality, as his mental index was 55 and developmental quotient (DQ) was 68. His respective DQ scores of language, adaptation, gross motor, fine motor, and personal social functioning in Gesell developmental scales were 25, 35, 40, 46, and 35, suggesting that speech impairment was the most prominent disability.
We performed trio-based whole-exome sequencing (WES) on the patient and his unaffected parents. Genomic DNA was extracted from ethylene diamine tetraacetic acid-treated peripheral blood. Library preparation was constructed with xGen Exome Research Panel v1.0 probe sequence capture array (Integrated Device Technology, Coralville, IA, United States). The captured DNA fragments were then sequenced by Illumina NovaSeq 6000 (Illumina, San Diego, CA, United States) at the Chigene Translational Medical Research Center (Beijing, China). The sequencing data were mapped to a reference genome (GRCh 37/hg 19) using Burrows-Wheeler Aligner software. Variant calling was performed using SAMtools and Pindel software. The sequence variants were functionally annotated and filtered using known populations and databases, including 1000 genomes, Single Nucleotide Polymorphism Database, Genome Aggregation Database, ClinVar, Human Gene Mutation Database, and Online Mendelian Inheritance in Man. We focused on nonsynonymous single nucleotide variants, insertions, deletions, and splice-site variants. Candidate variants were then evaluated in the context of clinical presentation and inheritance mode. Unreported nonsynonymous single nucleotide variants were predicted using in silico predictive algorithms such as MutationTaster (http://www.mutationtaster.org), Sorting Intolerant from Tolerant (http://sift.jcvi.org), Polymorphism Phenotyping v2 (http://genetics.bwh.harvard.edu/pph2/), and phyloP (http://compgen.bscb.cornell.edu/phast/) to evaluate any potentially damaging effects to the protein function. Putative pathogenic variants were subsequently validated by Sanger sequencing. The pathogenicity of variants was annotated according to the American College of Medical Genetics and Genomics (ACMG) standards and guidelines[5]. Mutation nomenclature is according to Human Genome Variation Society recommendations (http://varnomen.hgvs.org/).
The trio-based WES revealed a heterozygous nonsense variant c.1795G>T in exon 11 of the ASXL3 gene (NM_030632.2) in the proband, predicted to result in a stop codon at amino acid 599 (p.E599*) and generate a premature truncation. The variant was not reported previously. No variation was detected at this site in his parents, which was subsequently validated by Sanger sequencing (Figure 2). This novel de novo variant was classified as “pathogenic” according to the ACMG criteria (PVS1 + PS2 + PM2 + PP3). No other variant with clinical significance was identified by the trio-based WES.
Bainbridge-Ropers syndrome.
The patient received rehabilitation and nutritional intervention.
Nutritional intervention was performed for 1.5 years with poor effect except for weight, while rehabilitation training seemed to have a mild improvement. At the last follow-up, he was 4 years and 6 mo of age, weight was 15 kg [-2 SD to -1 SD (WHO)], length was 100 cm [-2 SD to -1 SD (WHO)], and the OFC was 46.5 cm [far below the 3rd percentile (WHO)] (growth charts are shown in Supplementary Figure 1). At the age of 4 years, his respective DQ scores of language, adaptation, gross motor, fine motor, and personal social functioning in Gesell developmental scales were 35, 38, 50, 50, and 40. At his last follow-up, the respective DQ scores of language, adaptation, gross motor, fine motor, and personal social functioning in Gesell developmental scales rose up to 39, 42, 55, 54, and 50.
