Published online Sep 15, 2023. doi: 10.4251/wjgo.v15.i9.1556
Peer-review started: May 5, 2023
First decision: July 9, 2023
Revised: July 21, 2023
Accepted: August 18, 2023
Article in press: August 18, 2023
Published online: September 15, 2023
Processing time: 131 Days and 8.7 Hours
The molecular mechanisms of colorectal cancer development and progression are far from being elucidated.
To investigate the role of microRNA-363-3p (miR-363-3p) in the progression of colorectal cancer.
Real-time polymerase chain reaction was performed to detect miRNA expression in human colorectal cancer tissues and paired normal colorectal tissues. PITA 6 was utilized to predict the targets of miR-363-3p. Dual-luciferase reporter system was used to validate the target of miR-363-3p. Plate colony formation assay and wound-healing assay were performed to evaluate cancer cells’ clonogenic survival ability and migration ability, respectively. Cell proliferation was examined by cell counting kit-8 assay. Immunohistochemical staining was used to determine the expression level of interferon-induced transmembrane protein 1 (IFITM1) in colorectal cancer tissues and adjacent tissues. The TCGA and GTEx databases were used to compare the expression levels of IFITM1 mRNA in colorectal cancer tissues and normal colorectal tissues and analyze the correlation between the expression levels of IFITM1 mRNA and overall survival and disease-free survival of patients. A colorectal cancer cell line with a deficiency of IFITM1 was constructed, and the regulation effect of IFITM1 on the clonogenic growth of colorectal cancer cells was clarified.
MiR-363-3p was decreased in colorectal cancer tissues compared to normal colorectal tissues. IFITM1 was characterized as a direct target of miR-363-3p. Overexpression of miR-363-3p led to decreased clonogenic survival, proliferation, and migration of colorectal cancer cells, which could be reversed by forced IFITM1 expression.
MiR-363-3p can constrain clonogenic survival, proliferation, and migration of colorectal cancer cells via targeting IFITM1.
Core Tip: MicroRNAs (miRNAs) have been implicated in almost all known cancer processes. Although many algorithms can predict target genes for miRNA, the exact regulatory relationships still need to be experimentally verified. In this study, we investigated the role of miR-363-3p in clonogenic survival, proliferation, and migration of colorectal cancer cells and interferon-induced transmembrane protein 1 (IFITM1) was identified as a direct target of miR-363-3p. These findings widen and deepen the understanding of the molecular function of miR-363-3p and IFITM1.
- Citation: Wang Y, Bai SK, Zhang T, Liao CG. MicroRNA-363-3p inhibits colorectal cancer progression by targeting interferon-induced transmembrane protein 1. World J Gastrointest Oncol 2023; 15(9): 1556-1566
- URL: https://www.wjgnet.com/1948-5204/full/v15/i9/1556.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i9.1556
Colorectal cancer is the third most common cancer type worldwide; in 2020, almost 2 million cases were diagnosed. Colorectal cancer is the second most common cause of cancer death, leading to almost 1 million deaths per year, accounting for approximately 10% of new tumor cases and deaths[1]. In 2019, China (607900), the United States (227242), and Japan (160211) had the highest number of new cases of colorectal cancer, and China (261777), India (79098), and the United States (84026) had the highest number of colorectal cancer deaths[2]. Low-dairy diets (15.6%), smoking (13.3%), low-calcium diets (12.9%), and alcohol consumption (9.9%) are important risk factors for colorectal cancer[2], but the molecular mechanisms of colorectal cancer development and progression are far from being elucidated.
MicroRNAs (miRNAs) are small endogenous non-coding RNAs (ncRNAs) of about 22 nucleotides in size. miRNAs play important roles in gene regulation via a posttranscriptional manner, and their dysregulation is implicated in various human diseases including cancer. It is estimated that miRNAs can target more than 60% of human protein-coding genes[3]. Mechanically, miRNAs prevent the translation of target mRNAs that are then sequestered into mRNA-processing bodies (P-bodies) and degraded. MiRNAs also contribute to the degradation of the target mRNAs without sequestration to P-bodies[4]. The specificity of miRNA - mRNA interaction is bestowed mainly by a miRNA’s first eight nucleotides (known as seed sequence)[5]. Over the past period, miRNAs have been implicated in almost all known cancer processes. Depending on the target gene and tumor type, some miRNAs typically negatively affect oncogenes encoding proteins, while some other miRNAs can inhibit known tumor suppressors, so miRNAs can act as onco-miRNAs or tumor suppressor miRNAs[6]. For example, miR-100 and miR-125b coordinately repressed five Wnt/β-catenin negative regulators, resulting in increased Wnt signaling in colorectal cancer[7]. MiR-146a targets PTGES2 and suppresses colorectal cancer[8]. Recently, miR-363-3p was reported to participate in the regulation of a variety of diseases. In addition, the downregulation of miR-363-3p is closely correlated with the degree of differentiation, tumor-node-metastasis stage, and lymph node metastasis in gastric cancer[9]. Overexpression of miR-363-3p is a strong predictor of favorable prognosis in adenocarcinoma of the uterine cervix[10]. MiR-363-3p suppresses tumor growth and metastasis of colorectal cancer via targeting sphingosine kinase 2[11] and SRY-related high-mobility-group box 4 (SOX4)[12]. In contrast, the expression of miR-363-3p was increased in glioma[13] and pediatric T-cell acute lymphoblastic leukemia[14]. MiR-363-3p functions as onco-miRNA promotes cell proliferation, protects against apoptosis, and enhances invasion by directly targeting PDHB in glioma[13] and PTEN and BIM in leukemic cells[14]. Based on previous studies, we speculated that miR-363-3p might exert an essential effect on colorectal cancer progression.
Interferon-induced transmembrane protein 1 (IFITM1), also known as DSPA2a and CD225, is a member of the interferon-induced transmembrane protein family. Friedman et al[15] first identified the IFITM1 gene in neuroblastoma cells. The IFITM1-coding gene is located at 11p15.5, and the IFITM1 protein consists of 125 amino acid residues with a molecular weight of about 13.96 kDa, including the C-terminal extracellular domain, two transmembrane domains, and N-terminal intracellular domains. Li et al[16] found higher levels of IFITM1 expression in gallbladder adenocarcinoma and adenosquamous cell carcinoma tissues with high tumor-node-metastasis (TNM) stage and with lymph node metastasis and invasion. In estrogen receptor (ER)-positive breast cancer, IFITM1 expression levels are associated with TNM staging and poor prognosis[17]. Therefore, IFITM1 is closely related to the occurrence and development of tumors, but the regulation and clinical significance of IFITM1 in colorectal cancer tissues still need to be studied in depth. In this study, we investigated the role and the underlying mechanisms of miR-363-3p in the clonogenic survival, proliferation, and migration of colorectal cancer cells. To our knowledge, this is the first study to identify IFITM1 as a direct target of miR-363-3p.
Human colorectal cancer cell lines SW480, SW1116, Colo320, and Caco-2 were obtained from American Type Culture Collection and cultured in RPMI 1640 medium (Thermo Fisher Scientific, Waltham, MA, United States) containing 10% fetal bovine serum (Thermo Fisher Scientific) and Penicillin-Streptomycin (Thermo Fisher Scientific). IFITM1 knockout SW480 cell line (SW480KO) was generated using the CRISPR/Cas9 system. gRNA targeting sequence was 5’-CCGCTGTGGTGTCCGGATGC-3’. SW480 cells were transfected with PX459 V2.0 containing the gRNA sequence using Lipofectamine 2000 (Invitrogen, Waltham, MA). After 48 h of transfection, the positive cells were selected with puromycin at 2 μg/mL for 5 d. The puromycin-resistant cells were seeded into a 96-well plate at one cell per well using CytoFLEX SRT (Beckman, Brea, CA). The knockout cells were confirmed by western blotting. All cultures were maintained at 5% CO2 and 37 °C.
Colorectal cancer tissues and adjacent normal tissues were obtained from patients at Tangdu Hospital, Air Force Medical University. All human individuals provided written informed consent. The study was approved by the Hospital Ethics Committee (202203-116). All participants (aged 42-76 years, 60% males, stages ranging from I to IVA) did not receive chemotherapy or radiation therapy before resection. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Agomir-363-3p, agomir-NC, antagomir-363-3p and antagomir-NC were obtained from RiboBio (Guangzhou, China). Cells in the logarithmic growth phase were trypsinized, resuspended, and seeded into 6-well plates. After being cultured overnight, cells were transfected with 75 pmol agomir or antagomir using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After 48 h, the cells were collected for subsequent analysis.
Total RNA was extracted using a Trizol reagent (Life Technologies, Carlsbad, CA) based on the supplier’s instruction. MiRNA was reversely transcribed using the miRNA 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China). mRNA was reversely transcribed using the SuperScript™ IV First-Strand Synthesis System with ezDNase™ Enzyme (Thermo Fisher Scientific). Quantitative polymerase chain reaction (qPCR) with specific primers was performed with the SYBR kit (TaKaRa, Shiga, Japan). Samples were normalized to housekeeping expression of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or snRNA U6 using the 2-ΔΔCt method. The sequences of primers were as follows: IFITM1 sense, 5’-CCAAGGTCCACCGTGATTAAC-3’; antisense, 5’-ACCAGTTCAAGAAGAGGGTGTT-3’; GAPDH sense, 5’-GCACCGTCAAGGCTGAGAAC-3’; antisense, 5’-TGGTGAAGACGCCAGTGGA-3’; miRNA363-3p sense, 5’-AATTGCACGGTATCCA-3’; antisense, 5’-AGTGCAGGGTCCGAGGTATT-3’; snRNA U6 sense, 5’-CTCGCTTCGGCAGCACA-3’; antisense, 5’-AACGCTTCACGAATTTGCGT-3’.
Cells were lysed with RIPA lysis buffer containing protease inhibitors. Protein concentration was determined using a BCA kit (Thermo Fisher Scientific). The proteins were separated using standard gel electrophoresis and blotted onto polyvinylidene fluoride membranes. Membranes were blocked with 5% nonfat milk in poly(butylene succinate-butylene terephthalate) (PBST) and incubated in primary antibody solution at 4 °C overnight. After being washed with PBST three times, the membranes were incubated with a secondary antibody at 24 °C for 45 min. Immunoblots were developed using an ECL-chemiluminescence Kit (Merck Millipore, Watford, United Kingdom), according to the manufacturer’s instructions. The primary antibody against IFITM1 (5B5E2) was obtained from Proteintech (Wuhan, China). α-tubulin was used as loading control and its antibody was purchased from Cell Signaling Technology (Danvers, MA). HRP-linked secondary antibodies were obtained from Thermo Fisher Scientific. The raw blots have been included in Supple
The predicted binding region of miR-363-3p in IFITM1 3’ untranslated region (UTR) (pGL3-wt) or mutated targeting sequence (pGL3-mt) was ligated into the pGL3-Basic vector. SW480 cells were transfected with pGL3 construct and pRL-TK (ratio of 50 to 1) using Lipofectamine 2000 (Invitrogen). Luciferase activity was measured using Dual-Luciferase Reporter Assay System (Promega, Madison, WI) according to the manufacturer’s instructions. The relative luciferase activity was calculated by dividing the results from the Firefly luciferase assay over the Renilla luciferase assay.
Cells were digested to prepare single-cell suspensions and seeded at 300 cells/2 mL in 6-well plates. Cells were cultured for about 2 wk until visible clones formed. The clones were fixed with 4% paraformaldehyde at 24 °C for 15 min. After being rinsed with phosphate buffered saline (PBS) three times, the clones were stained with Coomassie brilliant blue R250 at 24 °C for 20 min. The plates were washed with PBS several times to remove the residual dye. The clones in each well were counted.
Cells were seeded at 3 × 105 cells per well in 24-well plates. After the cells became confluent, a 10 μL pipette tip was used to wound the monolayer by scratching and the cells in suspension were removed by changing the medium. With a cell-free gap prepared, a series of time-lapse images were acquired as cells migrated into the cell-free gap. The change in the wound width (the average distance between the two margins of the scratch) was measured using ImageJ.
Cell proliferation was determined using cell counting kit-8 (CCK-8, Solarbio, Beijing, China). Cells were digested to prepare single-cell suspensions and seeded at a density of 2 × 103 cells/100 μL in 96-well plates. 10 μL of the CCK-8 solution was added to each well of the plate. After incubating the plate for 2 h in the incubator, the absorbance at 450 nm was measured using a microplate reader (Fluoroskan FL, Thermo Fisher Scientific). Subtraction of the blank well absorbance (absorbance of wells containing medium and CCK-8) was performed before analysis.
Sections 5 μm in thickness were prepared from formalin-fixed, paraffin-embedded tissues. Paraffin sections were deparaffinized, hydrated followed by a Tris-EDTA-based antigen retrieval step, and blocked against non-specific binding using normal goat serum (Cell Signaling Technology) followed by incubation using an anti-IFITM1 antibody (5B5E2, Proteintech) at 4 °C overnight. After being washed for 20 min, the sections were treated with universal biotinylated anti-mouse/rabbit/goat IgG derived from the horse (Vector Laboratories, Burlingame, CA) at 24 °C for 30 min. Signal development was performed using the NovaRED kit (Vector Laboratories) according to the manufacturer’s instructions.
Statistical analysis was performed using GraphPad Prism 8.0 (San Diego, CA, United States). Student’s t-test was used to compare the two groups. The One-Way ANOVA was used to compare the means across three or more groups. Survival analysis was performed using the Kaplan-Meier method and compared using the log-rank test. Immunohistochemical (IHC) score was analyzed using a χ2 test. Statistical significance was considered at P < 0.05.
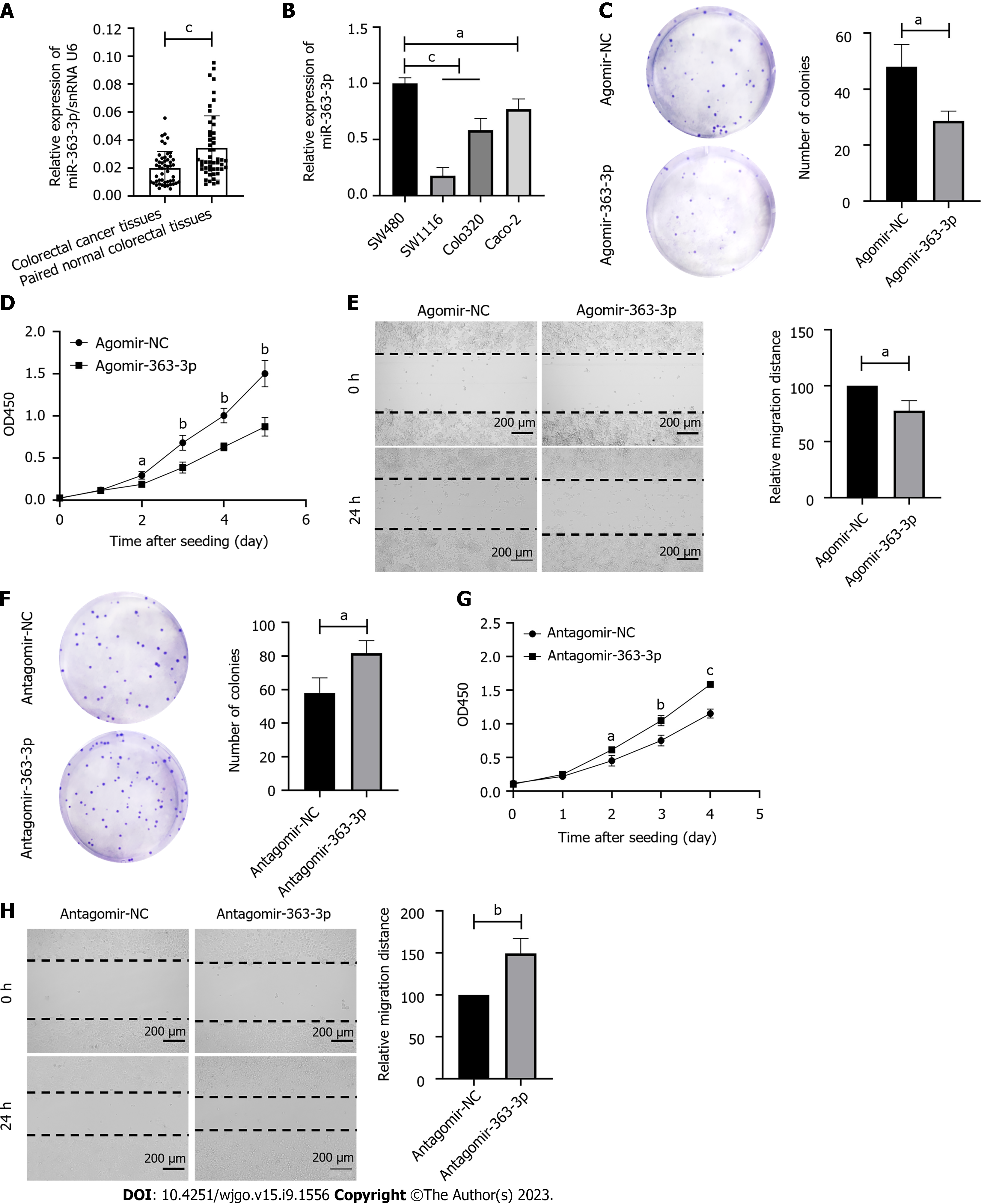
We first determined the expression pattern of miR-363-3p in 50 colorectal cancer tissues and paired normal colorectal tissues using real-time PCR. As shown in Figure 1A, we found that the expression of miR-363-3p was decreased in colorectal cancer tissues compared to the paired normal colorectal tissues.
To determine the effect of miR-363-3p on colorectal cancer progression, we first determined the expression level of miR-363-3p in human colorectal cancer cell lines (Figure 1B). As SW480 and SW1116 had the highest and lowest expression of miR-363-3p, respectively, the two cell lines were used in the subsequent experiments. We transfected SW1116 cells with agomir-363-3p and found that overexpression of miR-363-3p led to decreased clonogenic survival (Figure 1C), proliferation (Figure 1D), and migration (Figure 1E). In contrast, transfection of SW480 cells with antagomir-363-3p resulted in enhanced clonogenic survival (Figure 1F), proliferation (Figure 1G), and migration (Figure 1H). All these data suggest that miR-363-3p is a suppressive player involved in colorectal cancer progression.
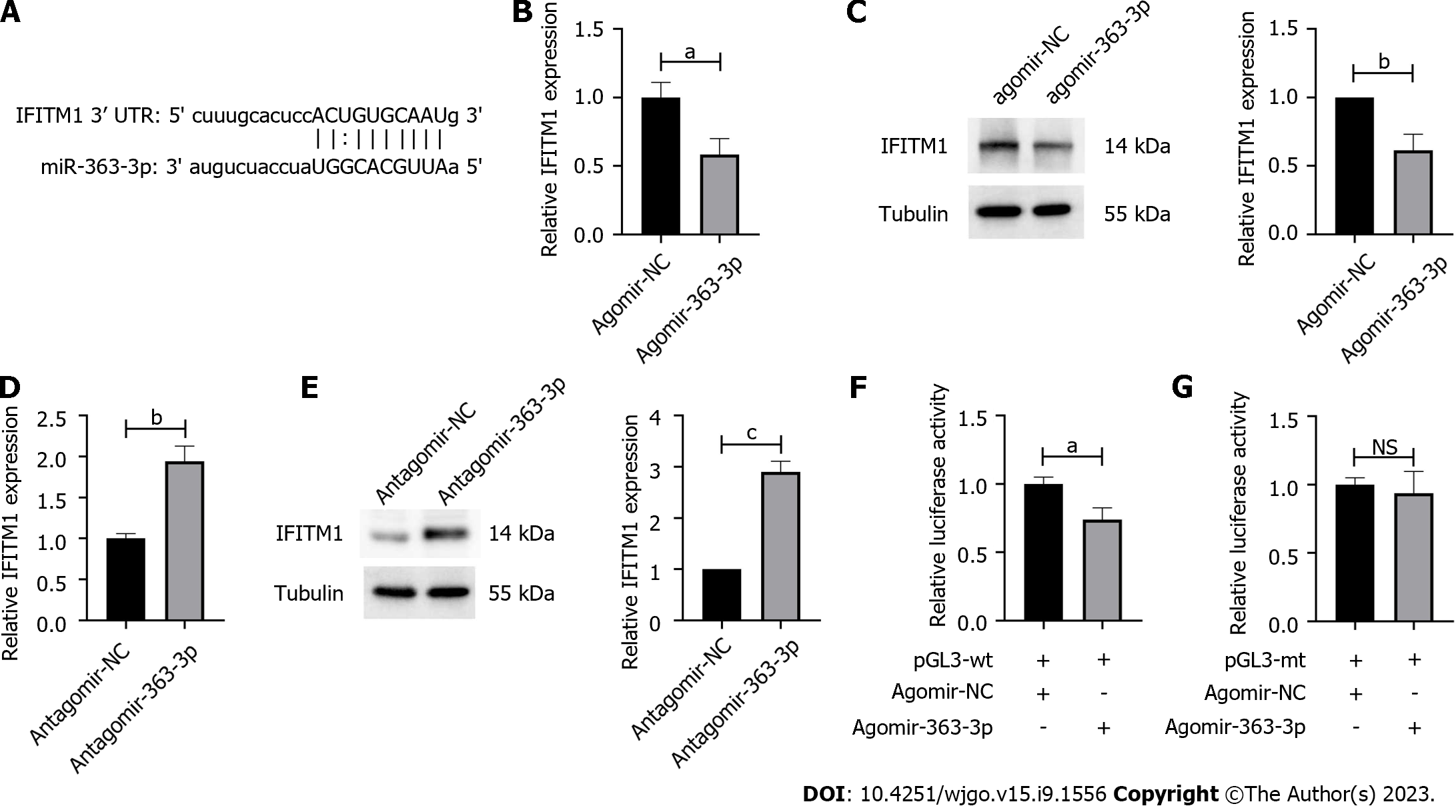
MiRNAs are supposed to regulate various cellular behaviors by targeting specific sites in mammalian mRNAs. Using PITA 6[18], IFITM1 was predicted as a promising target for miR-363-3p (Figure 2A). We found that overexpression of agomir-363-3p in SW1116 cells significantly decreased IFITM1 expression at both mRNA and protein levels (Figures 2B and C). As expected, inhibiting miR-363-3p via transfecting SW480 cells with antagomir-363-3p increased IFITM1 expression (Figures 2D and E). To examine the interaction between miR-363-3p and its targeting site in IFITM1 mRNA, luciferase reporter gene assays using constructs containing the predicted targeting sequence (pGL3-wt) and mutated targeting sequence (pGL3-mt) were performed. We found that co-transfection of agomir-363-3p and pGL3-wt in SW1116 cells led to decreased luciferase activity compared with the scramble control (Figure 2F), while co-transfection of agomir-363-3p and pGL3-mt in SW1116 cells showed luciferase activity comparable to that of the scramble control (Figure 2G). All these results demonstrate that IFITM1 is a direct target of miR-363-3p.
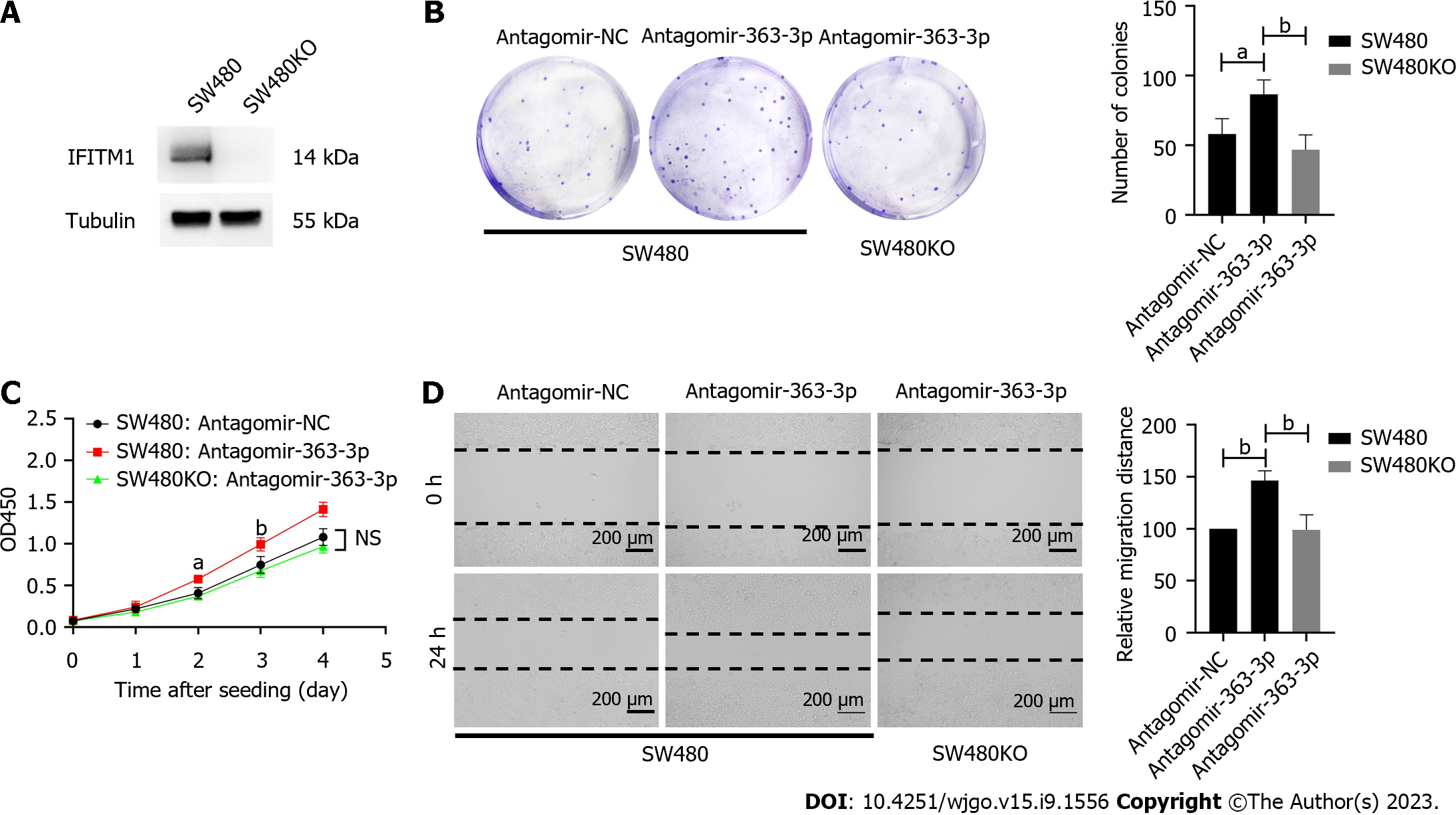
As miR-363-3p modulates IFITM1 expression, next we determined whether IFITM1 contributed to the regulatory effects of miR-363-3p on colorectal cancer progression. We generated IFITM1 knockout SW480 cell line (SW480KO) using the CRISPR-Cas9 system (Figure 3A) and inhibited miR-363-3p via transfecting antagomir-363-3p. As mentioned above, inhibiting miR-363-3p resulted in elevated clonogenic survival, proliferation, and migration in SW480 cells, in contrast, these effects became marginal in SW480KO cells (Figures 3B-D). These results suggest that IFITM1 contributes, at least partially, to the regulatory effects of miR-363-3p on clonogenic survival, proliferation, and migration of colorectal cancer cells.
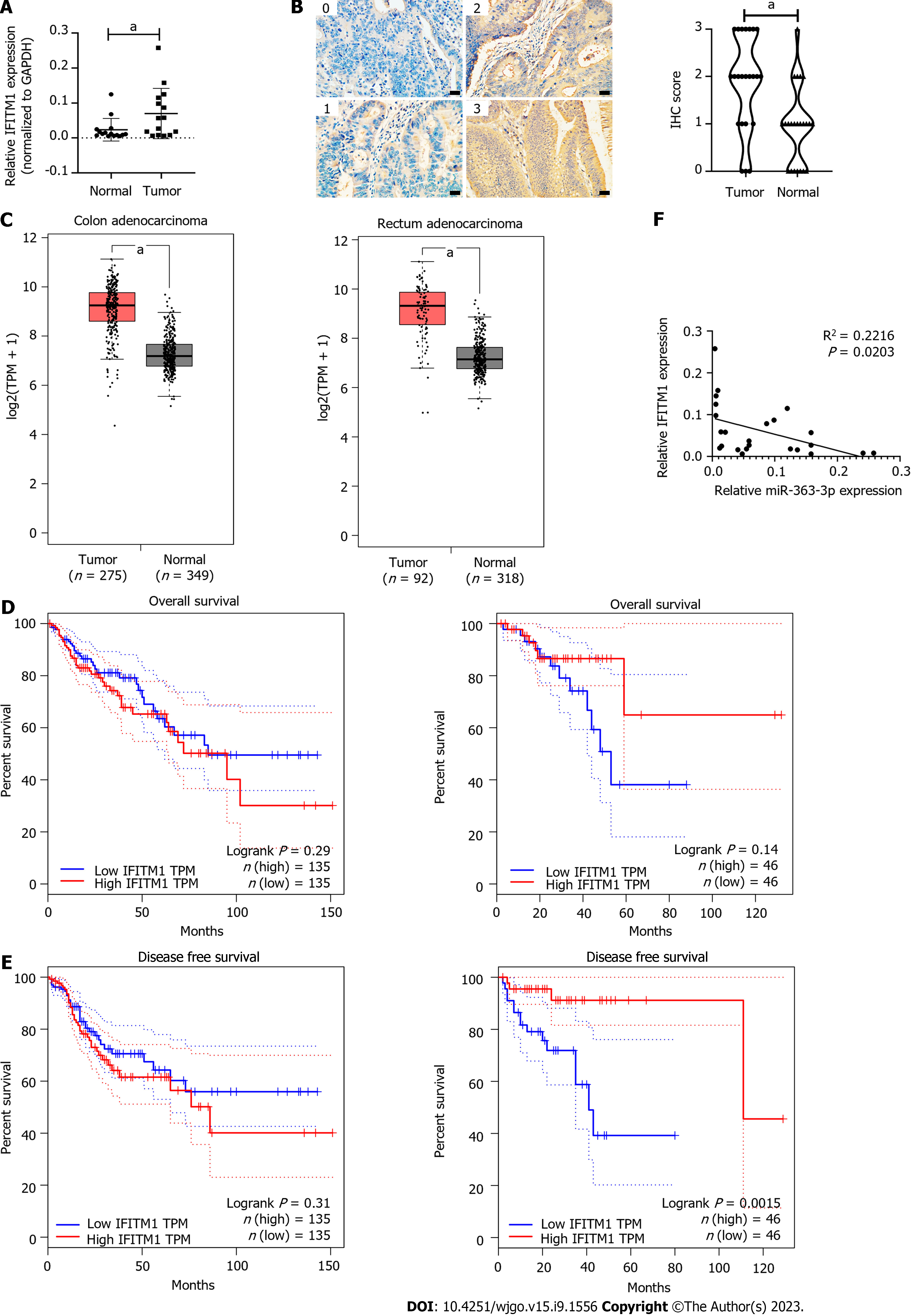
We identified IFITM1 as a direct target of miR-363-3p using colorectal cancer cell lines. To make this conclusion more solid, we evaluated the expression of miR-363-3p and IFITM1 in human colorectal cancer tissues. We found that the mRNA and protein expression levels of IFITM1 in colorectal cancer tissues were significantly higher than those in normal colorectal tissues (Figures 4A and B, Supplementary Table 1); TCGA data also showed that IFITM1 mRNA expression was increased in colorectal cancer tissues (Figure 4C) and was not associated with overall survival (OS) in patients with colorectal cancer (Figure 4D), but was positively correlated with disease-free survival (DFS) in patients with rectal cancer (Figure 4E). We also evaluated miR-363-3p and IFITM1 mRNA expression in 24 colorectal cancer tissues using qPCR. We found that IFITM1 mRNA expression was inversely correlated to miR-363-3p expression (R2 = 0.2216, Figure 4F). These results indicate that miR-363-3p negatively modulates IFITM1 expression in colorectal cancer tissues.
MiR-363-3p has been reported to be dysregulated and exert a promoting or inhibiting effect on tumor development and progression in many types of cancers. MiR-363-3p was significantly decreased in hepatocellular carcinoma (HCC)[19], papillary thyroid carcinoma[20], lung cancer[21], osteosarcoma[22], gastric cancer[9], CD133+ larynx cancer stem-like cells[23] and colorectal cancer[11]. However, the underlying mechanisms behind this dysregulation are far from clear. Li et al[24] reported that miR-363-3p is activated by its upstream transcription activator MYB in osteoporosis pathogenesis. MiR-363-3p could be sponged by circCTNNA1[25], circ_0002111[26], lncRNA NR2F1-AS1[27], lncRNA SNHG5[28], and lncRNA MALAT1[29] in colorectal cancer, papillary thyroid carcinoma, non-small cell lung cancer, and clear cell renal cell carcinoma.
MiR-363-3p inhibits tumorigenesis by directly targeting SOX4[19], high mobility group protein 2 (HMGA2)[30], USP28[31], and specificity protein 1[32] in HCC. MiR-363-3p inhibits tumor growth by targeting mouse double minute 2[33], proliferating cell nuclear antigen[34], HMGA2[35], neural precursor cell-expressed developmentally down-regulated 9 and SOX4[36] in lung cancer. miR-363-3p suppresses anoikis resistance via targeting integrin alpha 6 in papillary thyroid carcinoma[20]. MiR-363-3p is induced by hypoxia-inducible factor 2alpha to promote the stemness of melanoma cells via inhibiting p21[37]. MiR-363-3p markedly inhibits the proliferation, migration, and invasion of osteosarcoma cells via targeting SOX4[22]. This study provides another piece of evidence supporting miR-363-3p as a tumor suppressor. We confirmed that miR-363-3p is downregulated in human colorectal cancer tissues and inhibits clonogenic survival, proliferation, and migration of colorectal cancer cells.
IFITM1 belongs to a family of small homologous proteins, localized in the plasma and endolysosomal membranes, which regulate T cell differentiation and function and confer cellular resistance to many viruses[38]. There is mounting evidence that IFITM1 is an oncogene. IFITM1 expression is upregulated in gastric cancer[7], aromatase inhibitor-resistant breast cancer[8], triple-negative breast cancer[9], oral squamous cell carcinoma[10], and non-small cell lung cancer[11]. Furthermore, IFITM1 expression levels are closely related to patient outcomes[7,12]. IFITM1 regulates diverse aspects of tumorigenesis and progression, such as tumor cell proliferation, invasion, angiogenesis, metastasis, and therapeutic resistance, indicating that IFITM1 is a promising therapeutic target, and inhibiting IFITM1 (e.g., blocking IFITIM1 by antibody, suppressing IFITM1 expression by oligonucleotides, targeted IFITIM1 degradation using bifunctional small molecules) may be a promising strategy for cancer treatment. In colorectal cancer, the elevated IFITM1 expression significantly correlates with colorectal cancer lymph node and distance metastasis, a more advanced clinical stage as well as a shorter OS[39]. However, TCGA data showed that increased IFITM1 mRNA expression was not associated with OS in patients with colorectal cancer (Figure 4D), but was positively correlated with DFS in patients with rectal cancer (Figure 4E). He et al[40] reported that high expression of IFITM1 is associated with poor prognosis of rectal cancer, and no association was found between IFITM1 expression and the prognostic significance with patients with colon cancer. This discrepancy may be due to several factors. First, analysis using TCGA data focuses on IFITM1 mRNA expression, however, the level of mRNA expression is not exactly the same as the level of protein expression. Secondly, the antibodies used for IFITM1 detection and the scoring criteria for IHC staining are not exactly the same. Finally, patient survival is associated with many factors, and tumors are highly heterogeneous.
In vitro assays revealed that IFITIM1 promotes migration[41] and invasion[39] of human colorectal cancer via caveolin-1. Apc mutation induces the expression of IFITM1 and high expression of IFITM1 reduces the uptake of fibroblast extracellular vesicles[42]. In this study, we identified miR-363-3p as a new epigenetic modulator of IFITM1. We revealed the binding site of miR-363-3p in IFITM1 3’ UTR region and proved that the expression of IFITM1 can be efficiently inhibited by miR-363-3p and that the negative regulatory relationship exists in human colorectal cancer tissues. Moreover, a deficiency of IFITM1 can abolish the regulatory effects of miR-363-3p on clonogenic survival, proliferation, and migration of colorectal cancer cells. It would be interesting to further investigate whether the regulatory relationship between miR-363-3p and IFITM1 is common to other types of cancer, and the related lncRNA or circRNA.
Taken together, we identified that the expression of miR-363-3p and IFITM1 was downregulated and upregulated in colorectal cancer, respectively. Furthermore, IFITM1 is a direct target of miR-363-3p and the inhibitory effect of miR-363-3p on colorectal cancer progression is, at least partially, attributed to IFITM1 downregulation. We acknowledge several limitations in the present study. First, we didn’t determine the contribution of the miR-363-3p/IFITM1 axis to colorectal cancer progression using in vivo models. Second, whether the negative regulatory relationship between miR-363-3p and IFITM1 is prevalent in different kinds of tumors has to be further studied. Last, miR-363-3p is dysregulated in numerous tumors including colorectal cancer, however, the underlying mechanism was not further explored in the current study.
Colorectal cancer is the second most common cause of cancer death, however, the molecular mechanisms of tumorigenesis and development of colorectal cancer are far from being elucidated.
MicroRNAs play important roles in gene regulation and modulate numerous physical and pathological processes. The motivation of this study is to reveal the role of microRNA-363-3p (miR-363-3p) in the development of colorectal cancer and the underlying mechanisms.
Compare the expression of miR-363-3p between colorectal cancer tissues and adjacent normal tissues; clarify the role of miR-363-3p in clonogenic survival, migration, and proliferation of colorectal cancer cells; identify the direct target of miR-363-3p in colorectal cancer cells.
Real-time polymerase chain reaction was performed to detect miRNA expression. PITA 6 was utilized to predict the targets of miR-363-3p. Dual-luciferase reporter system was used to validate the target of miR-363-3p. Plate colony formation and wound-healing assays were performed to evaluate cancer cells’ clonogenic survival and migration ability, respectively. Cell proliferation was examined by cell counting kit-8 assay. Immunohistochemical staining was used to determine the expression level of interferon-induced transmembrane protein 1 (IFITM1).
MiR-363-3p was decreased in colorectal cancer tissues. IFITM1 was characterized as a direct target of miR-363-3p.
MiR-363-3p inhibits clonogenic survival, proliferation, and migration of colorectal cancer cells via targeting IFITM1.
MiR-363-3p/IFITM1 axis may represent a therapeutic target in colorectal cancer.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): D, D
Grade E (Poor): E
P-Reviewer: Chuang WL, Taiwan; Grassi G, Italy; Janvilisri T, Thailand; Sato H, Japan; Stojanovic B, Serbia S-Editor: Wang JJ L-Editor: A P-Editor: Chen YX
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64721] [Article Influence: 16180.3] [Reference Citation Analysis (177)] |
| 2. | GBD 2019 Colorectal Cancer Collaborators. Global, regional, and national burden of colorectal cancer and its risk factors, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Gastroenterol Hepatol. 2022;7:627-647. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 327] [Cited by in RCA: 374] [Article Influence: 124.7] [Reference Citation Analysis (0)] |
| 3. | Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5833] [Cited by in RCA: 6543] [Article Influence: 384.9] [Reference Citation Analysis (0)] |
| 4. | Budakoti M, Panwar AS, Molpa D, Singh RK, Büsselberg D, Mishra AP, Coutinho HDM, Nigam M. Micro-RNA: The darkhorse of cancer. Cell Signal. 2021;83:109995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 97] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 5. | Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2851] [Cited by in RCA: 2957] [Article Influence: 164.3] [Reference Citation Analysis (0)] |
| 6. | He B, Zhao Z, Cai Q, Zhang Y, Zhang P, Shi S, Xie H, Peng X, Yin W, Tao Y, Wang X. miRNA-based biomarkers, therapies, and resistance in Cancer. Int J Biol Sci. 2020;16:2628-2647. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 310] [Cited by in RCA: 394] [Article Influence: 78.8] [Reference Citation Analysis (0)] |
| 7. | Lu Y, Zhao X, Liu Q, Li C, Graves-Deal R, Cao Z, Singh B, Franklin JL, Wang J, Hu H, Wei T, Yang M, Yeatman TJ, Lee E, Saito-Diaz K, Hinger S, Patton JG, Chung CH, Emmrich S, Klusmann JH, Fan D, Coffey RJ. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/β-catenin signaling. Nat Med. 2017;23:1331-1341. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 219] [Cited by in RCA: 334] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 8. | Garo LP, Ajay AK, Fujiwara M, Gabriely G, Raheja R, Kuhn C, Kenyon B, Skillin N, Kadowaki-Saga R, Saxena S, Murugaiyan G. MicroRNA-146a limits tumorigenic inflammation in colorectal cancer. Nat Commun. 2021;12:2419. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 89] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 9. | Chen Z, Liu X, Hu Z, Wang Y, Liu M, Li H, Ji R, Guo Q, Zhou Y. Identification and characterization of tumor suppressor and oncogenic miRNAs in gastric cancer. Oncol Lett. 2015;10:329-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 10. | Park H, Lee MJ, Jeong JY, Choi MC, Jung SG, Joo WD, Lee C, An HJ. Dysregulated microRNA expression in adenocarcinoma of the uterine cervix: clinical impact of miR-363-3p. Gynecol Oncol. 2014;135:565-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 11. | Dong J, Geng J, Tan W. MiR-363-3p suppresses tumor growth and metastasis of colorectal cancer via targeting SphK2. Biomed Pharmacother. 2018;105:922-931. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 12. | Hu F, Min J, Cao X, Liu L, Ge Z, Hu J, Li X. MiR-363-3p inhibits the epithelial-to-mesenchymal transition and suppresses metastasis in colorectal cancer by targeting Sox4. Biochem Biophys Res Commun. 2016;474:35-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 58] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 13. | Xu DX, Guo JJ, Zhu GY, Wu HJ, Zhang QS, Cui T. MiR-363-3p modulates cell growth and invasion in glioma by directly targeting pyruvate dehydrogenase B. Eur Rev Med Pharmacol Sci. 2018;22:5230-5239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 14. | Drobna M, Szarzyńska B, Jaksik R, Sędek Ł, Kuchmiy A, Taghon T, Van Vlierberghe P, Szczepański T, Witt M, Dawidowska M. hsa-miR-20b-5p and hsa-miR-363-3p Affect Expression of PTEN and BIM Tumor Suppressor Genes and Modulate Survival of T-ALL Cells In Vitro. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 15. | Friedman RL, Manly SP, McMahon M, Kerr IM, Stark GR. Transcriptional and posttranscriptional regulation of interferon-induced gene expression in human cells. Cell. 1984;38:745-755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 508] [Cited by in RCA: 623] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 16. | Li D, Yang Z, Liu Z, Zou Q, Yuan Y. DDR2 and IFITM1 Are Prognostic Markers in Gallbladder Squamous Cell/Adenosquamous Carcinomas and Adenocarcinomas. Pathol Oncol Res. 2019;25:157-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 17. | Lui AJ, Geanes ES, Ogony J, Behbod F, Marquess J, Valdez K, Jewell W, Tawfik O, Lewis-Wambi J. IFITM1 suppression blocks proliferation and invasion of aromatase inhibitor-resistant breast cancer in vivo by JAK/STAT-mediated induction of p21. Cancer Lett. 2017;399:29-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 18. | Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278-1284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1820] [Cited by in RCA: 1891] [Article Influence: 105.1] [Reference Citation Analysis (0)] |
| 19. | Wang J, Tang Q, Lu L, Luo Z, Li W, Lu Y, Pu J. LncRNA OIP5-AS1 interacts with miR-363-3p to contribute to hepatocellular carcinoma progression through up-regulation of SOX4. Gene Ther. 2019;27:495-504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 20. | Pan Y, Zhu X, Wang K, Chen Y. MicroRNA-363-3p suppresses anoikis resistance in human papillary thyroid carcinoma via targeting integrin alpha 6. Acta Biochim Biophys Sin (Shanghai). 2019;51:807-813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 21. | Wang Y, Chen J, Lin Z, Cao J, Huang H, Jiang Y, He H, Yang L, Ren N, Liu G. Role of deregulated microRNAs in non-small cell lung cancer progression using fresh-frozen and formalin-fixed, paraffin-embedded samples. Oncol Lett. 2016;11:801-808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 22. | Wang K, Yan L, Lu F. miR-363-3p Inhibits Osteosarcoma Cell Proliferation and Invasion via Targeting SOX4. Oncol Res. 2019;27:157-163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 23. | Karatas OF, Suer I, Yuceturk B, Yilmaz M, Oz B, Guven G, Cansiz H, Creighton CJ, Ittmann M, Ozen M. Identification of microRNA profile specific to cancer stem-like cells directly isolated from human larynx cancer specimens. BMC Cancer. 2016;16:853. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 24. | Li M, Luo R, Yang W, Zhou Z, Li C. miR-363-3p is activated by MYB and regulates osteoporosis pathogenesis via PTEN/PI3K/AKT signaling pathway. In Vitro Cell Dev Biol Anim. 2019;55:376-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 25. | Zhang Y, Zheng S, Liao N, Huang H, Chen W, Wu Z, Wu D. CircCTNNA1 acts as a ceRNA for miR-363-3p to facilitate the progression of colorectal cancer by promoting CXCL5 expression. J Biol Res (Thessalon). 2021;28:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 26. | Zhang Z, Mo Y, Wu L, Wang X, Liao G, Tan W, Li D. Circ_0002111 modulates the growth process of papillary thyroid carcinoma cells by targeting the miR-363-3p/HMGB1 axis. Anticancer Drugs. 2022;33:923-934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 27. | Jin L, Chen C, Huang L, Sun Q, Bu L. Long noncoding RNA NR2F1-AS1 stimulates the tumorigenic behavior of non-small cell lung cancer cells by sponging miR-363-3p to increase SOX4. Open Med (Wars). 2022;17:87-95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 28. | Li WZ, Zou Y, Song ZY, Wei ZW, Chen G, Cai QL, Wang Z. Long non-coding RNA SNHG5 affects the invasion and apoptosis of renal cell carcinoma by regulating the miR-363-3p-Twist1 interaction. Am J Transl Res. 2020;12:697-707. [PubMed] |
| 29. | Xie JJ, Li WH, Li X, Ye W, Shao CF. LncRNA MALAT1 promotes colorectal cancer development by sponging miR-363-3p to regulate EZH2 expression. J Biol Regul Homeost Agents. 2019;33:331-343. [PubMed] |
| 30. | Wang J, Liang H, Ge H, Guo X, Gu D, Yuan Y. MicroRNA3633p inhibits hepatocarcinogenesis by targeting HMGA2 and is associated with liver cancer stage. Mol Med Rep. 2019;19:935-942. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 31. | Han H, Sun D, Li W, Shen H, Zhu Y, Li C, Chen Y, Lu L, Zhang J, Tian Y, Li Y. A c-Myc-MicroRNA functional feedback loop affects hepatocarcinogenesis. Hepatology. 2013;57:2378-2389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 75] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 32. | Ying J, Yu X, Ma C, Zhang Y, Dong J. MicroRNA-363-3p is downregulated in hepatocellular carcinoma and inhibits tumorigenesis by directly targeting specificity protein 1. Mol Med Rep. 2017;16:1603-1611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 33. | Rong H, Chen B, Wei X, Peng J, Ma K, Duan S, He J. Long non-coding RNA XIST expedites lung adenocarcinoma progression through upregulating MDM2 expression via binding to miR-363-3p. Thorac Cancer. 2020;11:659-671. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 34. | Wang Y, Chen T, Huang H, Jiang Y, Yang L, Lin Z, He H, Liu T, Wu B, Chen J, Kamp DW, Liu G. miR-363-3p inhibits tumor growth by targeting PCNA in lung adenocarcinoma. Oncotarget. 2017;8:20133-20144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 65] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 35. | Jiang C, Cao Y, Lei T, Wang Y, Fu J, Wang Z, Lv Z. microRNA-363-3p inhibits cell growth and invasion of nonsmall cell lung cancer by targeting HMGA2. Mol Med Rep. 2018;17:2712-2718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 36. | Chang J, Gao F, Chu H, Lou L, Wang H, Chen Y. miR-363-3p inhibits migration, invasion, and epithelial-mesenchymal transition by targeting NEDD9 and SOX4 in non-small-cell lung cancer. J Cell Physiol. 2020;235:1808-1820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 37. | Hao T, Li CX, Ding XY, Xing XJ. MicroRNA-363-3p/p21(Cip1/Waf1) axis is regulated by HIF-2α in mediating stemness of melanoma cells. Neoplasma. 2019;66:427-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 38. | Yánez DC, Ross S, Crompton T. The IFITM protein family in adaptive immunity. Immunology. 2020;159:365-372. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 105] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 39. | Yu F, Xie D, Ng SS, Lum CT, Cai MY, Cheung WK, Kung HF, Lin G, Wang X, Lin MC. IFITM1 promotes the metastasis of human colorectal cancer via CAV-1. Cancer Lett. 2015;368:135-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 67] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 40. | He J, Li J, Feng W, Chen L, Yang K. Prognostic significance of INF-induced transmembrane protein 1 in colorectal cancer. Int J Clin Exp Pathol. 2015;8:16007-16013. [PubMed] |
| 41. | Sari IN, Yang YG, Phi LT, Kim H, Baek MJ, Jeong D, Kwon HY. Interferon-induced transmembrane protein 1 (IFITM1) is required for the progression of colorectal cancer. Oncotarget. 2016;7:86039-86050. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 42. | Kelemen A, Carmi I, Oszvald Á, Lőrincz P, Petővári G, Tölgyes T, Dede K, Bursics A, Buzás EI, Wiener Z. IFITM1 expression determines extracellular vesicle uptake in colorectal cancer. Cell Mol Life Sci. 2021;78:7009-7024. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |