Published online Apr 16, 2022. doi: 10.12998/wjcc.v10.i11.3334
Peer-review started: August 10, 2021
First decision: October 20, 2021
Revised: October 29, 2021
Accepted: January 17, 2022
Article in press: January 17, 2022
Published online: April 16, 2022
Processing time: 240 Days and 20.6 Hours
Melanomas are malignant tumors that can occur in different body parts or tissues such as the skin, mucous membrane, uvea, and pia mater. Long non-coding RNAs (lncRNAs) are key factors in the occurrence and development of many malignant tumors, and are involved in the prognosis of some patients.
To identify autophagy-related lncRNAs in melanoma that are crucial for the diagnosis, treatment, and prognosis of melanoma patients.
We retrieved transcriptome expression profiles and clinical information of 470 melanoma patients from The Cancer Genome Atlas (TCGA) database. Then, we identified autophagy-related genes in the Human Autophagy Database. Using R, coexpression analysis of lncRNAs and autophagy-related genes was conducted to obtain autophagy-related lncRNAs and their expression levels. We also performed univariate and multivariate Cox proportional risk analyses on the obtained datasets, to systematically evaluate the prognostic value of autophagy-related lncRNAs in melanoma. Fifteen autophagy-related lncRNAs were identified and an autophagy-related prognostic signature for melanoma was established. The Kaplan-Meier and univariate and multivariate Cox regression analyses were used to calculate risk scores. Based on the risk scores, melanoma patients were randomly divided into high- and low-risk groups. Receiver operating characteristic curve analysis, dependent on time, was performed to assess the accuracy of the prognostic model. At the same time, we also downloaded the melanoma data sets GSE65904, GSE19234, and GSE78220 from the GENE EXPRESSION OMNIBUS database for model verification. Finally, we performed Gene Set Enrichment Analysis functional annotation, which showed that the low and the high-risk groups had different enriched pathways.
The co-expression network for autophagy-related genes was constructed using R, and 936 lncRNAs related to autophagy were identified. Then, 52 autophagy-related lncRNAs were significantly associated with TCGA melanoma patients’ survival by univariate Cox proportional risk analysis (P < 0.01). Further, the 52 autophagy-related lncRNAs mentioned above were analyzed by multivariate Cox analysis with R. Fifteen lncRNAs were selected: LINC01943, AC090948.3, USP30-AS1, AC068282.1, AC004687.1, AL133371.2, AC242842.1, PCED1B-AS1, HLA-DQB1-AS1, AC011374.2, LINC00324, AC018553.1, LINC00520, DBH-AS1, and ITGB2-AS1. The P values in all survival analyses using these 15 lncRNAs were < 0.05. These lncRNAs were used to build a risk model based on the risk score. Negative correlations were observed between risk scores and overall survival rate in melanoma patients over time. Additionally, the melanoma risk curve and scatter plot analyses showed that the death number increased along with the increase in the risk score. Overall, we identified and established a new prognostic risk model for melanoma using 15 autophagy-related lncRNAs. The risk model constructed with these lncRNAs can help and guide melanoma patient prognosis predictions and individualized treatments in the future.
Overall, the risk model developed based on the 15 autophagy-related lncRNAs can have important prognostic value and may provide autophagy-related clinical targets for melanoma treatment.
Core Tip: Long non-coding ribonucleic acids (lncRNAs) are key factors in the development of many malignant tumors and are involved in the prognosis of some patients. The expression of autophagy-associated lncRNAs was associated with survival in melanoma patients. We obtained 15 autophagy-related lncRNAs and established a melanoma prognosis model, which can predict the prognosis of melanoma patients and is more accurate than TNM stage, age, gender, and other clinical indicators, and may provide autophagy-related clinical targets for the treatment of melanoma.
- Citation: Qiu Y, Wang HT, Zheng XF, Huang X, Meng JZ, Huang JP, Wen ZP, Yao J. Autophagy-related long non-coding RNA prognostic model predicts prognosis and survival of melanoma patients. World J Clin Cases 2022; 10(11): 3334-3351
- URL: https://www.wjgnet.com/2307-8960/full/v10/i11/3334.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i11.3334
Melanomas are malignant tumors that can occur in different body parts or tissues such as the skin, mucous membrane, uvea, and pia mater. While the incidence of many tumors is declining, the incidence of melanoma continues to increase[1]. The global prevalence of melanoma is increasing by about 3% to 7% per year[2]. Although many melanoma patients have localized tumors at diagnosis that can be completely removed by surgery, many patients present signs of metastasis[3]. Early melanoma has a good prognosis, but advanced melanoma patients have a very poor prognosis, with a 5%-10% survival rate, even with treatment[4]. Autophagy, originally considered a process of lysosomal-dependent degradation of cytoplasmic components in response to starvation, has been shown to affect multiple homeostasis aspects and to constitute a prevention barrier to malignant transformation[5]. Therefore, the combination of autophagy-related factors and pathological classification for risk stratification in melanoma patients can be crucial to prognosis and treatment response predictions.
Long non-coding RNAs (lncRNAs) are transcribed RNAs with a length of more than 200 nucleotides that are not translated into proteins[6]. Many studies have shown that noncoding RNAs can be associated with tumor pathogenesis by epigenetic regulation, as well as transcriptional and/or post-transcriptional processes. Moreover, lncRNAs can be used as sensitive and specific cancer biomarkers[7]. Previous studies have shown that melanoma pathogenesis was associated with different lncRNAs, in vitro and in vivo, and that some of them can be potential melanoma therapy targets, such as UCA1, DSCAM-AS1, and miR155HG[8,9]. However, the lncRNAs involved in autophagy and their prognostic value were not previously investigated, and many mechanisms remain unclear.
Therefore, we retrieved human clinical melanoma datasets from The Cancer Genome Atlas (TCGA) database and screened and analyzed genes associated with melanoma prognosis. Finally, 15 autophagy-related lncRNAs were identified as melanoma prognosis biomarkers. Compared to other clinical indicators, these lncRNAs had higher accuracy to predict melanoma patients’ survival.
The data of 471 melanoma patients were retrieved from the TCGA database (https://cancergenome.nih.gov/). In addition, the GSE65904, GSE19234, and GSE78220 data sets were downloaded from the GENE EXPRESSION OMNIBUS (GEO) database, including the expression data and clinical information of a total of 265 melanoma patients (https://www.ncbi.nlm.nih.gov/geo/). In this study, the lncRNA expression data in the TCGA database included complete clinical and follow-up data of 349 patients, which were finally used for analyses. Patients’ clinical characteristics are detailed in Table 1. This study followed the TCGA published guidelines, and since the data used here were from the published TCGA database, Institutional Review Board approval was not required.
| Variable | Group | n | Mean | SD | t value | P value |
| Age | ≤ 65 | 228 | 1.25 | 0.788 | -2.14416 | 0.033 |
| > 65 | 121 | 1.488 | 1.081 | |||
| Gender | Female | 133 | 1.323 | 1.016 | -0.1461 | 0.884 |
| Male | 216 | 1.338 | 0.834 | |||
| Stage | I-II | 194 | 1.283 | 0.801 | -1.10172 | 0.272 |
| III-IV | 155 | 1.394 | 1.022 | |||
| T | 0-2 | 135 | 1.155 | 0.747 | -3.10656 | 0.002 |
| 3-4 | 214 | 1.444 | 0.978 | |||
| M | 0 | 338 | 1.336 | 0.912 | 0.502769 | 0.625 |
| 1 | 11 | 1.223 | 0.727 | |||
| N | 0 | 202 | 1.281 | 0.8 | -1.19357 | 0.234 |
| 1-3 | 147 | 1.403 | 1.033 |
The Human Autophagy Database was used to identify autophagy-related genes. Then, R was used to analyze the association of autophagy-related genes and lncRNAs (correlation coefficient > 0.3; P < 0.001), and a co-expression network was constructed. Finally, 936 melanoma autophagy-related lncRNAs were identified, and their expression levels were obtained.