BRPS is a rare autosomal dominant neurodevelopmental disorder caused by loss-of-function mutations in the ASXL3 gene. The relatively prevalent features, as observed in > 50% of 49 reported BRPS cases, were developmental delay, intellectual disability, poor or absent speech, generalized or trunk hypotonia, feeding difficulties, failure to thrive or poor growth, facial dysmorphism, autistic features, and microcephaly (Supplementary Table 1). The most common facial dysmorphism in the literature is high-arched palate, followed by arched eyebrows, anteverted nares, downslanting palpebral fissures, strabismus, and prominent forehead (Supplementary Table 1). Additionally, hand anomalies such as ulnar deviation of hands at rest (5/7, 71.4%) and hypertonic extremities (5/6, 83.3%) have been frequently reported in Chinese BRPS patients and were also seen in our patient[6-9]. The incidence of these hand anomalies might be underestimated. In our patient, poor feeding in the neonatal period was the most common initial symptom, while neurologic findings such as developmental delay/intellectual disability and microcephaly were the first reasons for pursuing medical care. Interestingly, our patient previously consulted with a dentist and ophthalmologist for his crowded teeth and strabismus, respectively. It has been implied that some patients with BRPS were initially misdiagnosed as dental or optical disorder and remain undiagnosed. Short stature and seizures were reported in around one-third of the patients[10,11], which was not observed in our patient. It was unknown whether our patient would be at risk of short stature and seizures; follow-up clinical surveillance would be important.
The phenotypic features of our patient largely conform to the description of reported BRPS patients. In the clinical diagnosis, BRPS was not easily distinguished from other conditions characterized by syndromic intellectual disability, especially Bohring-Opitz syndrome and Shashi-Pena syndrome. The overlap among the manifestations in the three different disorders, including developmental or intellectual impairments, distinct facial dysmorphisms, feeding difficulties and so on, could be due to the similarities in function of the ASXL gene family members[12]. Following a thorough review of all reported BRPS cases, we considered the remarkable features distinct from BRPS were facial nevus flammeus and macrocephaly[4,13]. However, because the main clinical features of BRPS are nonspecific and the total number of reported cases is limited, WES technology is increasingly used to identify the pathogenesis and establish a definite diagnosis[14-16]. We performed trio-based WES and identified a novel de novo nonsense mutation (c.1795G>T, p.E599*) of the ASXL3 gene in our patient. The variant was classified as “pathogenic” according to the ACMG criteria, supporting a genetic diagnosis of BRPS for the proband, with main complaints of progressive postnatal microcephaly and intellectual disability with severe speech impairment.
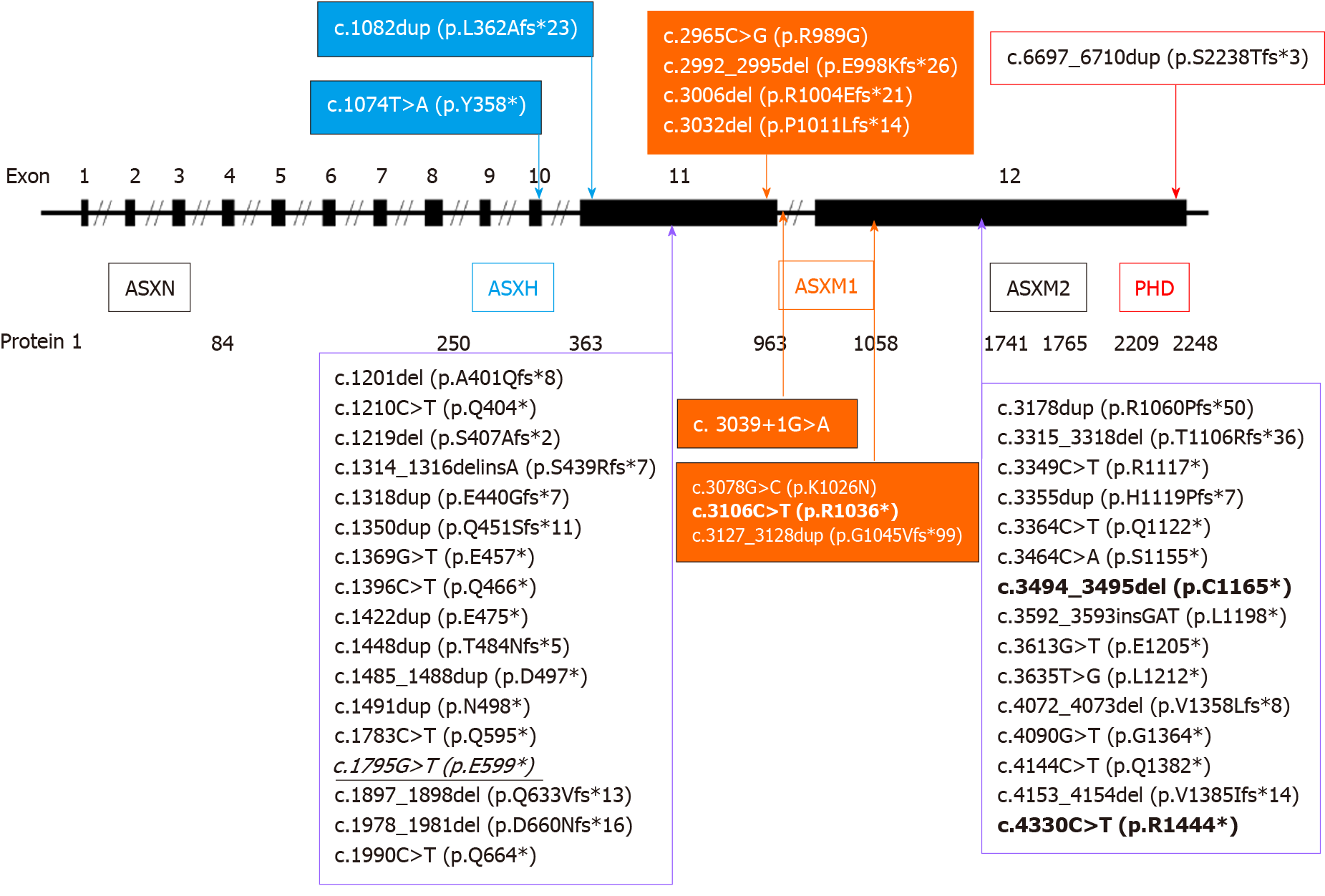
As previously reported, the ASXL3 gene contains 12 exons and encodes a 2248 amino-acid protein. Like other ASXL family members, the ASXL3 protein has a conserved domain structure: ASXN and ASXH domains in the N terminus; ASXM1 and ASXM2 domains in the middle region; and a plant homeodomain finger in the C terminus (Figure 3). The ASXN domain and plant homeodomain finger play a role in the regulation of gene transcription, representing putative DNA or histonerecognition sites; the region around the ASXH domain creates proteinprotein interaction sites for association with epigenetic regulators; the ASXM1 and ASXM2 domains are involved in proteinprotein interactions[12]. Between the ASXH and ASXM1 domains, there is the 5' mutational cluster region (MCR); while between the ASXM1 and ASXM2 domains, there is 3' MCR[2,3]. The novel null variant (c.1795G>T, p.E599*) detected in our patient is located in exon 11 and the known 5’ MCR. The truncated protein tended to give rise to an aberrant ASXL protein with intact ASXN and ASXH domains. To clarify whether haploinsufficiency or a dominant-negative effect of the novel variant may be a causative mechanism of BRPS[1] will require further functional studies.
Together with our case, 43 different loss-of-function ASXL3 variants in 48 unrelated individuals and two siblings[17] with BRPS are known. Of the detected pathogenic mutations, 16 (37.2%) were nonsense mutations, 12 (27.9%) were small insertions (including 11 duplications), 11 (25.6%) were small deletion, two (5%) were missense mutations (detected in compound heterogeneous state in a patient reported by Giri et al[10]), and the remaining two were consensus splice site mutation and small indel, respectively (Figure 3). The majority of the reported mutations were frameshift truncations. Consistent with the previous reports, the 43 different mutations are scattered over the two largest exons, which represent 84% of the entire ASXL3 protein-coding region, either located in the conserved domains or the MCRs[2,13] (Figure 3). The only one exception was a splice site mutation (located in intron 11) detected in two unrelated patients reported by Myers et al[11] and Hori et al[18], respectively. In addition to the recurrent splice site mutation, three other mutations were detected more than once in unrelated families, including c.3106C>T (p.R1036*)[7,8,11,13,17], c.3494_3495del (p.C1165*)[6,13], and c.4330C>T (p.R1444*)[2,3]. The mutation c.3106C>T (p.R1036*) was identified in five separate families in the literature, suggesting it is a likely mutational hotspot. More cases of BRPS from different ethnic populations are required for this to be validated.
We reported a novel nonsense mutation (c.1795G>T, p.E599*) in the ASXL3 gene in a Chinese boy with BRPS. This finding not only contributed to better genetic counseling and prenatal diagnosis for this family, but also expanded the spectrum of pathogenic mutations for BRPS. Combined with the literature review of BRPS to date, we believe that BRPS should be considered as a potential diagnosis in patients presenting with progressive postnatal microcephaly and intellectual disability with severe speech impairment as well as motor delay, hypotonia, early feeding difficulties, poor growth, autistic features, and dysmorphic features (e.g., prominent forehead, arched eyebrows, strabismus, downslanting palpebral fissures, anteverted nares, high-arched palate, and crowded teeth).
The authors thank the family members for providing blood samples, coordination with treatment, and agreeing to participate in this study.
Manuscript source: Unsolicited manuscript
Specialty type: Genetics and heredity
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cuevas-Covarrubias SA, Tanabe S S-Editor: Zhang L L-Editor: Filipodia P-Editor: Zhang YL
| 1. | Bainbridge MN, Hu H, Muzny DM, Musante L, Lupski JR, Graham BH, Chen W, Gripp KW, Jenny K, Wienker TF, Yang Y, Sutton VR, Gibbs RA, Ropers HH. De novo truncating mutations in ASXL3 are associated with a novel clinical phenotype with similarities to Bohring-Opitz syndrome. Genome Med. 2013;5:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 124] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 2. | Balasubramanian M, Willoughby J, Fry AE, Weber A, Firth HV, Deshpande C, Berg JN, Chandler K, Metcalfe KA, Lam W, Pilz DT, Tomkins S. Delineating the phenotypic spectrum of Bainbridge-Ropers syndrome: 12 new patients with de novo, heterozygous, loss-of-function mutations in ASXL3 and review of published literature. J Med Genet. 2017;54:537-543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 3. | Srivastava A, Ritesh KC, Tsan YC, Liao R, Su F, Cao X, Hannibal MC, Keegan CE, Chinnaiyan AM, Martin DM, Bielas SL. De novo dominant ASXL3 mutations alter H2A deubiquitination and transcription in Bainbridge-Ropers syndrome. Hum Mol Genet. 2016;25:597-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 4. | Shashi V, Pena LD, Kim K, Burton B, Hempel M, Schoch K, Walkiewicz M, McLaughlin HM, Cho M, Stong N, Hickey SE, Shuss CM; Undiagnosed Diseases Network; Freemark MS; Bellet JS; Keels MA; Bonner MJ; El-Dairi M; Butler M; Kranz PG; Stumpel CT; Klinkenberg S; Oberndorff K; Alawi M; Santer R; Petrovski S; Kuismin O; Korpi-Heikkilä S; Pietilainen O; Aarno P; Kurki MI; Hoischen A; Need AC; Goldstein DB; Kortüm F. De Novo Truncating Variants in ASXL2 Are Associated with a Unique and Recognizable Clinical Phenotype. Am J Hum Genet. 2016;99:991-999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 59] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 5. | Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405-424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19696] [Cited by in RCA: 22562] [Article Influence: 2256.2] [Reference Citation Analysis (0)] |
| 6. | Yang L, Guo B, Zhu W, Wang L, Han B, Che Y, Guo L. Bainbridge-ropers syndrome caused by loss-of-function variants in ASXL3: Clinical abnormalities, medical imaging features, and gene variation in infancy of case report. BMC Pediatr. 2020;20:287. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 7. | Zhang R, He XH, Lin HY, Yang XH. [Bainbridge-Ropers syndrome with ASXL3 gene variation in a child and literature review]. Zhonghua Er Ke Za Zhi. 2018;56:138-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 8. | Gou J, Zhou S, Cai H, Wang H. Bainbridge-Ropers syndrome: a case report and literature review. Linchuang Erke Zazhi. 2019;37:212-214. [DOI] [Full Text] |
| 9. | Qiao L, Liu Y, Ge J, Li T. Novel Nonsense Mutation in ASXL3 causing Bainbridge-Ropers Syndrome. Indian Pediatr. 2019;56:792-794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 10. | Giri D, Rigden D, Didi M, Peak M, McNamara P, Senniappan S. Novel compound heterozygous ASXL3 mutation causing Bainbridge-ropers like syndrome and primary IGF1 deficiency. Int J Pediatr Endocrinol. 2017;2017:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 11. | Myers KA, White SM, Mohammed S, Metcalfe KA, Fry AE, Wraige E, Vasudevan PC, Balasubramanian M, Scheffer IE. Childhood-onset generalized epilepsy in Bainbridge-Ropers syndrome. Epilepsy Res. 2018;140:166-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 12. | Katoh M. Functional proteomics of the epigenetic regulators ASXL1, ASXL2 and ASXL3: a convergence of proteomics and epigenetics for translational medicine. Expert Rev Proteomics. 2015;12:317-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 13. | Kuechler A, Czeschik JC, Graf E, Grasshoff U, Hüffmeier U, Busa T, Beck-Woedl S, Faivre L, Rivière JB, Bader I, Koch J, Reis A, Hehr U, Rittinger O, Sperl W, Haack TB, Wieland T, Engels H, Prokisch H, Strom TM, Lüdecke HJ, Wieczorek D. Bainbridge-Ropers syndrome caused by loss-of-function variants in ASXL3: a recognizable condition. Eur J Hum Genet. 2017;25:183-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 14. | Zhu X, Petrovski S, Xie P, Ruzzo EK, Lu YF, McSweeney KM, Ben-Zeev B, Nissenkorn A, Anikster Y, Oz-Levi D, Dhindsa RS, Hitomi Y, Schoch K, Spillmann RC, Heimer G, Marek-Yagel D, Tzadok M, Han Y, Worley G, Goldstein J, Jiang YH, Lancet D, Pras E, Shashi V, McHale D, Need AC, Goldstein DB. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet Med. 2015;17:774-781. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 219] [Cited by in RCA: 260] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 15. | Wayhelova M, Oppelt J, Smetana J, Hladilkova E, Filkova H, Makaturova E, Nikolova P, Beharka R, Gaillyova R, Kuglik P. Novel de novo frameshift variant in the ASXL3 gene in a child with microcephaly and global developmental delay. Mol Med Rep. 2019;20:505-512. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 16. | Ababneh F, Nashabat M, Alfadhel M. A new case of Bainbridge–Ropers syndrome (BRPS): delineating the phenotype and review of literature. JBC Genet. 2019: 65-69. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 17. | Koboldt DC, Mihalic Mosher T, Kelly BJ, Sites E, Bartholomew D, Hickey SE, McBride K, Wilson RK, White P. A de novo nonsense mutation in ASXL3 shared by siblings with Bainbridge-Ropers syndrome. Cold Spring Harb Mol Case Stud. 2018;4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 18. | Hori I, Miya F, Ohashi K, Negishi Y, Hattori A, Ando N, Okamoto N, Kato M, Tsunoda T, Yamasaki M, Kanemura Y, Kosaki K, Saitoh S. Novel splicing mutation in the ASXL3 gene causing Bainbridge-Ropers syndrome. Am J Med Genet A. 2016;170:1863-1867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |