Published online Jan 15, 2022. doi: 10.4251/wjgo.v14.i1.334
Peer-review started: March 22, 2021
First decision: June 14, 2021
Revised: July 3, 2021
Accepted: September 8, 2021
Article in press: September 8, 2021
Published online: January 15, 2022
Processing time: 294 Days and 9.9 Hours
Liver cancer is the fourth most significant cause of cancer-related death. Lack of early diagnosis strategy and a scarcity of efficient therapy constitute the main reasons for its lethality. Exosomes, which contain various bioactive molecules, are characterized by high biocompatibility, low immunogenicity, and high transport efficiency. As a result, exosomes have become a research hotspot and present significant potential for cancer diagnosis biomarkers, biotherapeutics, therapy targets, drug carriers and therapeutic agents.
To explore the potential of exosomes in the diagnosis and treatment of liver cancer.
We conducted a systematic literature search via PubMed and Web of Science. The following keywords were used: "exosomal biomarkers", "exosomal therapy", "exosomal therapy", and "liver cancer" or "HCC". The duplicate data were deleted by EndNote software. Literature search focused on full-texts and references of each article were carefully checked. One author (Xiao-Cui Wei) screened the literature that met the following inclusion criteria: (1) Detection of exosomes or their contents in clinical samples (body fluid or tissue); or (2) Exosomes served as drug carriers or therapeutic factors. Two authors (Xiao-Cui Wei and Li-Juan Liu) independently reviewed all retained literature and analyzed the information.
A total of 1295 studies were identified using the systematic literature search. Of these, 835 duplicate studies were removed. A further 402 irrelevant studies were excluded due to being irrelevant, including other diseases, review articles, the literature containing neither clinical samples nor animal experiments, exosome-independent studies, methods for detecting exosomes, or articles in Chinese. Finally, 58 published papers were retained and analyzed in the study. It showed a list of potential exosomal biomarkers that were upregulated in the blood samples of patients with liver cancer. Those downregulated in exosomes might serve as possible biotherapeutics. Some exosomes derived from cells in vitro were used for cytology or animal experiments to explore the mechanism of these exosome contents in disease. These contents might serve as potential targets for liver cancer. Additionally, we also discussed that exosomes serve as drug carriers or therapeutic factors.
Exosomes might serve as potential biomarkers or therapeutic biotargets in liver cancer and have the potential to act as drug carriers and self-treatment factors for liver cancer patients.
Core tip: We used a literature search to identify potential exosome diagnostic markers and novel therapeutic strategies for liver cancer. The latest literature was published in June 2021. Results were presented in tabular form, including 40 potential liver cancer biomarkers, 13 potential biotherapeutics, and 10 potential therapeutic targets for hepatocellular carcinoma. In addition, we also listed papers about exosomes as drug carriers and therapeutic factors.
- Citation: Wei XC, Liu LJ, Zhu F. Exosomes as potential diagnosis and treatment for liver cancer. World J Gastrointest Oncol 2022; 14(1): 334-347
- URL: https://www.wjgnet.com/1948-5204/full/v14/i1/334.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i1.334
Liver cancer is a common malignancy and the fourth leading cause of cancer death worldwide[1]. It is one of the most challenging cancers to treat. For patients with an early stage of liver cancer, surgical treatment is the standard of care. However, most patients with liver cancer are already in the advanced stage at the initial diagnosis, which results in a poor prognosis[2]. Currently, α-fetoprotein (AFP) is the most commonly used serum marker for liver cancer[3]. However, AFP has a sensitivity of 41%–64% and a specificity of 80%–94%, which is often missed diagnosis, especially in the early stages of liver cancer[4]. Therefore, it is vital to develop more sensitive and specific liver cancer biomarkers to improve patient survival.
Recent studies have shown that exosomes have potential as biomarkers for liver cancer[5]. Once considered cellular waste, exosomes are rich in bioactive molecules, such as proteins, lipids, and nucleic acids[6,7]. Almost all human cells can secrete exosomes. Tumor cells release more exosomes than normal cells, and the exosome contents of tumor cells are different from those of normal cells[8,9]. Additionally, the exosomal envelope protects proteins, nucleic acids, and other substances in exosomes from degradation by extramembrane enzymes[10]. The stability and abundance of exosome contents show the advantages of its unique liver cancer biomarkers.
Exosomes are widely involved in cell-to-cell communication. They can deliver their functional RNAs and proteins to recipient cells and affect their physiological functions[11]. Therefore, exosomes can also serve as drug delivery vehicles. Here, we summarize the potential of exosome contents in the diagnosis and treatment of liver cancer, provide new ideas for the diagnosis and treatment of liver cancer, and promote further research on the potential clinical applications of exosomes.
According to the conventional research methods of systematic review[12], a systematic literature search was conducted in PubMed and Web of Science using the following keywords: "exosomal biomarkers", "exosomal therapy", "exosomal therapy" and "liver cancer" or "HCC". The EndNote software was used to delete duplicate data[13]. The latest literature was published in June 2021. Literature search focused on full texts. Two reviewers independently screened the references of each article to remove the irrelevant studies according to our inclusion criteria. The inclusion criteria were as follows: (1) Detection of exosomes or their contents in clinical samples (body fluid or tissue); or (2) Exosomes served as drug carriers or therapeutic factors. Two authors (Xiao-Cui Wei and Li-Juan Liu) independently reviewed the full texts of all retained literature and analyzed the information.
The data collected from each study included the clinical sample, expression level, and application of exosomes divided into three major segments. The first part involved the exosomes isolated from the body fluid samples. The second part meant the data that were relevant to the detection of exosomal contents in the clinical tissue samples. The third part included the collection of data pertinent to the application of exosomes.