The univariate proportional risk, Kaplan-Meier survival, and multivariate risk analyses were conducted using R to calculate patients’ risk scores and to identify if these autophagy-related lncRNAs were involved in the melanoma prognosis. The risk score was calculated using the formula: Risk score = expr (lncRNA1) × coef (lncRNA1) + expr (lncRNA2) × coef (lncRNA2) + … + expr (lncRNAn) × coef (lncRNAn). Melanoma patients were divided into high- and low-risk groups, based on the median risk score. Finally, three data sets from the GEO database (GSE65904, GSE19234, and GSE78220) were used for verification analysis.
Univariate and multivariate regression analyses were used to systematically assess the relations between prognosis, clinicopathological factors, and risk scores in melanoma patients. To evaluate and verify the predictive accuracy of the prognostic model, we conducted receiver operating characteristic (ROC) analysis on the data from the TCGA database and the GSE78220 data set from the GEO database, and drew the ROC curve.
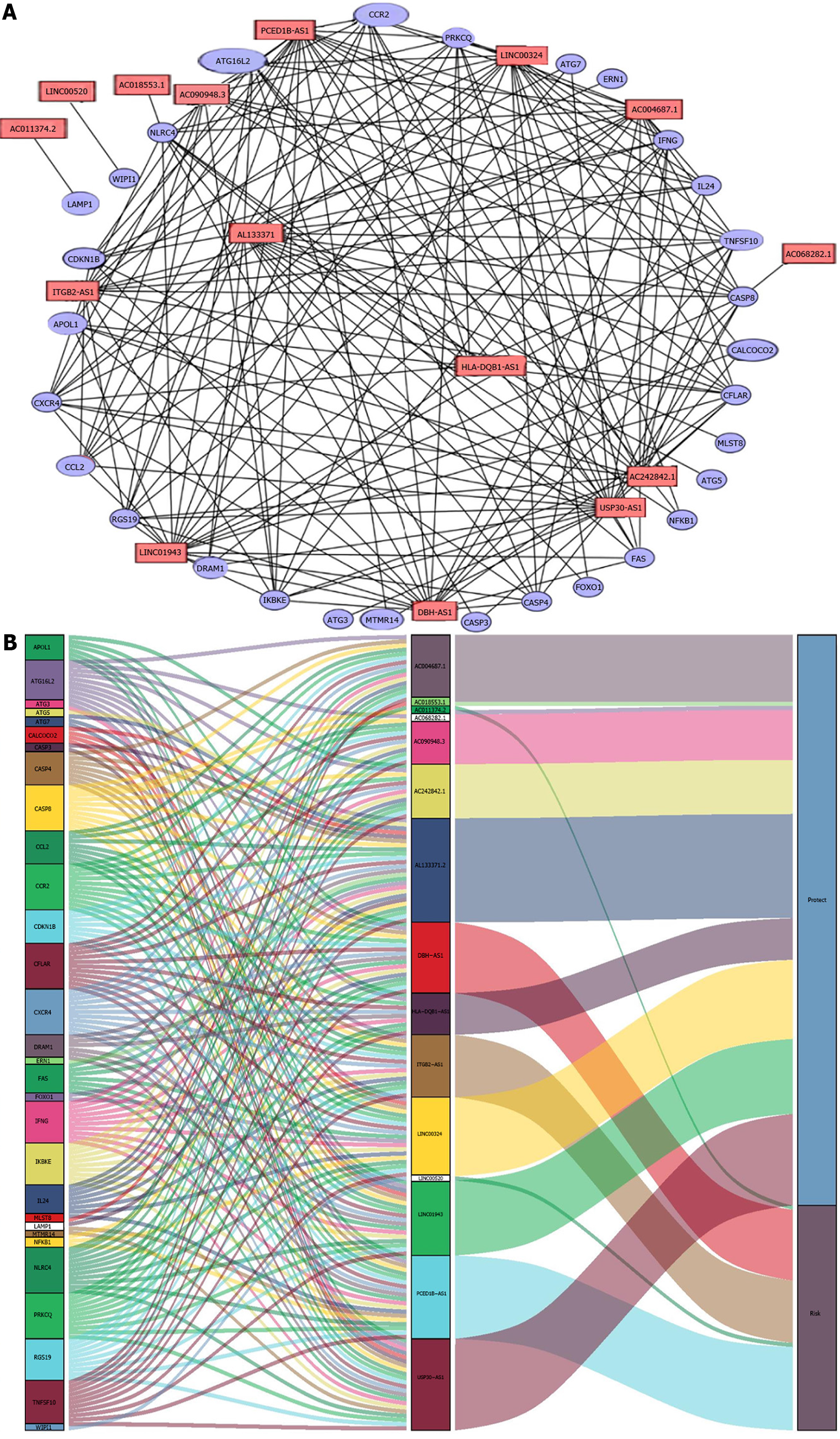
A co-expression network with 15 prognostic autophagy-related genes was constructed using R (version 4.0.4). The mulberries were mapped and the network was visualized using Cytoscape. Then, survival curves were drawn for these 15 lncRNAs, and risk survival and risk curves were drawn. Then, Gene Set Enrichment Analysis (GSEA) was employed for functional annotation. The GSEA focused not only on high-score genes but also on a range of genes related to biological processes that are associated with cancer pathogenesis, including stress response, transcription, and metabolic pathways. P < 0.05 was considered statistically significant[10].
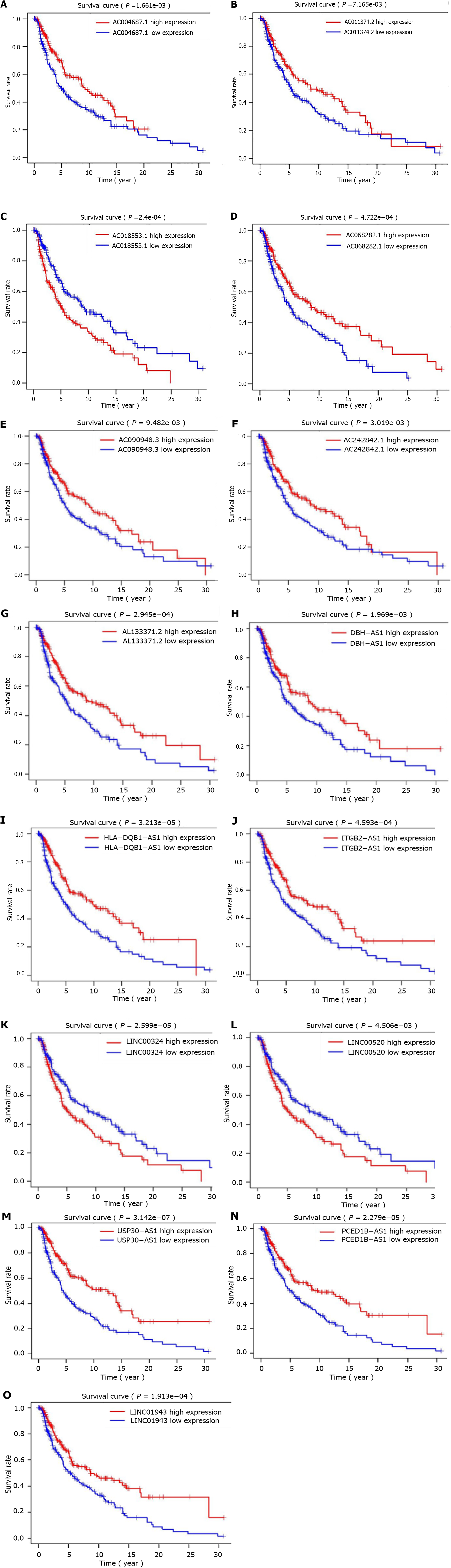
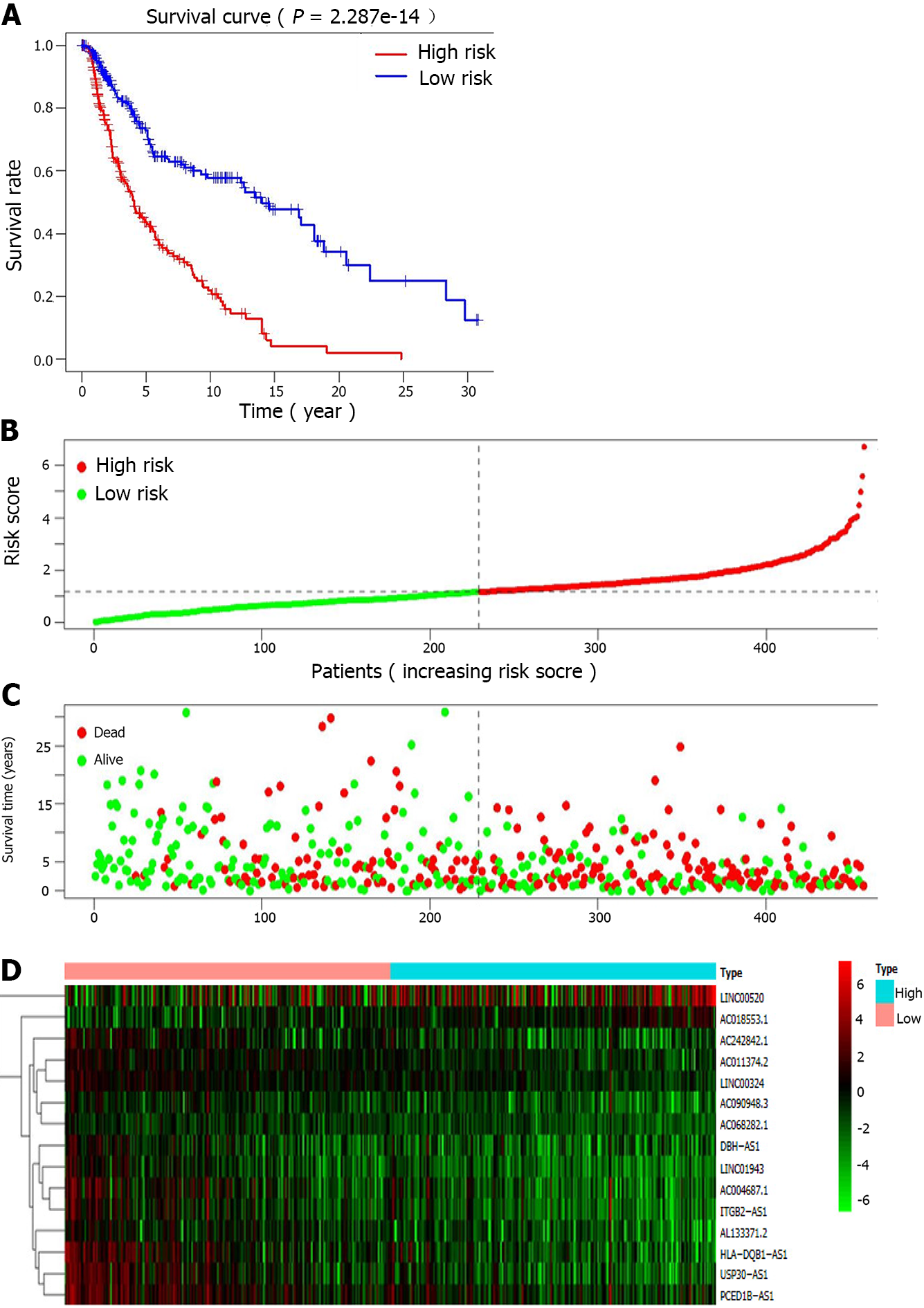
The co-expression network for autophagy-related genes was constructed using R, and 936 lncRNAs related to autophagy were identified. Then, 52 autophagy-related lncRNAs were significantly associated with TCGA melanoma patients’ survival by univariate Cox proportional risk analysis (P < 0.01). Among the 52 autophagy-related lncRNAs, four were related to a high risk (Table 2). Further, the 52 autophagy-related lncRNAs mentioned above were analyzed by multivariate Cox analysis with R. Fifteen lncRNAs were selected: LINC01943, AC090948.3, USP30-AS1, AC068282.1, AC004687.1, AL133371.2, AC242842.1, PCED1B-AS1, HLA-DQB1-AS1, AC011374.2, LINC00324, AC018553.1, LINC00520, DBH-AS1, and ITGB2-AS1 (Table 3). They were subsequently used to construct a melanoma prognostic risk model (Figure 1). Then, we assigned the enrolled melanoma patients into two groups based on their median risk scores. The P values in all survival analyses using these 15 lncRNAs were < 0.05 (Figure 2). To investigate the survival rates in these groups, other survival analyses were performed. Negative correlations were observed between risk scores and overall survival rate in melanoma patients over time (Figure 3A). Additionally, the melanoma risk curve and scatter plot analyses showed that the death number increased along with the increase in the risk score. Therefore, this demonstrated that the melanoma patients’ mortality was related to the risk score (Figure 3B-D). The heat map produced with these 15 autophagy-related lncRNAs shows that LINC00520 and AC018553.1 were highly expressed in the high-risk group. High expression of others lncRNAs was observed in patients with a low-risk score. In order to verify the accuracy and stability of the model, we decided to use the GSE65904, GSE19234, and GSE78220 data sets from the GEO database to verify, and conduct survival, risk curve, scatter plot, and heat map analyses, and the results are basically consistent with those of the above analysis (Supplementary Figure 1). Overall, we screened 15 pairs of lncRNAs with prognostic significance in melanoma, paving the way for the subsequent melanoma prognostic model establishment.
| LncRNA | HR | Lower limit (95%CI) | Upper limit (95%CI) | P value | Risk |
| C5orf56 | 0.478 | 0.348 | 0.656 | 0.000 | Low |
| AC011899.2 | 0.588 | 0.437 | 0.792 | 0.000 | Low |
| AC098613.1 | 0.595 | 0.456 | 0.776 | 0.000 | Low |
| AC068282.1 | 0.617 | 0.448 | 0.851 | 0.003 | Low |
| LINC01943 | 0.627 | 0.500 | 0.786 | 0.000 | Low |
| AP002807.1 | 0.630 | 0.464 | 0.856 | 0.003 | Low |
| AL590764.1 | 0.634 | 0.496 | 0.811 | 0.000 | Low |
| LINC00324 | 0.635 | 0.517 | 0.779 | 0.000 | Low |
| ZEB1-AS1 | 0.659 | 0.531 | 0.817 | 0.000 | Low |
| VIM-AS1 | 0.659 | 0.533 | 0.816 | 0.000 | Low |
| LINC02328 | 0.662 | 0.504 | 0.870 | 0.003 | Low |
| MMP25-AS1 | 0.674 | 0.544 | 0.835 | 0.000 | Low |
| AC015911.3 | 0.674 | 0.553 | 0.821 | 0.000 | Low |
| AL662844.4 | 0.688 | 0.526 | 0.901 | 0.007 | Low |
| AL137003.2 | 0.689 | 0.544 | 0.871 | 0.002 | Low |
| MIAT | 0.701 | 0.575 | 0.854 | 0.000 | Low |
| AL133371.2 | 0.707 | 0.587 | 0.852 | 0.000 | Low |
| AC022706.1 | 0.713 | 0.569 | 0.895 | 0.003 | Low |
| U62317.1 | 0.714 | 0.580 | 0.879 | 0.002 | Low |
| AC090948.3 | 0.727 | 0.574 | 0.921 | 0.008 | Low |
| AC011374.2 | 0.735 | 0.608 | 0.888 | 0.001 | Low |
| AC242842.1 | 0.741 | 0.639 | 0.859 | 0.000 | Low |
| AC004918.1 | 0.756 | 0.629 | 0.908 | 0.003 | Low |
| TRG-AS1 | 0.763 | 0.643 | 0.905 | 0.002 | Low |
| DBH-AS1 | 0.777 | 0.664 | 0.910 | 0.002 | Low |
| AC018755.4 | 0.779 | 0.670 | 0.906 | 0.001 | Low |
| AC060766.7 | 0.782 | 0.658 | 0.929 | 0.005 | Low |
| AC090559.1 | 0.789 | 0.680 | 0.916 | 0.002 | Low |
| USP30-AS1 | 0.803 | 0.732 | 0.880 | 0.000 | Low |
| PAXIP1-AS2 | 0.806 | 0.706 | 0.919 | 0.001 | Low |
| AL365361.1 | 0.814 | 0.719 | 0.922 | 0.001 | Low |
| AC012236.1 | 0.835 | 0.741 | 0.941 | 0.003 | Low |
| HLA-DQB1-AS1 | 0.835 | 0.765 | 0.912 | 0.000 | Low |
| TRBV11-2 | 0.838 | 0.738 | 0.950 | 0.006 | Low |
| AC093726.1 | 0.844 | 0.772 | 0.923 | 0.000 | Low |
| AL157871.2 | 0.847 | 0.759 | 0.945 | 0.003 | Low |
| MIR155HG | 0.848 | 0.775 | 0.928 | 0.000 | Low |
| AC243960.1 | 0.854 | 0.774 | 0.942 | 0.002 | Low |
| AC083799.1 | 0.871 | 0.817 | 0.927 | 0.000 | Low |
| LINC02446 | 0.873 | 0.809 | 0.941 | 0.000 | Low |
| ITGB2-AS1 | 0.875 | 0.793 | 0.966 | 0.008 | Low |
| AC004687.1 | 0.881 | 0.813 | 0.955 | 0.002 | Low |
| PCED1B-AS1 | 0.914 | 0.865 | 0.964 | 0.001 | Low |
| WAC-AS1 | 0.916 | 0.876 | 0.959 | 0.000 | Low |
| LINC01871 | 0.918 | 0.880 | 0.957 | 0.000 | Low |
| PSMB8-AS1 | 0.944 | 0.920 | 0.969 | 0.000 | Low |
| THCAT158 | 0.952 | 0.923 | 0.981 | 0.001 | Low |
| HCP5 | 0.975 | 0.964 | 0.986 | 0.000 | Low |
| KU-MEL-3 | 1.006 | 1.002 | 1.010 | 0.003 | High |
| LINC00520 | 1.011 | 1.005 | 1.017 | 0.001 | High |
| AC100791.3 | 1.186 | 1.067 | 1.318 | 0.001 | High |
| AC018553.1 | 1.255 | 1.129 | 1.396 | 0.000 | High |
| LncRNA | Coefficient | HR | Risk |
| LINC01943 | -0.265 | 0.768 | Low |
| AC090948.3 | -0.282 | 0.754 | Low |
| USP30-AS1 | -0.206 | 0.814 | Low |
| AC068282.1 | -0.253 | 0.776 | Low |
| AC004687.1 | -0.152 | 0.859 | Low |
| AL133371.2 | -0.253 | 0.777 | Low |
| AC242842.1 | -0.209 | 0.812 | Low |
| PCED1B-AS1 | 0.229 | 1.258 | High |
| HLA-DQB1-AS1 | -0.096 | 0.909 | Low |
| AC011374.2 | -0.218 | 0.804 | Low |
| LINC00324 | -0.231 | 0.794 | Low |
| ITGB2-AS1 | 0.188 | 1.206 | High |
| AC018553.1 | 0.121 | 1.128 | High |
| LINC00520 | 0.01 | 1.01 | High |
| DBH-AS1 | 0.259 | 1.296 | High |
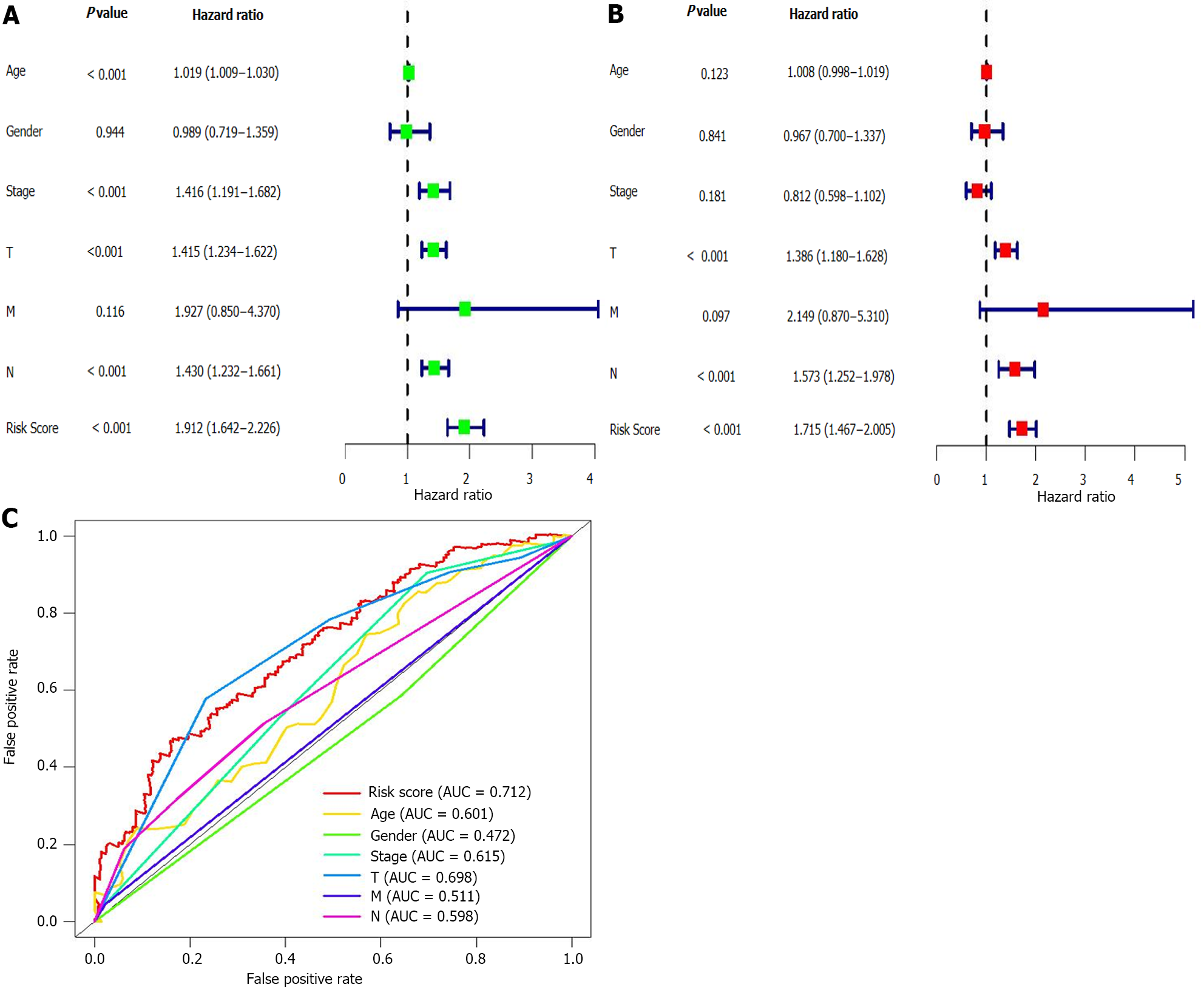
The above analyses identified 15 lncRNAs and established a prognostic risk model for the melanoma patients. Next, multivariate and univariate Cox regression analyses were employed to confirm if this model could be used for melanoma prognosis prediction. Results showed that the hazard ratio (HR) was 1.912 in the univariate analysis and 1.715 in the multivariate. Additionally, the 95% confidence interval of the risk score was 1.643-2.226 (P < 0.001) and 1.467-2.005 (P < 0.001) in the univariate and multivariate analyses, respectively (Figure 4A and B). These results suggested that in our model, the most important prognostic factors for melanoma were not age, gender, or TMN stage, but the 15 lncRNAs. To verify the sensitivity and specificity of the risk model, ROC curves of risk scores and other clinical indicators were plotted. The area under the risk score curve (AUC) was 0.712, exceeding the AUC of the other clinical factors (Figure 4C). In addition, we also verified the above analysis in three datasets from the GEO database, and the analysis results are consistent with those of the TCGA database (Supplementary Figure 2). These results suggested that the risk model was more accurate than other clinicopathological factors in predicting melanoma patients’ prognosis.
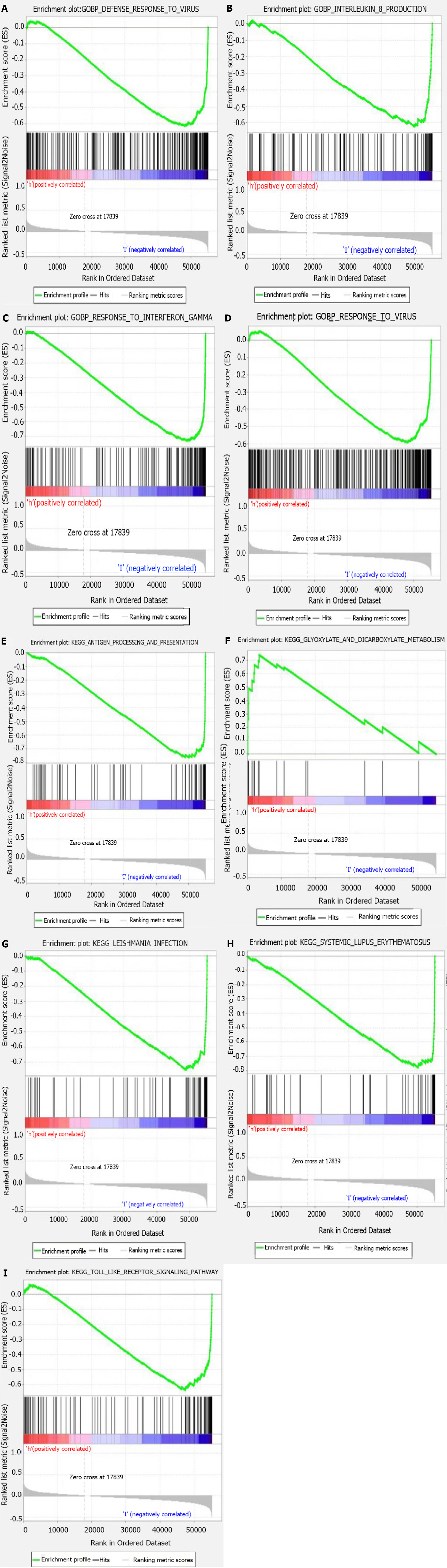
GSEA software (version 4.1.0) was used for functional annotation. The differentially expressed genes of the high-risk group were mainly enriched in glyoxylate and dicarboxylate metabolism, while in the low-risk group, pathways such as antigen processing and presentation, Toll-like receptor signaling, and cytokine production positive regulation were enriched (Figure 5).
As an aggressive tumor, melanoma has a poor prognosis and increasing incidence[11]. The main reason for the high mortality rate is late diagnosis[12]. Studies have found that the survival rate of melanoma patients with distant metastases was only 5%-10%[13]. Therefore, early detection and diagnosis of melanoma are crucial to improving the survival rate. At present, TNM staging is commonly used to evaluate the melanoma prognosis. However, some studies have found that TNM staging methods might predict different overall survival rates[14]. Increasing studies have shown that tumor TNM staging also has clinical limitations[15,16]. Therefore, new melanoma diagnosis and prognosis methods are required to improve melanoma patients’ prognosis and survival rate. The promotive and suppressive effects of autophagy on the development of tumors were observed by many previous studies[17,18]. In other words, autophagy is a dual process regarding tumors. For example, Mgrditchian et al[19] reported that autophagy can recruit natural killer cell infiltration into tumor tissues, and subsequently reduce the melanoma growth in vivo[19]. Luan et al[20] showed that POL can downregulate miR1290, leading to BECN1 upregulation and enhanced autophagy, thereby reducing the survival of melanoma cells. However, Xiao et al[21] showed that beclin-1 expression and LC3-II/I ratio were significantly increased by miR-24-1-5p. Meanwhile, they also observed the promotive effect of miR-24-1-5p on autophagy and subsequently inhibitory function in melanoma cell proliferation[21]. The studies listed above showed both positive and negative effects of autophagy on tumor development, and that autophagy plays a crucial role in this process.
Additionally, the roles of lncRNAs in the pathogenesis of different tumors have been identified by other researchers, including breast cancer, lung cancer, and hepatocellular carcinoma[22,23]. In this study, we analyzed and identified 15 lncRNAs that can be related to melanoma survival and prognosis, and established a risk model for melanoma prognosis. We consulted many previous studies and found that only LINC00520 and DBH-AS1 have been linked to melanoma. USP30-AS1, PCED1B-AS1, LINC00324, ITGB2-AS1, LINC00520, and DBH-AS1 have also been reported in other tumors. The remaining lncRNAs (LINC01943, AC090948.3, AC068282.1, AC004687.1, AL133371.2, AC242842.1, HLA-DQB1-AS1, AC011374.2, and AC018553.1) have not yet been reported. LICN00520 can promote melanoma’s proliferative and metastatic capabilities by competitively targeting miR-125b-5p and acting on the miR-125b-5p/eIF5A2 axis in vivo[24]. Also, LINC00520 can be involved in the progressive processes of many tumors. For example, Luan et al[24] pointed out that through MiR-3175 suppression, LINC00520 was involved in lung cancer progression[25]. Jin et al[26] found that LINC00520 can act as a miR-577 competitive suppressor to enhance HSP27 expression, leading to colorectal cancer progression[26]. Besides, LINC00520 can promote thyroid papillary carcinoma, nasopharyngeal carcinoma, breast cancer, and non-small cell lung cancer progression, being a poor prognostic factor for these tumors[27-31]. Chen et al[32] found that DBH-AS1 can enhance the glycolytic activity of melanoma cells, thereby blocking the miR-223-3p/EGFR/AKT axis[32]. Crucial roles of the lncRNA DBH-AS1 were also observed in osteosarcoma, diffuse large b-cell lymphoma, liver cancer, and non-small cell lung cancer[33-36]. In our analyses, both DBH-AS1 and LINC00520 were high-risk autophagy-related lncRNAs. Their elevated expression in melanoma patients indicated a poor prognosis. Through sequestering miR-229-3p and subsequently enhancing the expression of PTP4A1[37], USP30-AS1 expression can promote cervical cancer malignant progression. The lncRNA pCED1B-AS1 can activate glioma cell proliferation and limit apoptosis, working with the miR-194-5p/ pCED1B axis[38]. pCED1B-AS1 can also induce hepatocellular carcinoma immunosuppression through PD-1 and PD-L1 regulation via sponging has-miR-1945p[39]. Other studies have shown that the lncRNA pcED1B-AS1 can also promote clear cell renal carcinoma and pancreatic ductal adenocarcinoma progression[40,41]. Important roles of the lncRNAs LINC00324 and A ITGB2-AS1 have been observed during different tumors progressions. For example, the proliferative and invasive capabilities of osteosarcoma were promoted by LINC00324 through WDR66 regulation. Also, the promotive effects of LNC00324 on papillary thyroid carcinoma progression were found, due to its regulatory effect on Notch-related pathways[42,43]. The lncRNA ITGB2-AS1 can upregulate ITGB2 and promote breast cancer cell migration and invasion. Besides, ITGB2-AS1 can also promote pancreatic ductal adenocarcinoma growth and metastasis, acting on the miR-4319/RAF1 axis[44,45].
The mechanisms of the remaining nine lncRNAs identified in our study (LINC01943, AC090948.3, AC068282.1, AC004687.1, AL133371.2, AC242842.1, HLA-DQB1-AS1, AC011374.2, and AC018553.1) have not been yet reported and remain to be explored. Five of them, including PCED1B-AS1, ITGB2-AS1, AC018553.1, LINC00520, and DBH-AS1, were risk-related lncRNAs, and their increased expression levels suggested a poor prognosis. The remaining ten lncRNAs (LINC01943, AC090948.3, USP30-AS1, AC068282.1, AC004687.1, AL133371.2, AC242842.1, HLA-DQB1-AS1, AC011374.2, and LINC00324) were protective lncRNAs, and their elevated expression levels in melanoma patients predicted a good prognosis. The survival analysis (P = 2.287e - 14) suggested a significant difference between the survival rate of the high- and low-risk score patients. Meanwhile, we also observed negative correlations between the risk scores and overall survival rate in melanoma patients. Finally, we constructed the ROC curve, and the AUC value was 0.712, higher than the AUC of all clinicopathological indicators analyzed. Not only that, we also downloaded 265 melanoma samples from GEO database to verify the accuracy of this model, and the results are consistent with those of the previous analysis. These results demonstrated that the prognostic model established based on autophagy-related lncRNAs in melanoma patients had a higher prognostic accuracy than other clinical indicators. Altogether, these results suggested that the risk model established based on the 15 autophagy-related lncRNAs could better predict melanoma patients’ prognosis and survival.
The GSEA analysis revealed a difference in signaling pathway enrichment between the high- and low-risk melanoma patients. In low-risk populations, the main significantly enriched pathways were immune-related, such as antigen processing and presentation, Toll-like receptor signaling pathway, systemic lupus erythematosus, and autoimmune thyroid disease. The high-risk populations were mainly enriched in metabolism-related pathways, such as glyoxylate and dicarboxylate signaling pathways. These results indicated that the immunity improvement might be related to melanoma patients’ prognosis improvement and that a poor prognosis can be associated with glyoxylate and dicarboxylate metabolic pathways.
Recently, increasing studies have explored the significance of lncRNAs in cell autophagy, and their functional effect in tumors have attracted researchers’ attention. However, the function, mechanisms, and value of these lncRNAs in melanoma prognosis remain unclear. Our current study proposed a new melanoma risk model composed of 15 lncRNAs involved in autophagy that can be helpful for future melanoma treatment and prognosis evaluation. However, our research also has limitations. Although the data and analyses used have been verified for their accuracy in different studies, our study was not verified experimentally. The molecular mechanism of autophagy-related lncRNAs has not yet been elucidated, and some lncRNAs have never even been reported in the literature. Therefore, further experimental studies are required to verify our results.
Overall, we have identified and established a new prognostic risk model for melanoma using 15 autophagy-related lncRNAs: LINC01943, AC090948.3, USP30-AS1, AC068282.1, AC004687.1, AL133371.2, AC242842.1, PCED1B-AS1, HLA-DQB1-AS1, AC011374.2, LINC00324, ITGB2-AS1, AC018553.1, LINC00520, and DBH-AS1. The risk model constructed with these lncRNAs can help guide prognosis prediction and individualized treatment in melanoma patients in the future.
At present, melanoma is mainly treated by surgical resection, but many patients have tumor metastasis. Patients with advanced melanoma have a very poor prognosis, so a new method is needed to predict and evaluate the prognosis and survival of patients. Autophagy, originally thought to be a process of lysosomal dependent degradation of cytoplasmic components in response to starvation, has been shown to influence multiple dynamic equilibria and to constitute a barrier against malignant transformation. However, long non-coding RNAs (lncRNAs) involved in autophagy and their prognostic value have not been studied before, and many mechanisms remain unclear. Therefore, risk stratification of melanoma patients based on autophagy-associated lncRNAs combined with pathological classification is crucial to predict prognosis and treatment response.
The main purpose of this study was to identify autophagy lncRNAs associated with melanoma prognosis and establish a risk model to predict survival and prognosis. Among the 15 autophagy-related lncRNAs analyzed in this study, the mechanisms of action of some lncRNAs are still unclear and have not been reported in the literature, so further studies are needed to explore the role of these lncRNAs. The solution of this major problem will help us to have a deeper understanding of melanoma and make it possible to completely cure melanoma.
The main objective of this study was to establish a more accurate method for predicting and evaluating the prognosis and survival of melanoma patients.
First, data from The Cancer Genome Atlas and GENE EXPRESSION OMNIBUS (GEO) databases were processed, and then R was used to analyze the correlation between autophagy-related genes and lncRNAs (correlation coefficient > 0.30; P < 0.001), and a co-expression network was constructed. In order to evaluate the relationship between autophagy-related lncRNAs and melanoma prognosis, univariate proportional risk, Kaplan-Meier survival, and multivariate risk analyses were performed. R software was used to calculate the risk score of each patient, and the calculation formula is as follows: Risk score = expR (lncRNA1) × COEF (lncRNA1) + expr (lncRNA2) × COEF (lncRNA2) +... + expr (lncRNAn) × COEF (lncRNAn). In order to assess the stability of risk models, univariate and multivariate regression analyses and receiver operating characteristic analyses were also performed. Finally, Gene Set Enrichment Analysis was used for functional annotation and GEO data was used for further validation to ensure the accuracy of the results.
Our current study proposed a new melanoma risk model composed of 15 lncRNAs involved in autophagy that can be helpful for future melanoma treatment and prognosis evaluation. However, our research also has limitations. Although the data and analyses used have been verified for their accuracy in different studies, our study was not verified experimentally. The molecular mechanisms of phagocytosis-related lncRNAs have not yet been elucidated, and some lncRNAs have never even been reported in the literature. Therefore, further experimental studies are required to verify our results.
This study identified new and unreported lncRNAs through a series of analyses, which are closely related to the prognosis of melanoma patients, providing a new direction for future research on melanoma-related lncRNAs.
In the future, we can conduct further experimental verification on these 15 lncRNAs to understand the specific mechanism of action and specific role of these 15 lncRNAs in melanoma.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Wang CY S-Editor: Fan JR L-Editor: Wang TQ P-Editor: Cai YX
| 1. | MacKie RM, Hauschild A, Eggermont AM. Epidemiology of invasive cutaneous melanoma. Ann Oncol. 2009;20 Suppl 6:vi1-vi7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 261] [Cited by in RCA: 264] [Article Influence: 16.5] [Reference Citation Analysis (2)] |
| 2. | Rigel DS. Epidemiology of melanoma. Semin Cutan Med Surg. 2010;29:204-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 107] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 3. | Duncan LM. The classification of cutaneous melanoma. Hematol Oncol Clin North Am. 2009;23:501-513, ix. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 38] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 4. | Ugurel S, Röhmel J, Ascierto PA, Flaherty KT, Grob JJ, Hauschild A, Larkin J, Long GV, Lorigan P, McArthur GA, Ribas A, Robert C, Schadendorf D, Garbe C. Survival of patients with advanced metastatic melanoma: the impact of novel therapies-update 2017. Eur J Cancer. 2017;83:247-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 216] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 5. | Li S, Song Y, Quach C, Guo H, Jang GB, Maazi H, Zhao S, Sands NA, Liu Q, In GK, Peng D, Yuan W, Machida K, Yu M, Akbari O, Hagiya A, Yang Y, Punj V, Tang L, Liang C. Transcriptional regulation of autophagy-lysosomal function in BRAF-driven melanoma progression and chemoresistance. Nat Commun. 2019;10:1693. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 121] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 6. | Fazal FM, Chang HY. lncRNA Structure: Message to the Heart. Mol Cell. 2016;64:1-2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 7. | Li X, Li Y, Yu X, Jin F. Identification and validation of stemness-related lncRNA prognostic signature for breast cancer. J Transl Med. 2020;18:331. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 76] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 8. | Chen X, Gao J, Yu Y, Zhao Z, Pan Y. RETRACTED: Long non-coding RNA UCA1 targets miR-185-5p and regulates cell mobility by affecting epithelial-mesenchymal transition in melanoma via Wnt/β-catenin signaling pathway. Gene. 2018;676:298-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 9. | Peng L, Chen Z, Chen Y, Wang X, Tang N. MIR155HG is a prognostic biomarker and associated with immune infiltration and immune checkpoint molecules expression in multiple cancers. Cancer Med. 2019;8:7161-7173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 108] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 10. | Li X, Jin F, Li Y. A novel autophagy-related lncRNA prognostic risk model for breast cancer. J Cell Mol Med. 2021;25:4-14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 90] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 11. | Liu N, Liu Z, Liu X, Chen H. Comprehensive Analysis of a Competing Endogenous RNA Network Identifies Seven-lncRNA Signature as a Prognostic Biomarker for Melanoma. Front Oncol. 2019;9:935. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 12. | Huang R, Mao M, Lu Y, Yu Q, Liao L. A novel immune-related genes prognosis biomarker for melanoma: associated with tumor microenvironment. Aging (Albany NY). 2020;12:6966-6980. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 73] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 13. | Sun L, Guan Z, Wei S, Tan R, Li P, Yan L. Identification of Long Non-coding and Messenger RNAs Differentially Expressed Between Primary and Metastatic Melanoma. Front Genet. 2019;10:292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 14. | Perakis SO, Thomas JE, Pichler M. Non-coding RNAs Enabling Prognostic Stratification and Prediction of Therapeutic Response in Colorectal Cancer Patients. Adv Exp Med Biol. 2016;937:183-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 15. | Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471-1474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5537] [Cited by in RCA: 6465] [Article Influence: 431.0] [Reference Citation Analysis (0)] |
| 16. | Galon J, Pagès F, Marincola FM, Thurin M, Trinchieri G, Fox BA, Gajewski TF, Ascierto PA. The immune score as a new possible approach for the classification of cancer. J Transl Med. 2012;10:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 415] [Cited by in RCA: 491] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 17. | Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990-995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2022] [Cited by in RCA: 2069] [Article Influence: 98.5] [Reference Citation Analysis (0)] |
| 18. | White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15:5308-5316. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 907] [Cited by in RCA: 908] [Article Influence: 56.8] [Reference Citation Analysis (0)] |
| 19. | Mgrditchian T, Arakelian T, Paggetti J, Noman MZ, Viry E, Moussay E, Van Moer K, Kreis S, Guerin C, Buart S, Robert C, Borg C, Vielh P, Chouaib S, Berchem G, Janji B. Targeting autophagy inhibits melanoma growth by enhancing NK cells infiltration in a CCL5-dependent manner. Proc Natl Acad Sci U S A. 2017;114:E9271-E9279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 182] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 20. | Luan W, Qian Y, Ni X, Chanda TK, Xia Y, Wang J, Yan Y, Xu B. Polygonatum odoratum lectin promotes BECN1 expression and induces autophagy in malignant melanoma by regulation of miR1290. Onco Targets Ther. 2017;10:4569-4577. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 21. | Xiao Y, Diao Q, Liang Y, Peng Y, Zeng K. MicroRNA 24 1 5p promotes malignant melanoma cell autophagy and apoptosis via regulating ubiquitin D. Mol Med Rep. 2017;16:8448-8454. [RCA] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 22. | Xiong H, Ni Z, He J, Jiang S, Li X, Gong W, Zheng L, Chen S, Li B, Zhang N, Lyu X, Huang G, Chen B, Zhang Y, He F. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36:3528-3540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 299] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 23. | Liu XW, Xiao ZD, Han L, Zhang JX, Lee SW, Wang WQ, Lee H, Zhuang L, Chen JJ, Lin HK, Wang J, Liang H, Gan BY. LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat Cell Biol. 2016;18:431-442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 198] [Cited by in RCA: 242] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 24. | Luan W, Ding Y, Yuan H, Ma S, Ruan H, Wang J, Lu F, Bu X. Long non-coding RNA LINC00520 promotes the proliferation and metastasis of malignant melanoma by inducing the miR-125b-5p/EIF5A2 axis. J Exp Clin Cancer Res. 2020;39:96. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 25. | Xia G, Li X, Chen F, Shao Z. LncRNA LINC00520 Predicts Poor Prognosis and Promotes Progression of Lung Cancer by Inhibiting MiR-3175 Expression. Cancer Manag Res. 2020;12:5741-5748. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 26. | Jin XH, Hong YG, Li P, Hao LQ, Chen M. Long noncoding RNA LINC00520 accelerates the progression of colorectal cancer by serving as a competing endogenous RNA of microRNA-577 to increase HSP27 expression. Hum Cell. 2020;33:683-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 27. | Sun Y, Shi T, Ma Y, Qin H, Li K. Long noncoding RNA LINC00520 accelerates progression of papillary thyroid carcinoma by serving as a competing endogenous RNA of microRNA-577 to increase Sphk2 expression. Cell Cycle. 2020;19:787-800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 28. | Wang JF, Xi ZN, Su HJ, Bao Z, Qiao YH. SP1-induced overexpression of LINC00520 facilitates non-small cell lung cancer progression through miR-577/CCNE2 pathway and predicts poor prognosis. Hum Cell. 2021;34:952-964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 29. | Chen X, Chen H, Liu M, Xiong J, Song Z. Long noncoding RNA LINC00520 accelerates lung adenocarcinoma progression via miR-1252-5p/FOXR2 pathway. Hum Cell. 2021;34:478-490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 30. | Xie T, Pi G, Yang B, Ren H, Yu J, Ren Q, Zhou X, Hu D, Zhang H, Zhang Q, Hu L, Li Y, Zhou F. Long non-coding RNA 520 is a negative prognostic biomarker and exhibits pro-oncogenic function in nasopharyngeal carcinoma carcinogenesis through regulation of miR-26b-3p/USP39 axis. Gene. 2019;707:44-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 31. | Henry WS, Hendrickson DG, Beca F, Glass B, Lindahl-Allen M, He L, Ji Z, Struhl K, Beck AH, Rinn JL, Toker A. LINC00520 is induced by Src, STAT3, and PI3K and plays a functional role in breast cancer. Oncotarget. 2016;7:81981-81994. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 32. | Chen XX, Zhang N, Fu XF, Jiang Y, Wang MY. LncRNA DBH-AS1 facilitates the tumorigenesis of melanoma by targeting miR-233-3p via IGF-1R/Akt signaling. Eur Rev Med Pharmacol Sci. 2020;24:7698-7708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 33. | Huang JL, Ren TY, Cao SW, Zheng SH, Hu XM, Hu YW, Lin L, Chen J, Zheng L, Wang Q. HBx-related long non-coding RNA DBH-AS1 promotes cell proliferation and survival by activating MAPK signaling in hepatocellular carcinoma. Oncotarget. 2015;6:33791-33804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 86] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 34. | Song Y, Gao F, Peng Y, Yang X. Long non-coding RNA DBH-AS1 promotes cancer progression in diffuse large B-cell lymphoma by targeting FN1 via RNA-binding protein BUD13. Cell Biol Int. 2020;44:1331-1340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 35. | Liu ZB, Wang JA, Lv RQ. Downregulation of long non-coding RNA DBH-AS1 inhibits osteosarcoma progression by PI3K-AKT signaling pathways and indicates good prognosis. Eur Rev Med Pharmacol Sci. 2019;23:1418-1427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 36. | Bao J, Chen X, Hou Y, Kang G, Li Q, Xu Y. LncRNA DBH-AS1 facilitates the tumorigenesis of hepatocellular carcinoma by targeting miR-138 via FAK/Src/ERK pathway. Biomed Pharmacother. 2018;107:824-833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 37. | Chen M, Chi Y, Chen H, Zhao L. Long non-coding RNA USP30-AS1 aggravates the malignant progression of cervical cancer by sequestering microRNA-299-3p and thereby overexpressing PTP4A1. Oncol Lett. 2021;22:505. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 38. | Yang J, Yu D, Liu X, Changyong E, Yu S. LncRNA PCED1B-AS1 activates the proliferation and restricts the apoptosis of glioma through cooperating with miR-194-5p/PCED1B axis. J Cell Biochem. 2020;121:1823-1833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 39. | Fan F, Chen K, Lu X, Li A, Liu C, Wu B. Dual targeting of PD-L1 and PD-L2 by PCED1B-AS1 via sponging hsa-miR-194-5p induces immunosuppression in hepatocellular carcinoma. Hepatol Int. 2021;15:444-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 75] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 40. | Qin J, Zhu T, Wu W, Chen H, He Y. Long Non-Coding RNA PCED1B-AS1 Promotes the Progression of Clear Cell Renal Cell Carcinoma Through miR-484/ZEB1 Axis. Onco Targets Ther. 2021;14:393-402. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 41. | Zhang Y, Ma H, Chen C. Long non-coding RNA PCED1B-AS1 promotes pancreatic ductal adenocarcinoma progression by regulating the miR-411-3p/HIF-1α axis. Oncol Rep. 2021;46:134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 42. | Wu S, Gu Z, Wu Y, Wu W, Mao B, Zhao S. LINC00324 accelerates the proliferation and migration of osteosarcoma through regulating WDR66. J Cell Physiol. 2020;235:339-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 43. | Wan JF, Wan JY, Dong C, Li L. Linc00324 promotes the progression of papillary thyroid cancer via regulating Notch signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24:6818-6824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 44. | Yang M, Qin Q, Zhu J, Guo Y, Yin T, Wu H, Wang C. Long noncoding RNA ITGB2-AS1 promotes growth and metastasis through miR-4319/RAF1 axis in pancreatic ductal adenocarcinoma. J Cell Physiol. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 45. | Liu M, Gou L, Xia J, Wan Q, Jiang Y, Sun S, Tang M, He T, Zhang Y. LncRNA ITGB2-AS1 Could Promote the Migration and Invasion of Breast Cancer Cells through Up-Regulating ITGB2. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 57] [Article Influence: 8.1] [Reference Citation Analysis (0)] |