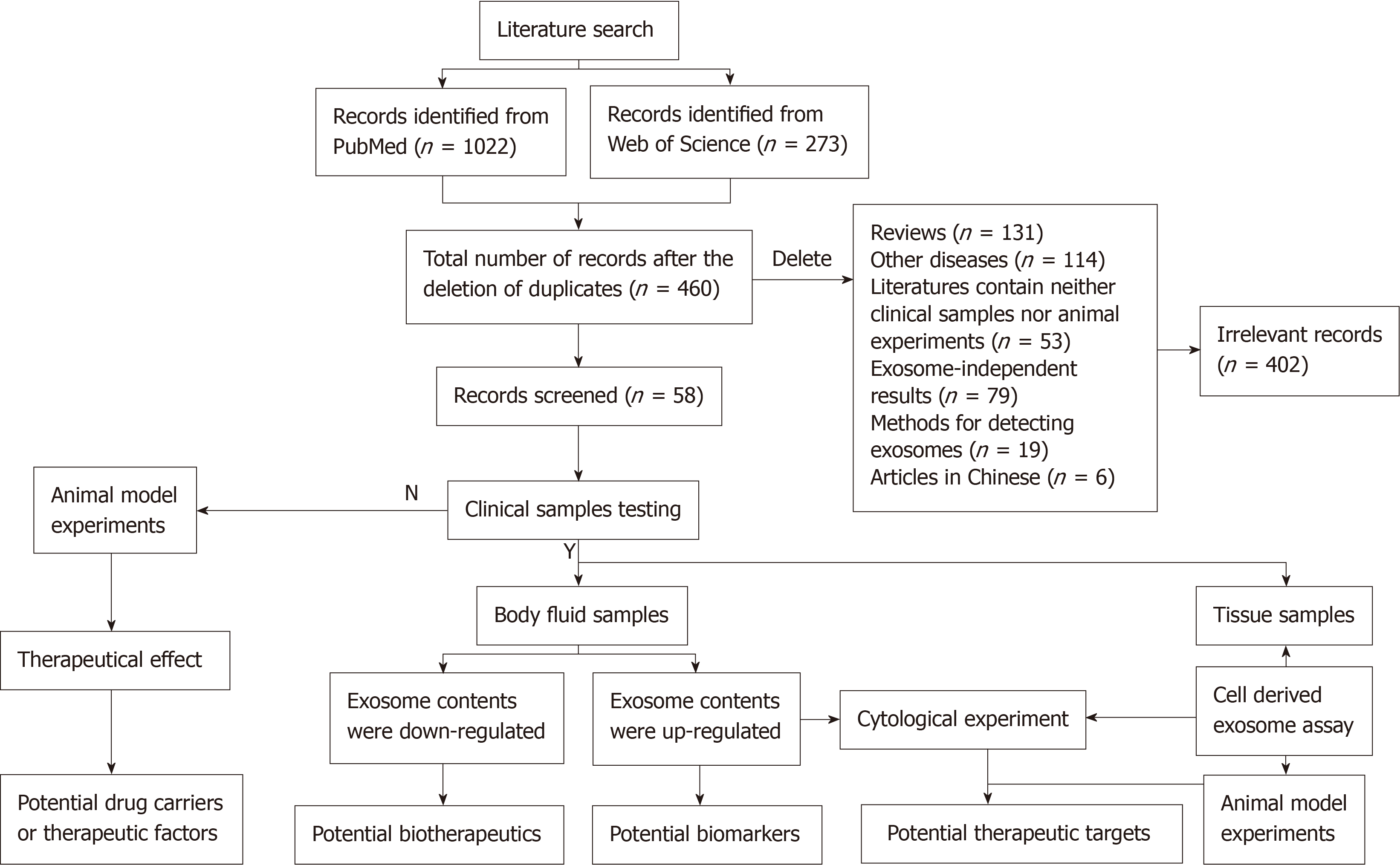
A total of 1295 studies were identified using the systematic literature search. After 835 duplicate studies were found and omitted, 460 were screened by two independent reviewers. A further 402 irrelevant studies were excluded, including review articles, other diseases, records containing neither clinical samples nor animal experiments, exosome-independent studies, methods for detecting exosome or articles in Chinese. Finally, 58 published papers were included in the study (Figure 1).
In some literature, exosomes were isolated from liver cancer patients’ blood samples. Then, the level of exosomal molecular contents was detected. Table 1[14-46] lists the potential biomarkers for liver cancer. In these studies, exosomal contents that were upregulated in blood exosomes might be potential exosomal biomarkers.
Table 2[22,35,39,47-56] includes potential biotherapeutics of exosomal contents for liver cancer. Those downregulated exosomal contents in blood liver cancer samples might serve as possible biotherapeutic drugs.
| Exosomal content | Sample | Expression | Isolation of exosomes | Content detection | Function | Ref. | Direction | |
| HCC | ||||||||
| Proteins | ||||||||
| ANGPT2 | Serum (n = 93) | Up | SBI | Immunoblotting and ELISA | Induces tumor angiogenesis | [14] | Potential targets | |
| mRNAs | ||||||||
| hnRNPH1 | Serum (n = 223) | Up | Total exosome isolation reagent (Thermo Fisher Scientific Co.) | qRT-PCR | Associated with the Child–Pugh classification, portal vein tumor emboli, lymph node metastasis, TNM stage, and OS | [15] | ||
| LDH-C4 | Serum (n = 212) | Up | exoRNeasy Serum/Plasma Midi Kit (Qiagen) | qRT-PCR | Related to treatments and recurrence prediction of HCC patients | [16] | ||
| miRNAs | ||||||||
| miR-10b-5p | Serum (n = 37) | Up | Ultracentrifugation | qRT-PCR | Respectively, associated with early diagnosis and prognosis of HCC | [17] | ||
| miR-1247-3p | Serum (n = 135) | Up | Ultracentrifugation | qRT–PCR | Shows a positive correlation with lung metastasis in HCC patients | [18] | Potential targets | |
| miR-125b | Serum (n = 218) | Up | SBI | qRT-PCR | Discriminate HCC patients with a high risk of recurrence and poor prognosis | [19] | ||
| miR-182 | Serum and ascitic fluid | Up | exoRNeasy Serum/Plasma Midi Kit (Qiagen) | qRT-PCR | Up-regulated in NASH-induced liver cirrhosis with HCC compared to NASH-induced liver cirrhosis without HCC | [20] | ||
| miR-21 | Serum (n = 79) | Up | SBI | qRT-PCR | Related to TNM stage and other prognostic factors | [21] | ||
| Plasma (n = 150) | Up | SBI | qRT-PCR | Significantly higher in patients with HCC compared with cirrhotic patients and the control group | [22] | |||
| Serum (n = 90) | Up | Total Exosome Isolation Reagent (Invitrogen) | qRT-PCR | Positively correlated with cirrhosis and tumor stage | [23] | |||
| Serum (n = 95) | Up | Ultracentrifugation | qRT–PCR | Shows a positive correlation with survival in HCC patients | [24] | Potential targets | ||
| miR-215-5p | Serum (n = 37) | Up | Ultracentrifugation | qRT-PCR | Respectively, associated with early diagnosis and prognosis of HCC | [17] | ||
| miR-224 | Serum (n = 139) | Up | Total Exosome Isolation Kit | qRT–PCR | Related to tumor size and differentiate HCC patients from healthy controls | [25] | Potential targets | |
| miR23-a/b | Serum (n = 50) | Up | Ultracentrifugation | qRT–PCR | A promising target for future treatment of HCC | [26] | Potential targets | |
| miR-301a | Serum and ascitic fluid (n = 52) | Up | exoRNeasy Serum/Plasma Midi Kit (Qiagen) | qRT-PCR | Up-regulated in NASH-induced liver cirrhosis with HCC compared to NASH-induced liver cirrhosis without HCC | [20] | ||
| miR-373 | Serum and ascitic fluid (n = 52) | Up | exoRNeasy Serum/Plasma Midi Kit (Qiagen) | qRT-PCR | Up-regulated in NASH-induced liver cirrhosis with HCC compared to NASH-induced liver cirrhosis without HCC | [20] | ||
| miR-4661-5p | Serum (n = 720) | Up | SBI | qRT-PCR | Associated with the prognosis of patients with HCC | [27] | ||
| miR-638 | Serum (n = 54) | Up | Ultracentrifugation | qRT–PCR | Promising for surveillance of HCC recurrence | [28] | Potential targets | |
| miR-665 | Serum (n = 40) | Up | SBI | qRT–PCR | Associated with tumor size, invasion, and clinical stage of HCC patients | [29] | Potential targets | |
| miR-92a-3p | Plasma (n = 42) | Up | Ultracentrifugation | qRT–PCR | Shows a positive correlation with metastasis in HCC patients | [30] | Potential targets | |
| miR-92b | Serum (n = 121) | Up | SBI | qRT-PCR | Prediction of posttransplant HCC early recurrence | [31] | ||
| miR-93 | Serum (n = 108) | Up | Total Exosome Isolation Reagent (Invitrogen) | qRT–PCR | Correlated with stage, tumor size and predict patients' survival rate of HCC patients | [32] | Potential targets | |
| miRNA-96 | Plasma (n = 150) | Up | SBI | qRT-PCR | Significantly higher in patients with HCC compared with cirrhotic patients and the control group | [22] | ||
| lncRNAs | ||||||||
| lncRNA-ATB | Serum (n = 79) | Up | SBI | qRT-PCR | Related to TNM stage and other prognostic factors | [21] | ||
| DANCR | Serum (n = 183) | Up | SBI | Digital droplet PCR (DDPCR) | Positively associated with HCV-HCC recurrence | [33] | ||
| lncRNA FAL1 | Serum (n = 60) | Up | SBI | qRT-PCR | Play an oncogenic role in HCC | [34] | Potential targets | |
| lnc-FAM72D-3 | Serum (n = 180) | Up | Ultracentrifugation | qRT-PCR | Functions as an oncogene in HCC | [35] | Potential targets | |
| lncRNA Jpx | Plasma (n = 100) | Up | SBI | qRT-PCR | Promising biomarkers for female patients with HCC | [36] | ||
| LINC00161 | Serum (n = 112) | Up | Total Exosome Isolation Kit (Invitrogen) | qRT-PCR | A significant prediction of tumor growth and metastasis in HCC | [37] | ||
| Serum (n = ?) | Up | - | qRT-PCR | Promote HCC tumorigenesis | [38] | Potential targets | ||
| lncRNA-RP11-583F2.2 | Serum (n = 120) | Up | exoRNeasy Serum/Plasma Midi Kit (Qiagen) | qRT-PCR | Up-regulated in the serum of hepatocellular carcinoma patients as compared with hepatitis C virus patients and normal good health control | [39] | ||
| ENSG00000248932.1 ENST00000440688.1 ENST00000457302.2 | Serum (n = 600) | Up | SBI | qRT-PCR | Potential fingerprints for the tumorigenesis prediction | [40] | ||
| circRNAs | ||||||||
| circ_0070396 | Plasma (n = 273) | Up | exoEasy Maxi Kit (QIAGEN) | qRT-PCR | Discriminate HCC individuals from patients with chronic hepatitis B and liver cirrhosis | [41] | ||
| circAKT3 | Serum (n = 224) | Up | SBI | qRT-PCR | Associated with HCC recurrence and mortality | [42] | ||
| circ-DB | Plasma (n = 40) | Up | Ultracentrifugation | qRT-PCR | Promote the tumor growth | [43] | Potential targets | |
| circPTGR1 | Serum (n = 129) | Up | SBI | qRT-PCR | Promote HCC progression | [44] | Potential targets | |
| circUHRF1 | Serum (n = 643) | Up | SBI | qRT-PCR | Drive resistance to anti-PD1 immunotherapy | [45] | Potential targets | |
| HB | ||||||||
| miRNAs | ||||||||
| miR-21 | Serum (n = 64) | Up | SBI | qRT-PCR | Significantly higher in patients with HB | [46] | ||
| Exosomal content | Sample | Expression | Isolation of exosomes | Content detection | Function | Ref. |
| HCC | ||||||
| miRNAs | ||||||
| miR-122 | Serum (n = 75) | Down | SBI | qRT-PCR | Reflect the liver damage and residual liver function levels | [47] |
| Plasma (n = 150) | Down | SBI | qRT-PCR | Significantly lower in patients with HCC compared with cirrhotic patients and the control group | [22] | |
| miRNA-1298 | Serum (n = 120) | Down | exoRNeasy Serum/Plasma MidiKit (Qiagen) | qRT-PCR | Down-regulated in patients of hepatocellular carcinoma compared with patients of hepatitis C virus and normal good health control | [39] |
| miR-320a | Serum (n = 209) | Down | SBI | qRT-PCR | Associated with lymph node metastasis, vein invasion, TNM stage, and survival of HCC patients | [48] |
| miR-320d | Serum (n = 150) | Down | Total Exosome Isolation Kit (Invitrogen) | qRT-PCR | Associated with clinicopathological parameters and prognosis of HCC patients | [49] |
| miR-638 | Serum (n = 147) | Down | Total Exosome Isolation Kit (Invitrogen) | qRT-PCR | Influence liver carcinogenesis | [50] |
| miR-718 | Serum (n = 59) | Down | Ultracentrifugation | qRT-PCR | Significantly different expression of HCC cases with recurrence after LT compared with those without recurrence | [51] |
| miR-744 | Serum (n = 20) | Down | Ultracentrifugation | qRT–PCR | Facilitates the propagation and drug resistance of HCC cells | [52] |
| miR-9-3p | Serum (n = ?) | Down | Ultracentrifugation | qRT-PCR | A potential therapeutic target for HCC | [53] |
| lncRNAs | ||||||
| lnc-EPC1-4 | Serum (n = 180) | Down | Ultracentrifugation | qRT-PCR | Function as a tumor suppressor gene | [35] |
| SENP3-EIF4A1 | Serum (n = 6) | Down | SBI | qRT-PCR | Block HCC progression | [54] |
| circRNAs | ||||||
| circ-0051443 | Plasma (n = 120) | Down | SBI | qRT-PCR | Suppress HCC progression | [55] |
| HB | ||||||
| miRNAs | ||||||
| miR-34s | Serum (n = 152) | Down | SBI | qRT-PCR | Significantly lower in patients with HB compared with the control group | [56] |
The expression of exosomal contents was detected in liver cancer clinical tissue samples, and cytology or animal experiments were used to identify the role of exosomal contents. Upregulated exosomal contents might enhance hepatocellular carcinoma (HCC) progression, angiogenesis, and drug resistance, while downregulated exosomal contents might attenuate angiogenesis. In Table 3[57-66], all these abnormally expressed exosomal contents may become novel therapeutic targets for liver cancer.
| Exosomal content | Sample | Expression | Content identification | Animal model (Yes/No) | Function | Ref. |
| HCC | ||||||
| Proteins | ||||||
| ENO1 | Cancer cells-exosomes, tissue (n = 94) | Up | IHC staining | Y | Promotes HCC growth, metastasis, and further patient deterioration | [57] |
| miRNAs | ||||||
| miR-125a/b | TAMs-exosomes Tissue (n = 6) | Down | qRT-PCR | N | A possible therapeutic target in HCC | [58] |
| miR-150-3p | Fibroblasts-exosomes, tissues (n = 82) | Down | qRT–PCR | N | Abrogate HCC migration and invasiveness | [59] |
| miR-32-5p | Bel/5-FU-exosomes, tissue (n = 72) | Up | qRT–PCR | Y | Induce multidrug resistance in HCC | [60] |
| miR-320a | Cancer cells-exosomes, tissue (n = 6) | Down | qRT–PCR | Y | Mediates HCC tumor progression | [61] |
| miR-3682-3p | Cancer cells-exosomes, tissue (n = 8) | Down | qRT–PCR | Y | Attenuate angiogenesis and provides novel potential targets for liver cancer therapy | [62] |
| miR-378b | Cancer cells-exosomes, tissue (n = 105) | Up | qRT–PCR | Y | Enhance HCC cell progression and angiogenesis | [63] |
| lncRNAs | ||||||
| ASMTL-AS1 | Cancer cells-exosomes, tissues (n = 70) | Up | qRT–PCR | Y | Aggravate the malignancy in residual HCC | [64] |
| PCED1B-AS1 | Cancer cells-exosomes, tissues (n = 45) | Up | qRT–PCR | Y | Induce immunosuppression in HCC | [65] |
| circRNAs | ||||||
| circRNA Cdr1as | Cancer cells-exosomes, tissues (n = 42) | Up | qRT–PCR | Y | Promote the progression of HCC by sponging miR-1270 to upregulate AFP level | [66] |
Table 4[67-69] focuses on the carrier roles of exosomes in HCC. Drug-carrying exosomes were injected into tumor-prone mice to observe the effects of the drugs. These studies indicated that exosomes could serve as drug carriers that made cancer cells sensitive to antitumor drugs or enhanced their antitumor efficacy.
Table 5[70,71] shows the self-derived exosomes from dendritic cells as potential therapeutic factors. Data showed exosomes isolated from dendritic cells could inhibit tumor growth and improve the immune response. This indicated that exosomes serve as potential therapeutic factors.
Liver cancer is a global disease with high morbidity and mortality[72]. Despite the continuous development of novel treatment options, the 5-year survival rate of liver cancer patients is still low because of the delayed diagnosis[73,74]. Scientists are still trying to find new markers for early diagnosis and individualized treatments.
Over the past decade, exosomes have received widespread attention. Many studies have found that the differential expression of exosome proteins and RNAs has diagnostic significance for various cancers. Previous studies have suggested that exosomes may serve as liquid biopsies to help diagnose malignancies such as breast, pancreatic and lung cancer, and glioblastoma[75-78]. Here, we listed exosomal contents that have been identified as possible biomarkers for liver cancer in recent years. We found multiple research reports about miR-21[21-24,46] and LINC00161[37,38]. There are five papers on exosomal miR-21. These studies indicate that expression level of miR-21 in serum exosomes of liver cancer patients is higher than that of healthy people, suggesting that it is the most likely marker for early liver cancer screening. Among the contents of liver cancer serum with downregulated exosomal expression, miR-122 has been reported most often. These studies suggest that miR-122 may be the most likely biotherapeutic drug for liver cancer[22,47].
In addition to serving as disease markers in patients’ serum, exosomes are involved in the occurrence, development and prognosis of various cancers[79]. Bai et al[80] have shown that exosomes secreted by gastric cancer cells deliver miR-135b to tumor cells and promote angiogenesis by negatively regulating intracellular forkhead box O1. This study provides a potential target for antiangiogenic therapy. Huang and his collaborators demonstrated that colon cancer cells secrete Wnt4-rich exosomes delivered to normoxic cells to activate β-catenin signaling and enhance their metastatic behavior. They found that β-catenin inhibitors ICG-001 can inhibit this metastatic behavior, which provides a new target for treating metastatic colon cancer[81]. In this paper, we listed the previous studies on the mechanism of exosomal contents involved in the development of liver cancer. Therefore, developing drugs targeting these exosomal contents may be a potential therapy for liver cancer.
As drug carriers, exosomes have the characteristics of stability in circulation, good biocompatibility, low immunogenicity, and low toxicity[82,83]. Liang et al[84] have shown that exosomes loaded with 5-fluorouracil and miR-21 inhibitors can effectively improve cancer cell drug resistance and colon cancer treatment efficiency. Zhang and his group also found that HEK293T-cell-derived exosomes deliver exogenous si-c-Met to gastric cancer cells and enhance gastric cancer cell sensitivity to cisplatin[85]. In this paper, we reviewed recent studies on the therapeutic effect of exosomes as carriers in HCC.
In addition to being carriers, some researchers have reported the therapeutic effect of exosomes. As early as 1998, Zitvogel et al[86] found that dendritic-cell-derived exosomes (DEXs) could activate tumor-specific cytotoxic T lymphocyte response and inhibit tumor growth in vivo. DEXs have been used in several clinical trials. Researchers have processed DEXs derived from melanoma patients, loaded them with melanoma antigens, and observed an enhanced antimelanoma immunity after self-inoculation[87]. Another trial indicated that DEX therapy increases natural killer cells (NKs) lytic activity in patients with non-small cell lung cancer (NSCLC)[88]. Besse’s group has conducted phase II clinical trials in NSCLC and confirmed the capacity of DEXs to boost the NK cell arm of antitumor immunity in patients with advanced NSCLC[89]. In addition to injecting DEXs, Dai and colleagues have found that the immunotherapy of colorectal cancer (CRC) with ascites-derived exosomes in combination with granulocyte–macrophage colony-stimulating factor can serve as a choice for immunotherapy of advanced CRC[90]. In liver cancer, however, there have been no such clinical trials.
Although exosomes present good application value, there are still problems with their clinical application. Firstly, the separation and purification of exosomes are complex. Secondly, the contents in exosomes are not unique. Thirdly, not all exosomes secreted by cells are suitable for use as carriers. Although there are currently small-scale clinical trials, the actual application of exosomes in the clinical diagnosis and treatment of liver cancer still needs more in-depth studies.
| Drugs | Source of exosomes | Animal model (Yes/No) | Clinical sample (Yes/No) | Functions | Ref. |
| Norcantharidin | BMSCs-exosomes | Y | N | Induce cell cycle arrest, reduced tumor cell proliferation, increased apoptosis | [67] |
| siGRP78 | BMSCs-exosomes | Y | N | Sensitize Sorafenib resistant cancer cells to Sorafenib | [68] |
| miR-214 | hCEC-exosomes | N | Y (n = 6) | Enhances the anti-tumor efficacy of oxaliplatin and sorafenib on HCC cells | [69] |
Exosomes are composed of a lipid bilayer membrane structure, which has the advantages of rich content, high stability, ability to reflect the state of disease, and cellular communication. These features make them a research hotspot for liver cancer for potential biomarkers, biotherapeutics, therapeutic targets, drug carriers, and therapeutic factors.
Liver cancer is one of the most common malignant tumors with high morbidity and mortality because of lacking early diagnosis and treatment. Exosomes have been a newly discovered cellular communication tool with high biocompatibility, low immunogenicity, and high transport efficiency. They show great potential for cancer diagnosis and therapy.
This review aimed to consolidate the evidence on exosomes as biomarkers for the diagnosis and therapeutics for liver cancer in a systematic fashion.
The main result that the authors are concerned about is discovering the great potential of exosomes in the diagnosis and treatment of liver cancer.
| Cargos | Source of exosomes | Animal model (Yes/No) | Clinical sample (Yes/No) | Functions | Ref. |
| Exosomes plus microwave ablation | DCs-exosomes | Y | N | Inhibit tumor growth and improve the immune microenvironment | [70] |
| Exosomes | DCs-exosomes | Y | N | Elicited strong antigen-specific immune responses and resulted in tumor growth retardation and prolonged survival rates in mice with ectopic | [71] |
A systematic literature search was performed using PubMed and Web of Science. The latest literature was published in June 2021.
Fifty-eight studies were included in this systematic review. Blood-derived exosomes could be biomarkers or biotherapeutics. Cell-derived exosomes, which were used to explore underlying mechanisms of differentially expressed exosome contents in clinical tissue samples, might serve as potential therapeutic targets for liver cancer. Exosomes might also serve as drug carriers or therapeutic factors.
Existing studies show that exosomes have great potential for clinical application as potential novel diagnostic and therapeutic markers of liver cancer.
This present review might be helpful as a reference for clinical research on exosomes in liver cancer.
We are grateful to Wang Ying for her skillful statistical analysis guidance.
Provenance and peer review: Invited article; Externally peer reviewed.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: de Melo FF S-Editor: Ma YJ L-Editor: Kerr C P-Editor: Yu HG
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics, 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55777] [Article Influence: 7968.1] [Reference Citation Analysis (132)] |
| 2. | Hollebecque A, Malka D, Ferté C, Ducreux M, Boige V. Systemic treatment of advanced hepatocellular carcinoma: from disillusions to new horizons. Eur J Cancer. 2015;51:327-339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 55] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 3. | Johnson PJ. Role of alpha-fetoprotein in the diagnosis and management of hepatocellular carcinoma. J Gastroenterol Hepatol. 1999;14 Suppl:S32-S36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 76] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 4. | Gupta S, Bent S, Kohlwes J. Test characteristics of alpha-fetoprotein for detecting hepatocellular carcinoma in patients with hepatitis C. A systematic review and critical analysis. Ann Intern Med. 2003;139:46-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 295] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 5. | Liu H, Li B. The functional role of exosome in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2018;144:2085-2095. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 6. | Wortzel I, Dror S, Kenific CM, Lyden D. Exosome-Mediated Metastasis: Communication from a Distance. Dev Cell. 2019;49:347-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 484] [Cited by in RCA: 898] [Article Influence: 179.6] [Reference Citation Analysis (0)] |
| 7. | Roma-Rodrigues C, Fernandes AR, Baptista PV. Exosome in tumour microenvironment: overview of the crosstalk between normal and cancer cells. Biomed Res Int. 2014;2014:179486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 188] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 8. | Henderson MC, Azorsa DO. The genomic and proteomic content of cancer cell-derived exosomes. Front Oncol. 2012;2:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 138] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 9. | Bebelman MP, Smit MJ, Pegtel DM, Baglio SR. Biogenesis and function of extracellular vesicles in cancer. Pharmacol Ther. 2018;188:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 592] [Article Influence: 84.6] [Reference Citation Analysis (1)] |
| 10. | Li W, Li C, Zhou T, Liu X, Li X, Chen D. Role of exosomal proteins in cancer diagnosis. Mol Cancer. 2017;16:145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 193] [Cited by in RCA: 320] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 11. | Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, Ratajczak MZ. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847-856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1312] [Cited by in RCA: 1246] [Article Influence: 65.6] [Reference Citation Analysis (0)] |
| 12. | Muka T, Glisic M, Milic J, Verhoog S, Bohlius J, Bramer W, Chowdhury R, Franco OH. A 24-step guide on how to design, conduct, and successfully publish a systematic review and meta-analysis in medical research. Eur J Epidemiol. 2020;35:49-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 241] [Cited by in RCA: 311] [Article Influence: 62.2] [Reference Citation Analysis (0)] |
| 13. | Bramer WM, Milic J, Mast F. Reviewing retrieved references for inclusion in systematic reviews using EndNote. J Med Libr Assoc. 2017;105:84-87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 152] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 14. | Xie JY, Wei JX, Lv LH, Han QF, Yang WB, Li GL, Wang PX, Wu SB, Duan JX, Zhuo WF, Liu PQ, Min J. Angiopoietin-2 induces angiogenesis via exosomes in human hepatocellular carcinoma. Cell Commun Signal. 2020;18:46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 80] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 15. | Xu H, Dong X, Chen Y, Wang X. Serum exosomal hnRNPH1 mRNA as a novel marker for hepatocellular carcinoma. Clin Chem Lab Med. 2018;56:479-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 96] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 16. | Cui Z, Li Y, Gao Y, Kong L, Lin Y, Chen Y. Cancer-testis antigen lactate dehydrogenase C4 in hepatocellular carcinoma: a promising biomarker for early diagnosis, efficacy evaluation and prognosis prediction. Aging (Albany NY). 2020;12:19455-19467. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 17. | Cho HJ, Eun JW, Baek GO, Seo CW, Ahn HR, Kim SS, Cho SW, Cheong JY. Serum Exosomal MicroRNA, miR-10b-5p, as a Potential Diagnostic Biomarker for Early-Stage Hepatocellular Carcinoma. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 97] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 18. | Fang T, Lv H, Lv G, Li T, Wang C, Han Q, Yu L, Su B, Guo L, Huang S, Cao D, Tang L, Tang S, Wu M, Yang W, Wang H. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun. 2018;9:191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 407] [Cited by in RCA: 747] [Article Influence: 106.7] [Reference Citation Analysis (0)] |
| 19. | Liu W, Hu J, Zhou K, Chen F, Wang Z, Liao B, Dai Z, Cao Y, Fan J, Zhou J. Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma. Onco Targets Ther. 2017;10:3843-3851. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 124] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 20. | Muhammad Yusuf AN, Raja Ali RA, Muhammad Nawawi KN, Mokhtar NM. Potential biomarkers in NASH-induced liver cirrhosis with hepatocellular carcinoma: A preliminary work on roles of exosomal miR-182, miR-301a, and miR-373. Malays J Pathol. 2020;42:377-384. [PubMed] |
| 21. | Lee YR, Kim G, Tak WY, Jang SY, Kweon YO, Park JG, Lee HW, Han YS, Chun JM, Park SY, Hur K. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int J Cancer. 2019;144:1444-1452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 202] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 22. | Wang S, Yang Y, Sun L, Qiao G, Song Y, Liu B. Exosomal MicroRNAs as Liquid Biopsy Biomarkers in Hepatocellular Carcinoma. Onco Targets Ther. 2020;13:2021-2030. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 23. | Wang H, Hou L, Li A, Duan Y, Gao H, Song X. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. Biomed Res Int. 2014;2014:864894. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 180] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 24. | Zhou Y, Ren H, Dai B, Li J, Shang L, Huang J, Shi X. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J Exp Clin Cancer Res. 2018;37:324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 329] [Cited by in RCA: 353] [Article Influence: 50.4] [Reference Citation Analysis (0)] |
| 25. | Cui Y, Xu HF, Liu MY, Xu YJ, He JC, Zhou Y, Cang SD. Mechanism of exosomal microRNA-224 in development of hepatocellular carcinoma and its diagnostic and prognostic value. World J Gastroenterol. 2019;25:1890-1898. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 60] [Cited by in RCA: 80] [Article Influence: 13.3] [Reference Citation Analysis (1)] |
| 26. | Liu Y, Tan J, Ou S, Chen J, Chen L. Adipose-derived exosomes deliver miR-23a/b to regulate tumor growth in hepatocellular cancer by targeting the VHL/HIF axis. J Physiol Biochem. 2019;75:391-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 79] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 27. | Cho HJ, Baek GO, Seo CW, Ahn HR, Sung S, Son JA, Kim SS, Cho SW, Jang JW, Nam SW, Cheong JY, Eun JW. Exosomal microRNA-4661-5p-based serum panel as a potential diagnostic biomarker for early-stage hepatocellular carcinoma. Cancer Med. 2020;9:5459-5472. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 28. | Yokota Y, Noda T, Okumura Y, Kobayashi S, Iwagami Y, Yamada D, Tomimaru Y, Akita H, Gotoh K, Takeda Y, Tanemura M, Murakami T, Umeshita K, Doki Y, Eguchi H. Serum exosomal miR-638 is a prognostic marker of HCC via downregulation of VE-cadherin and ZO-1 of endothelial cells. Cancer Sci. 2021;112:1275-1288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 91] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 29. | Qu Z, Wu J, Ji A, Qiang G, Jiang Y, Jiang C, Ding Y. Exosomal miR-665 as a novel minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis. Oncotarget. 2017;8:80666-80678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 104] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 30. | Yang B, Feng X, Liu H, Tong R, Wu J, Li C, Yu H, Chen Y, Cheng Q, Chen J, Cai X, Wu W, Lu Y, Hu J, Liang K, Lv Z, Zheng S. High-metastatic cancer cells derived exosomal miR92a-3p promotes epithelial-mesenchymal transition and metastasis of low-metastatic cancer cells by regulating PTEN/Akt pathway in hepatocellular carcinoma. Oncogene. 2020;39:6529-6543. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 143] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 31. | Nakano T, Chen IH, Wang CC, Chen PJ, Tseng HP, Huang KT, Hu TH, Li LC, Goto S, Cheng YF, Lin CC, Chen CL. Circulating exosomal miR-92b: Its role for cancer immunoediting and clinical value for prediction of posttransplant hepatocellular carcinoma recurrence. Am J Transplant. 2019;19:3250-3262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 90] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 32. | Xue X, Wang X, Zhao Y, Hu R, Qin L. Exosomal miR-93 promotes proliferation and invasion in hepatocellular carcinoma by directly inhibiting TIMP2/TP53INP1/CDKN1A. Biochem Biophys Res Commun. 2018;502:515-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 95] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 33. | Wang SC, Li CY, Chang WT, Cheng WC, Yen CH, Tu WY, Lin ZY, Lin CC, Yeh ML, Huang CF, Huang JF, Dai CY, Chuang WL, Chen YL, Yu ML. Exosome-derived differentiation antagonizing non-protein coding RNA with risk of hepatitis C virus-related hepatocellular carcinoma recurrence. Liver Int. 2021;41:956-968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 34. | Li B, Mao R, Liu C, Zhang W, Tang Y, Guo Z. LncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells. Life Sci. 2018;197:122-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 176] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 35. | Yao Z, Jia C, Tai Y, Liang H, Zhong Z, Xiong Z, Deng M, Zhang Q. Serum exosomal long noncoding RNAs lnc-FAM72D-3 and lnc-EPC1-4 as diagnostic biomarkers for hepatocellular carcinoma. Aging (Albany NY). 2020;12:11843-11863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 36. | Ma X, Yuan T, Yang C, Wang Z, Zang Y, Wu L, Zhuang L. X-inactive-specific transcript of peripheral blood cells is regulated by exosomal Jpx and acts as a biomarker for female patients with hepatocellular carcinoma. Ther Adv Med Oncol. 2017;9:665-677. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 37. | Sun L, Su Y, Liu X, Xu M, Chen X, Zhu Y, Guo Z, Bai T, Dong L, Wei C, Cai X, He B, Pan Y, Sun H, Wang S. Serum and exosome long non coding RNAs as potential biomarkers for hepatocellular carcinoma. J Cancer. 2018;9:2631-2639. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 105] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 38. | You LN, Tai QW, Xu L, Hao Y, Guo WJ, Zhang Q, Tong Q, Zhang H, Huang WK. Exosomal LINC00161 promotes angiogenesis and metastasis via regulating miR-590-3p/ROCK axis in hepatocellular carcinoma. Cancer Gene Ther. 2021;28:719-736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 39. | Matboli M, Labib ME, Nasser HE, El-Tawdi AHF, Habib EK, Ali-Labib R. Exosomal miR-1298 and lncRNA-RP11-583F2.2 Expression in Hepato-cellular Carcinoma. Curr Genomics. 2020;21:46-55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 40. | Lu Y, Duan Y, Xu Q, Zhang L, Chen W, Qu Z, Wu B, Liu W, Shi L, Wu D, Yang Y, Sun D, Chen X. Circulating exosome-derived bona fide long non-coding RNAs predicting the occurrence and metastasis of hepatocellular carcinoma. J Cell Mol Med. 2020;24:1311-1318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 51] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 41. | Lyu L, Yang W, Yao J, Wang H, Zhu J, Jin A, Liu T, Wang B, Zhou J, Fan J, Yang X, Guo W. The diagnostic value of plasma exosomal hsa_circ_0070396 for hepatocellular carcinoma. Biomark Med. 2021;15:359-371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 42. | Luo Y, Liu F, Gui R. High expression of circulating exosomal circAKT3 is associated with higher recurrence in HCC patients undergoing surgical treatment. Surg Oncol. 2020;33:276-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 43. | Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, Fan Q, Li J, Ning T, Tian F, Li H, Sun W, Ying G, Ba Y. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38:2844-2859. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 227] [Cited by in RCA: 272] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 44. | Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo J, Zhang Y, Li H, Zhang Q, Yang Y, Chen G. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine. 2019;40:432-445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 183] [Cited by in RCA: 194] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 45. | Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, Cai JB, Ke AW. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer. 2020;19:110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 391] [Cited by in RCA: 399] [Article Influence: 79.8] [Reference Citation Analysis (0)] |
| 46. | Liu W, Chen S, Liu B. Diagnostic and prognostic values of serum exosomal microRNA-21 in children with hepatoblastoma: a Chinese population-based study. Pediatr Surg Int. 2016;32:1059-1065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 47. | Suehiro T, Miyaaki H, Kanda Y, Shibata H, Honda T, Ozawa E, Miuma S, Taura N, Nakao K. Serum exosomal microRNA-122 and microRNA-21 as predictive biomarkers in transarterial chemoembolization-treated hepatocellular carcinoma patients. Oncol Lett. 2018;16:3267-3273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 48. | Hao X, Xin R, Dong W. Decreased serum exosomal miR-320a expression is an unfavorable prognostic factor in patients with hepatocellular carcinoma. J Int Med Res. 2020;48:300060519896144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 49. | Li W, Ding X, Wang S, Xu L, Yin T, Han S, Geng J, Sun W. Downregulation of serum exosomal miR-320d predicts poor prognosis in hepatocellular carcinoma. J Clin Lab Anal. 2020;34:e23239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 50. | Shi M, Jiang Y, Yang L, Yan S, Wang YG, Lu XJ. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J Cell Biochem. 2018;119:4711-4716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 136] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 51. | Sugimachi K, Matsumura T, Hirata H, Uchi R, Ueda M, Ueo H, Shinden Y, Iguchi T, Eguchi H, Shirabe K, Ochiya T, Maehara Y, Mimori K. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br J Cancer. 2015;112:532-538. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 212] [Cited by in RCA: 242] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 52. | Wang G, Zhao W, Wang H, Qiu G, Jiang Z, Wei G, Li X. Exosomal MiR-744 Inhibits Proliferation and Sorafenib Chemoresistance in Hepatocellular Carcinoma by Targeting PAX2. Med Sci Monit. 2019;25:7209-7217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 53. | Tang J, Li Y, Liu K, Zhu Q, Yang WH, Xiong LK, Guo DL. Exosomal miR-9-3p suppresses HBGF-5 expression and is a functional biomarker in hepatocellular carcinoma. Minerva Med. 2018;109:15-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 54. | Wang J, Pu J, Zhang Y, Yao T, Luo Z, Li W, Xu G, Liu J, Wei W, Deng Y. Exosome-transmitted long non-coding RNA SENP3-EIF4A1 suppresses the progression of hepatocellular carcinoma. Aging (Albany NY). 2020;12:11550-11567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 55. | Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L, Chen L, Liu Q, He P, Ye Y. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020;475:119-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 212] [Article Influence: 42.4] [Reference Citation Analysis (0)] |
| 56. | Jiao C, Jiao X, Zhu A, Ge J, Xu X. Exosomal miR-34s panel as potential novel diagnostic and prognostic biomarker in patients with hepatoblastoma. J Pediatr Surg. 2017;52:618-624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 57. | Jiang K, Dong C, Yin Z, Li R, Mao J, Wang C, Zhang J, Gao Z, Liang R, Wang Q, Wang L. Exosome-derived ENO1 regulates integrin α6β4 expression and promotes hepatocellular carcinoma growth and metastasis. Cell Death Dis. 2020;11:972. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 58. | Wang Y, Wang B, Xiao S, Li Y, Chen Q. miR-125a/b inhibits tumor-associated macrophages mediated in cancer stem cells of hepatocellular carcinoma by targeting CD90. J Cell Biochem. 2019;120:3046-3055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 90] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 59. | Yugawa K, Yoshizumi T, Mano Y, Itoh S, Harada N, Ikegami T, Kohashi K, Oda Y, Mori M. Cancer-associated fibroblasts promote hepatocellular carcinoma progression through downregulation of exosomal miR-150-3p. Eur J Surg Oncol. 2021;47:384-393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 77] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 60. | Fu X, Liu M, Qu S, Ma J, Zhang Y, Shi T, Wen H, Yang Y, Wang S, Wang J, Nan K, Yao Y, Tian T. Exosomal microRNA-32-5p induces multidrug resistance in hepatocellular carcinoma via the PI3K/Akt pathway. J Exp Clin Cancer Res. 2018;37:52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 193] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 61. | Zhang Z, Li X, Sun W, Yue S, Yang J, Li J, Ma B, Wang J, Yang X, Pu M, Ruan B, Zhao G, Huang Q, Wang L, Tao K, Dou K. Loss of exosomal miR-320a from cancer-associated fibroblasts contributes to HCC proliferation and metastasis. Cancer Lett. 2017;397:33-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 240] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 62. | Dong SS, Dong DD, Yang ZF, Zhu GQ, Gao DM, Chen J, Zhao Y, Liu BB. Exosomal miR-3682-3p Suppresses Angiogenesis by Targeting ANGPT1 via the RAS-MEK1/2-ERK1/2 Pathway in Hepatocellular Carcinoma. Front Cell Dev Biol. 2021;9:633358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 63. | Chen W, Huang L, Liang J, Ye Y, He S, Niu J. Hepatocellular carcinoma cells-derived exosomal microRNA-378b enhances hepatocellular carcinoma angiogenesis. Life Sci. 2021;273:119184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 64. | Ma D, Gao X, Liu Z, Lu X, Ju H, Zhang N. Exosome-transferred long non-coding RNA ASMTL-AS1 contributes to malignant phenotypes in residual hepatocellular carcinoma after insufficient radiofrequency ablation. Cell Prolif. 2020;53:e12795. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 65. | Fan F, Chen K, Lu X, Li A, Liu C, Wu B. Dual targeting of PD-L1 and PD-L2 by PCED1B-AS1 via sponging hsa-miR-194-5p induces immunosuppression in hepatocellular carcinoma. Hepatol Int. 2021;15:444-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 75] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 66. | Su Y, Lv X, Yin W, Zhou L, Hu Y, Zhou A, Qi F. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging (Albany NY). 2019;11:8183-8203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 170] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 67. | Liang L, Zhao L, Wang Y. Treatment for Hepatocellular Carcinoma Is Enhanced When Norcantharidin Is Encapsulated in Exosomes Derived from Bone Marrow Mesenchymal Stem Cells. Mol Pharm. 2021;18:1003-1013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 64] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 68. | Li H, Yang C, Shi Y, Zhao L. Exosomes derived from siRNA against GRP78 modified bone-marrow-derived mesenchymal stem cells suppress Sorafenib resistance in hepatocellular carcinoma. J Nanobiotechnology. 2018;16:103. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 112] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 69. | Semaan L, Zeng Q, Lu Y, Zhang Y, Zreik MM, Chamseddine MB, Chopp M, Zhang ZG, Moonka D. MicroRNA-214 enriched exosomes from human cerebral endothelial cells (hCEC) sensitize hepatocellular carcinoma to anti-cancer drugs. Oncotarget. 2021;12:185-198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 70. | Zhong X, Zhou Y, Cao Y, Ding J, Wang P, Luo Y, Liu H, Zhu Z, Jing X. Enhanced antitumor efficacy through microwave ablation combined with a dendritic cell-derived exosome vaccine in hepatocellular carcinoma. Int J Hyperthermia. 2020;37:1210-1218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 71. | Lu Z, Zuo B, Jing R, Gao X, Rao Q, Liu Z, Qi H, Guo H, Yin H. Dendritic cell-derived exosomes elicit tumor regression in autochthonous hepatocellular carcinoma mouse models. J Hepatol. 2017;67:739-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 307] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 72. | Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301-1314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2800] [Cited by in RCA: 4092] [Article Influence: 584.6] [Reference Citation Analysis (6)] |
| 73. | Anwanwan D, Singh SK, Singh S, Saikam V, Singh R. Challenges in liver cancer and possible treatment approaches. Biochim Biophys Acta Rev Cancer. 2020;1873:188314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 762] [Cited by in RCA: 896] [Article Influence: 179.2] [Reference Citation Analysis (2)] |
| 74. | De Stefano F, Chacon E, Turcios L, Marti F, Gedaly R. Novel biomarkers in hepatocellular carcinoma. Dig Liver Dis. 2018;50:1115-1123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 99] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 75. | Corcoran C, Friel AM, Duffy MJ, Crown J, O'Driscoll L. Intracellular and extracellular microRNAs in breast cancer. Clin Chem. 2011;57:18-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 170] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 76. | Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470-1476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4100] [Cited by in RCA: 3940] [Article Influence: 231.8] [Reference Citation Analysis (0)] |
| 77. | Cui S, Cheng Z, Qin W, Jiang L. Exosomes as a liquid biopsy for lung cancer. Lung Cancer. 2018;116:46-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 126] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 78. | Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D, Kalluri R. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2108] [Cited by in RCA: 2196] [Article Influence: 219.6] [Reference Citation Analysis (0)] |
| 79. | Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208-1215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 944] [Cited by in RCA: 1415] [Article Influence: 157.2] [Reference Citation Analysis (0)] |
| 80. | Bai M, Li J, Yang H, Zhang H, Zhou Z, Deng T, Zhu K, Ning T, Fan Q, Ying G, Ba Y. miR-135b Delivered by Gastric Tumor Exosomes Inhibits FOXO1 Expression in Endothelial Cells and Promotes Angiogenesis. Mol Ther. 2019;27:1772-1783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 81. | Huang Z, Yang M, Li Y, Yang F, Feng Y. Exosomes Derived from Hypoxic Colorectal Cancer Cells Transfer Wnt4 to Normoxic Cells to Elicit a Prometastatic Phenotype. Int J Biol Sci. 2018;14:2094-2102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 76] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 82. | Das CK, Jena BC, Banerjee I, Das S, Parekh A, Bhutia SK, Mandal M. Exosome as a Novel Shuttle for Delivery of Therapeutics across Biological Barriers. Mol Pharm. 2019;16:24-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 176] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 83. | Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nat Nanotechnol. 2007;2:751-760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6522] [Cited by in RCA: 6047] [Article Influence: 335.9] [Reference Citation Analysis (0)] |
| 84. | Liang G, Zhu Y, Ali DJ, Tian T, Xu H, Si K, Sun B, Chen B, Xiao Z. Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J Nanobiotechnology. 2020;18:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 182] [Cited by in RCA: 433] [Article Influence: 86.6] [Reference Citation Analysis (0)] |
| 85. | Zhang Q, Zhang H, Ning T, Liu D, Deng T, Liu R, Bai M, Zhu K, Li J, Fan Q, Ying G, Ba Y. Exosome-Delivered c-Met siRNA Could Reverse Chemoresistance to Cisplatin in Gastric Cancer. Int J Nanomedicine. 2020;15:2323-2335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 86. | Zitvogel L, Regnault A, Lozier A, Wolfers J, Flament C, Tenza D, Ricciardi-Castagnoli P, Raposo G, Amigorena S. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat Med. 1998;4:594-600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1514] [Cited by in RCA: 1697] [Article Influence: 62.9] [Reference Citation Analysis (0)] |
| 87. | Escudier B, Dorval T, Chaput N, André F, Caby MP, Novault S, Flament C, Leboulaire C, Borg C, Amigorena S, Boccaccio C, Bonnerot C, Dhellin O, Movassagh M, Piperno S, Robert C, Serra V, Valente N, Le Pecq JB, Spatz A, Lantz O, Tursz T, Angevin E, Zitvogel L. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: results of thefirst phase I clinical trial. J Transl Med. 2005;3:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 763] [Cited by in RCA: 1006] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 88. | Morse MA, Garst J, Osada T, Khan S, Hobeika A, Clay TM, Valente N, Shreeniwas R, Sutton MA, Delcayre A, Hsu DH, Le Pecq JB, Lyerly HK. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J Transl Med. 2005;3:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 678] [Cited by in RCA: 882] [Article Influence: 44.1] [Reference Citation Analysis (0)] |
| 89. | Besse B, Charrier M, Lapierre V, Dansin E, Lantz O, Planchard D, Le Chevalier T, Livartoski A, Barlesi F, Laplanche A, Ploix S, Vimond N, Peguillet I, Théry C, Lacroix L, Zoernig I, Dhodapkar K, Dhodapkar M, Viaud S, Soria JC, Reiners KS, Pogge von Strandmann E, Vély F, Rusakiewicz S, Eggermont A, Pitt JM, Zitvogel L, Chaput N. Dendritic cell-derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. Oncoimmunology. 2016;5:e1071008. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 387] [Cited by in RCA: 602] [Article Influence: 60.2] [Reference Citation Analysis (0)] |
| 90. | Dai S, Wei D, Wu Z, Zhou X, Wei X, Huang H, Li G. Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol Ther. 2008;16:782-790. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 529] [Cited by in RCA: 639] [Article Influence: 37.6] [Reference Citation Analysis (0)] |