Published online Aug 15, 2021. doi: 10.4251/wjgo.v13.i8.943
Peer-review started: April 8, 2021
First decision: May 24, 2021
Revised: May 27, 2021
Accepted: July 9, 2021
Article in press: July 9, 2021
Published online: August 15, 2021
Processing time: 128 Days and 2.5 Hours
Esophageal cancer (ESCA) is a heterogeneous cancer with variable outcomes that are challenging to predict. MicroRNA (miR)-1269a is a newly discovered non-coding RNA that shows promising prognostic prediction in other cancers, but its clinical value in ESCA remains unclear.
To explore the relationship between miR-1269a and its clinical value and to develop a nomogram to succinctly display this relationship.
We analyzed the expression of miR-1269a in 125 ESCA tissue samples with complete clinical data and 52 normal tissue samples. We determined the prog
The expression of miR-1269a was significantly higher in ESCA patients than healthy controls. Patients with high expression of miR-1269a showed poor prog
miR-1269a can be used as a potential indicator for the prognosis of ESCA patients. We developed an easy-to-use nomogram with excellent ESCA prognostic predic
Core Tip: MicroRNA-1269a expression levels, along with cancer stage and age, were shown to have significant predictive capacity for overall survival and cancer-specific survival in esophageal cancer. Using these results, we developed an easy-to-use nomo
- Citation: Yu Y, Ren KM. Development of a prognostic prediction model based on microRNA-1269a in esophageal cancer. World J Gastrointest Oncol 2021; 13(8): 943-958
- URL: https://www.wjgnet.com/1948-5204/full/v13/i8/943.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v13.i8.943
Esophageal cancer (ESCA) is one of the common gastrointestinal cancers in China, ranking eighth in terms of morbidity and sixth with regard to mortality among all cancers[1]. According to cancer statistics, China has 477900 new cases of ESCA and 375000 deaths each year[2]. The high mortality rate of ESCA brings a heavy burden on patients and health care system. Esophagogastroduodenoscopy and endoscopic ultrasound as the main assessment methods of esophageal lesions have made remar
Non-coding RNAs, which do not encode proteins, have attracted interest in cancer. Long non-coding RNA and microRNAs (miR) have aroused much attention because they regulate protein expression at the post transcriptional level[10-12]. MicroRNAs are short-chain non-coding, highly conserved RNAs approximately 21–25 nt in length. They play a powerful role in regulating various cellular activities, including cell growth, development, proliferation and apoptosis[13,14]. Accumulating evidence suggests that microRNAs not only act as regulators in cancer progression but also have great potential as biomarkers because they are stable, easily detected and highly tissue-specific[15,16]. Among them, miR-1269a has been found to be involved in multiple types of cancer.
Studies have revealed that late-stage colorectal cancers (CRCs) have higher miR-1269a expression levels than early-stage CRCs, and high miR-1269a expression is associated with relapse and metastasis in stage II CRC patients[17]. The high miR-1269a levels of low-grade glioma are significantly correlated with poorer OS[18]. Moreover, miR-1269a from serum exosomes can be detected as a diagnostic biomarker to differ patients with lung cancer from healthy subjects, with a diagnostic efficacy of 0.878[19]. Using The Cancer Genome Atlas databases, Jang et al[20] established a risk score model for assessing the OS and recurrence of ESCA with three miRNAs (miR-223, miR-1269a and miR-886) whose expression is significantly associated with OS and recurrence-free survival. Furthermore, this model is an independent risk prediction for OS and cancer recurrence in multivariable analysis[20]. However, the role of miR-1269a in ESCA has never been investigated. Therefore, there is a need to clarify the correlation between miR-1269a expression and prognosis in ESCA.
Against this background, we determined miR-1269a expression in ESCA, evaluated its prognostic value in OS and cancer-specific survival (CSS), and assessed the association between miR-1269a levels and clinical variables. Univariate and multi
Generally, we determined the expression value and prognostic roles of miR-1269a and developed an easy-to-use nomogram for prognostic prediction, which can be used as promising prognostic markers for ESCA treatment.
We recruited 125 patients diagnosed with ESCA and treated in the Shengjing Hospital of China Medical University between January 2012 and January 2015. They were en
Total RNA from frozen tissue was extracted using RNAiso Plus reagent (TaKaRa, Japan) and was soluble in 10 μL RNase-free water. We determined the concentration and quality of total RNA in each samples using NanoPhotometer 50 (Thermo Fisher Scientific, USA). For reverse transcription, we synthesize cDNA with 1.0 μg total RNA according to the manufacturer’s instructions of miRNA-specific stem-loop RT primer and the PrimeScript RT Reagent Kit (TaKaRa). Briefly, the condition of 20 μL reactions were shown in the following order: 15 min at 42 ℃, 5 s at 85 ℃, and cooled to 4 ℃. The resulting cDNA were stored at -20 ℃ for next assays. Then, we amplified miR-1269a expression through quantitative real-time polymerase chain reaction (qRT-PCR). The amplification system included 2 μL cDNA, 1 μL each of forward and reverse primers, 10 μL SYBR® Premix Ex TaqII (2×), and 0.4 µL ROX Reference DyeII (50×); ddH2O was added to make the volume 20 μL in total (TaKaRa). The ABI 7500 fast RT-PCR system (Thermo Fisher Scientific, USA) was applied to run the reactions with the following amplification conditions. In briefly, the initial denaturation process started at 95 ℃ for 30 s, followed by 40 cycles of 95 ℃ for 5 s and 60 ℃ for 30 s. A melt curve was auto
Complete clinicopathological data were extracted for each patient, including age at diagnosis, gender, smoking history, alcohol consumption, tumor location, histologic grade, tumor stage, lymph stage, metastatic stage, and AJCC stage. The primary endpoints of our research are OS and CSS. OS refers to the period from the date of diagnosis until the date of death. CSS is defined as death from the diagnosis until the date of death for ESCA and has a much greater relevance to cancer biology and therapeutic impact. All patients were followed-up mainly by outpatient visits or telephone as well as re-examination at 3, 6, 9, 12 mo in the 1st year, every 4 mo from the 2nd to 3rd years, every 6 mo from the 4th to the last day of follow-up.
To detect the role of miR-1269a in ESCA, we assessed its expression and clinical correlation statistically and evaluated its prognostic value through Kaplan-Meier analysis for OS and CSS. We constructed a receiver operating characteristic (ROC) curves to evaluate the diagnostic value of miR-1269a in ESCA. Area under curve (AUC) was applied to determine its diagnostic performance. The closer the AUC was to 1.0, the better was the diagnostic performance of miR-1269a. Additionally, we estimated independent risk factors for OS and CSS using univariate and multivariate Cox analysis, respectively and compared the diagnostic performance of miR-1269a with clinical variables including age, histologic grade, and AJCC stage both in OS and CSS. Furthermore, a nomogram was established to succinctly display the predictive relationships among age, AJCC stage, and miR-1269a expression both in OS and CSS. Kaplan-Meier analysis was applied to assess the prognostic ability of our nomogram for both OS and CSS. ROC analysis was performed to evaluate the 3- and 5-year diagnostic ability of our nomogram for both OS and CSS.
Target genes of miR-1269a were predicted using three databases including miRTar
In this study, GraphPad 7 software and R software (version 3.6.1) were applied to analyze and draw figures. Briefly, we analyzed the expression level of miR-1269a through a two-tailed Student t-test, and expressed the results by mean ± SD. The χ2 test was performed to detect the connection between clinicopathological features and miR-1269a expression levels. We calculated the statistic difference between groups through the Student t-test and compared the statistic difference among multiple using one-way ANOVA. A Kaplan-Meier test and a log-rank T-test were applied to calculate the statistical significance of OS and CSS. We used univariate and multivariate Cox analyses to test the independent risk factors of prognosis through R software with “Survival” packages[27]. We constructed a nomogram using R software with “Sur
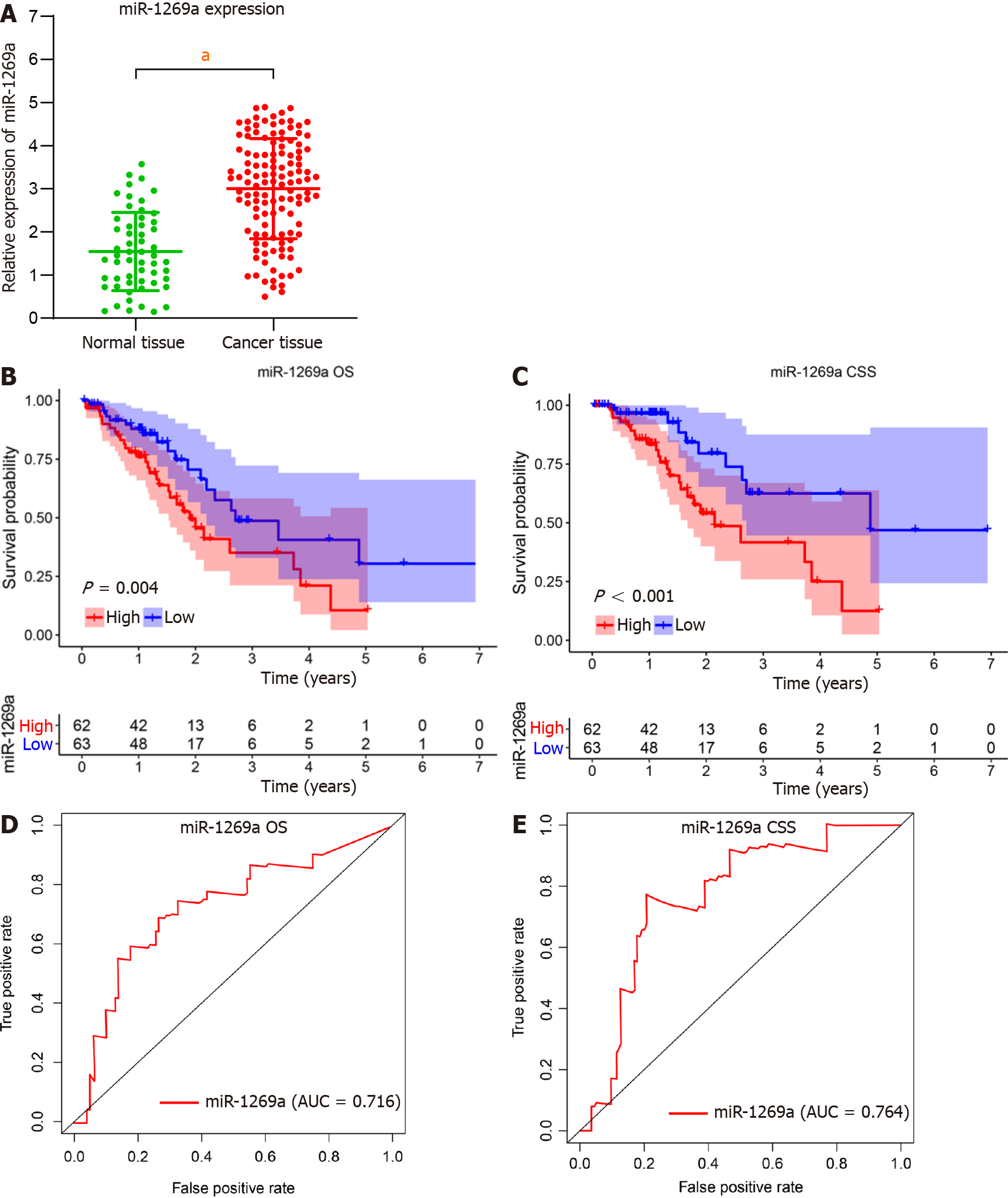
A total of 125 ESCA patients and 52 healthy subjects were enrolled in our research. There is no statistical significance between two groups including age, gender, smoking history, and alcohol consumption (Table 1). As shown in Figure 1, miR-1269a expre
| Characteristics | Normal group, n = 52 | ESCA group, n = 125 | P value |
| Age in yr | 58.7 ± 4.8 | 60 ± 5.4 | 0.857 |
| Gender | 0.962 | ||
| Female | 21 (40.4) | 50 (53.6) | |
| Male | 31 (59.6) | 75 (46.4) | |
| Smoking history | 0.383 | ||
| Yes | 32 (61.5) | 68 (54.4) | |
| No | 20 (38.5) | 57 (45.6) | |
| Alcohol consumption | 0.567 | ||
| Yes | 18 (34.6) | 49 (39.2) | |
| No | 34 (65.4) | 76 (60.8) | |
| Location | |||
| Upper area | 16 (12.8) | ||
| Middle area | 41 (32.8) | ||
| Lower area | 68 (54.4) | ||
| Histologic grade | |||
| G 1 | 38 (30.4) | ||
| G 2 + G 3 | 87 (69.6) | ||
| AJCC stage | |||
| Stage I + II | 74 (59.2) | ||
| Stage III + IV | 51 (40.8) | ||
| Tumor stage | |||
| T1 + T2 | 52 (41.6) | ||
| T3 + T4 | 73 (58.4) | ||
| Lymph stage | |||
| N0 | 59 (48.8) | ||
| N1 + N2 + N3 | 66 (51.2) | ||
| Metastatic stage | |||
| M0 | 119 (95.2) | ||
| M1 | 6 (4.8) |
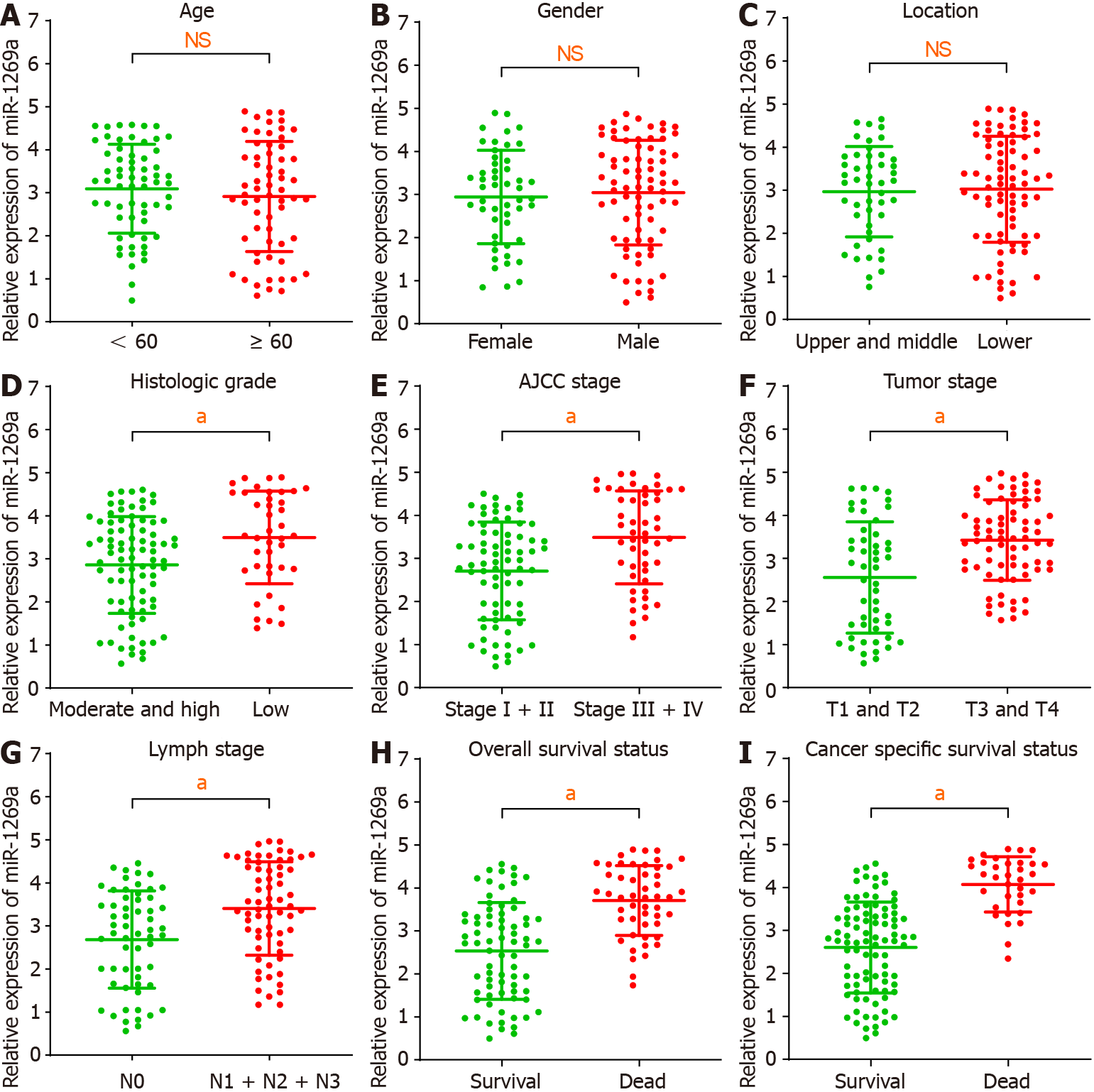
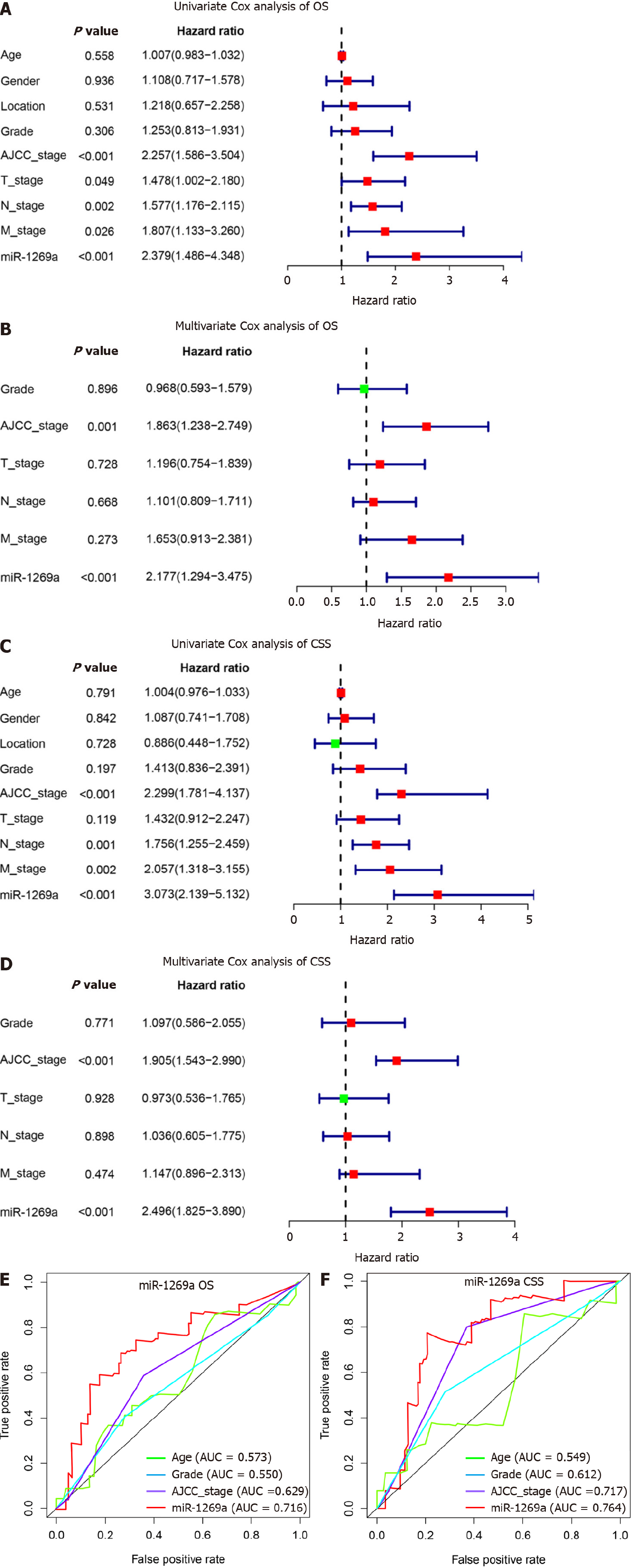
We investigated the relationship between miR-1269a expression and clinicopathological data (Table 2). As depicted in Figure 2, there are no statistical differences with age, gender, or tumor location (Figure 2A-C). High miR-1269a expression levels were significantly associated with lower histologic grade, higher tumor stage, positive lymph stage, and higher AJCC stage (Figure 2D-G). Notably, there was high miR-1269a expression levels in patients with poorer prognosis (Figure 2H and I), which suggests a close correlation between miR-1269a expression and OS and CSS. Therefore, univariate and multivariate Cox analyses were applied to detect the relationship between miR-1269a expression and OS and CSS. As displayed in Figure 3A and B, high expression of miR-1269a showed a significantly lower OS rate than those with low expression by univariate analysis [hazard ratio (HR)[30] in 2.379; P < 0.001] and multivariate analysis (HR = 2.177; P < 0.001) and a significantly lower CSS rate by univariate analysis (HR= 3.073; P < 0.001) and multivariate analysis (HR = 2.496; P < 0.001). Furthermore, we performed univariate and multivariate Cox analysis to inves
| Characteristics | High expression, n = 62 | Low expression, n = 63 | P value |
| Age in yr | 0.531 | ||
| < 60 | 33 (53.2) | 30 (47.6) | |
| ≥ 60 | 29 (46.7) | 33 (52.3) | |
| Gender | 0.77 | ||
| Female | 24 (38.7) | 26 (41.3) | |
| Male | 38 (61.2) | 37 (58.7) | |
| Location | 0.8 | ||
| Upper + middle area | 24 (38.7) | 23 (36.5) | |
| Lower area | 38 (61.2) | 40 (63.5) | |
| Histologic grade | 0.003 | ||
| G 1 | 27 (38.7) | 12 (38.7) | |
| G 2 + G 3 | 35 (38.7) | 51 (38.7) | |
| AJCC stage | 0.001 | ||
| Stage I + II | 21 (33.9) | 53 (84.1) | |
| Stage III + IV | 41 (66.1) | 10 (15.9) | |
| Tumor stage | 0.004 | ||
| T1 + T2 | 18 (29.0) | 34 (54.0) | |
| T3 + T4 | 44 (70.0) | 29 (46.0) | |
| Lymph stage | 0.009 | ||
| N0 | 22 (35.5) | 37 (58.7) | |
| N1 + N2 + N3 | 40 (64.5) | 26 (41.3) | |
| Metastatic stage | 0.011 | ||
| M0 | 56 (0.0) | 63 (100.0) | |
| M1 | 6 (100.0) | 0 (0.0) |
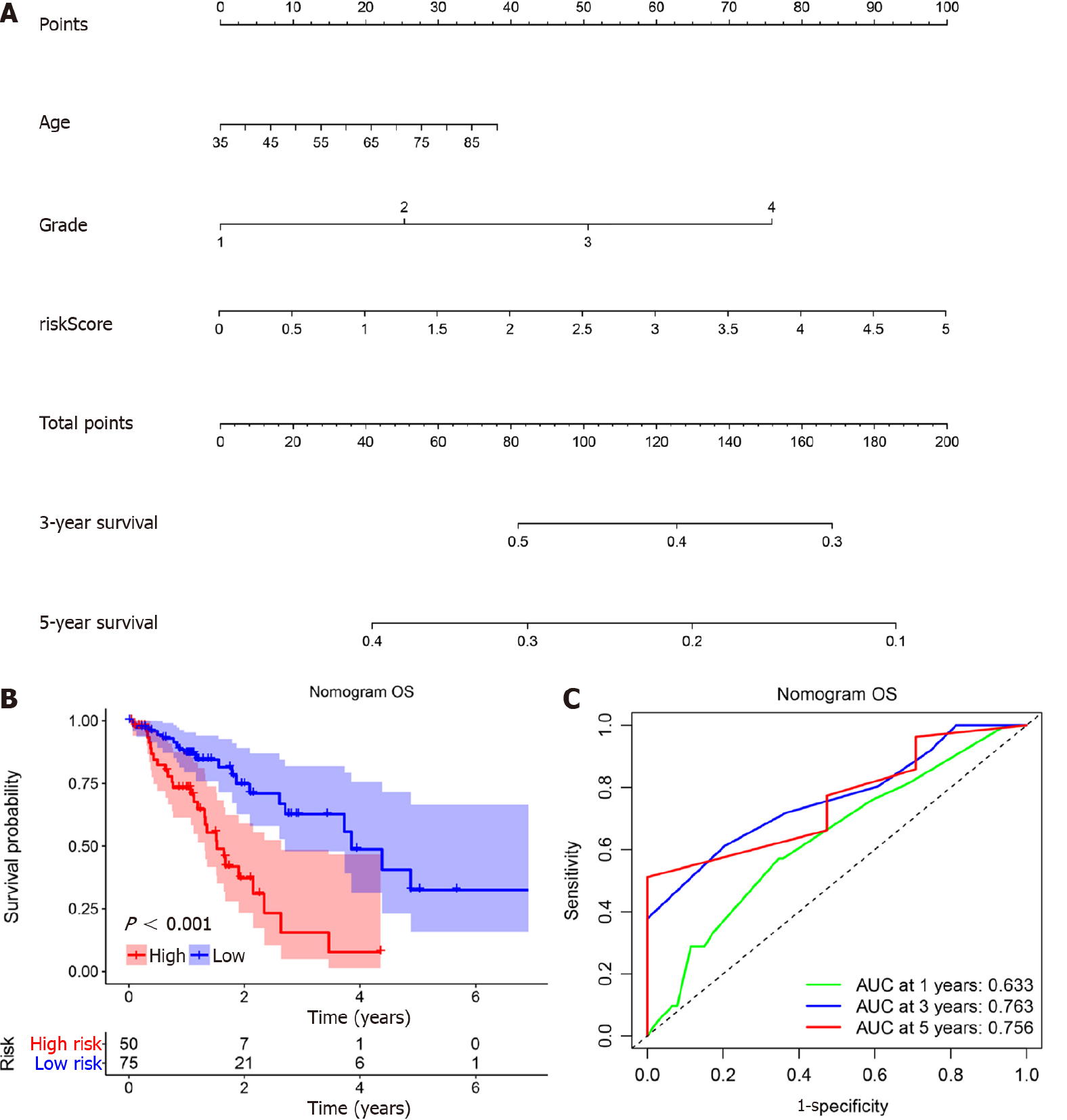
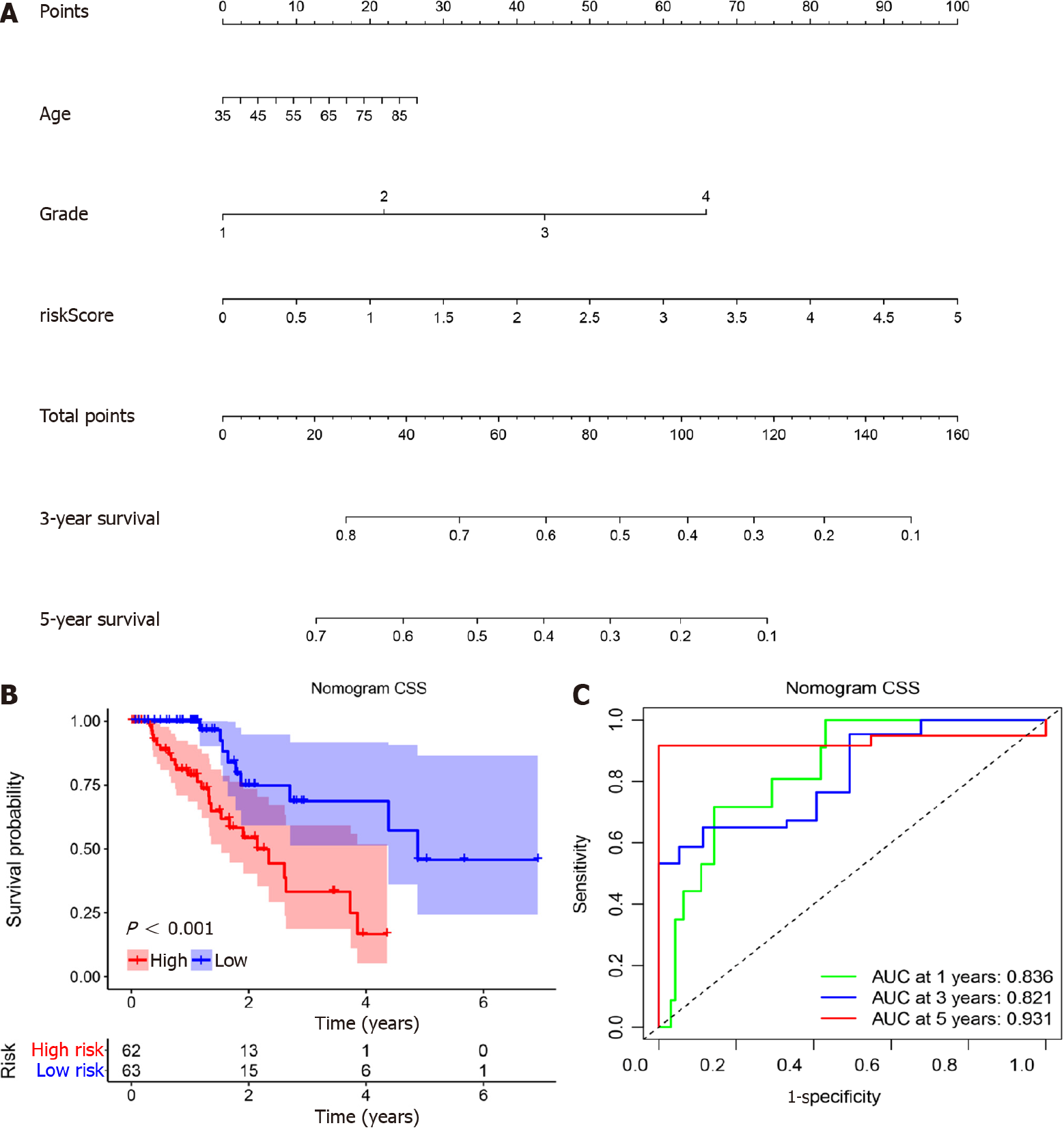
According to the results above, miR-1269a and AJCC stage had strong predictive capacity for prognosis. Based on our clinical practice, age also plays a central role in a patient’s prognosis. Therefore, a nomogram was established to display the predictive relationships among age, AJCC stage, and miR-1269a with OS and CSS (Figure 4A and Figure 5A). Additionally, we developed a risk assessment model for OS and CSS prediction. As shown in Figure 4B, our nomogram had distinguished differences in OS between the high risk and low-risk models (P < 0.001). The AUC values indicated the 1-, 3-, and 5-year OS of our nomogram were 0.633, 0.763, and 0.756, respectively, which is better than miR-1269a and AJCC stage alone (Figure 4C). The same or slightly better results were found for CSS predictions. The AUC values indicated the 1-, 3-, and 5-year CSS of our nomogram were 0.836, 0.821, and 0.931, respectively. As depicted in Figure 5C, our nomogram had a significant difference for CSS (P < 0.001). Generally, our nomogram including age had a higher discriminative capacity for predicting OS and CSS as compared to miR-1269a with AJCC stage.
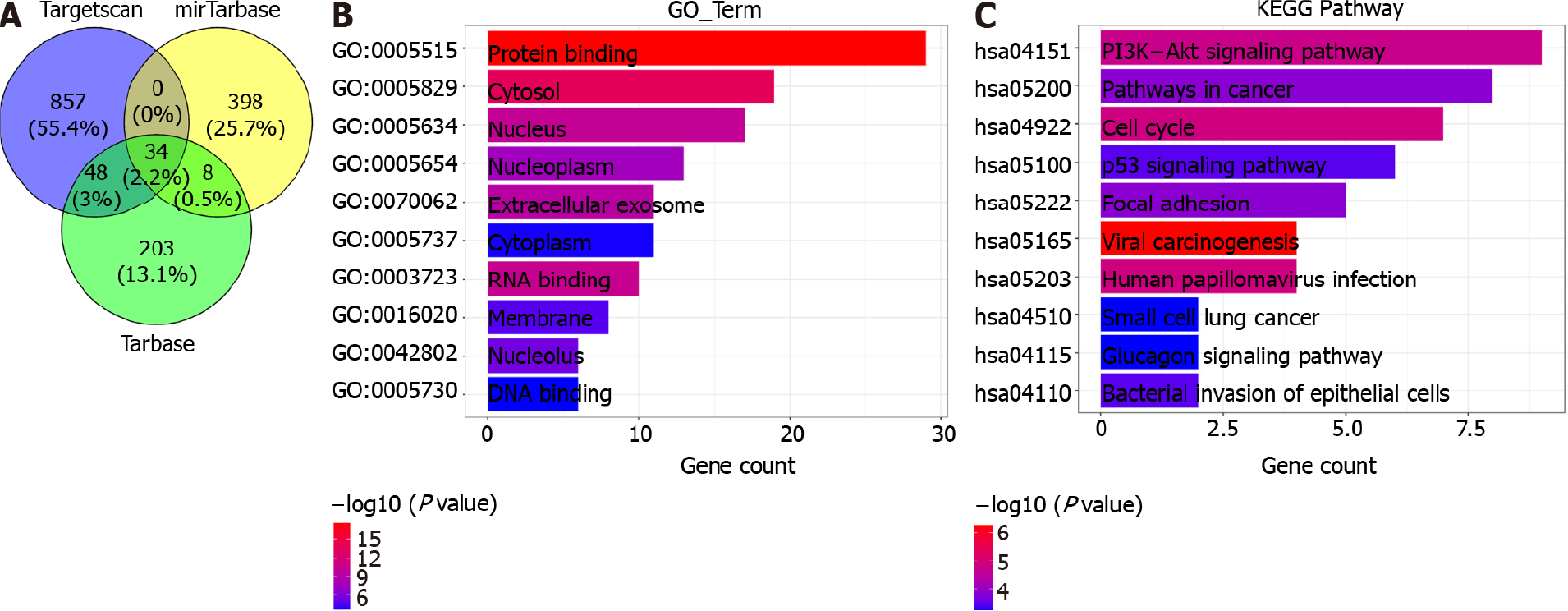
There are 937 target genes for miR-1269a in Targetscan, 440 target genes in miRTar
| ID | Term | Gene | Count | P value |
| GO:0005515 | Protein binding | CLTC, WDR6, PUM2, MATR3, AKAP11, SMARCA4, UBN1, YWHAZ, BSDC1, SMG1, ZNF146, BACH2, KHSRP, TPM3, TAGLN2, PTBP1, PHKG2, TNPO2, MAP1A, ZBTB4, HMGCR, AP3D1, FN1, PTMA, TBP, CCND2, NCL, CDK6, SEC61A1 | 29 | 3.58E-11 |
| GO:0005829 | Cytosol | YWHAZ, MAP1A, CCND2, SMG1, WDR1, PTMA, ZBTB4, BACH2, KHSRP, TPM3, CLTC, SEC61A1, PUM2, WDR6, CDK6, AKAP11, TAGLN2, PHKG2, ZNF146 | 19 | 3.79E-09 |
| GO:0005634 | Nucleus | YWHAZ, ZNF146, CCND2, SMG1, MATR3, PTMA, ZBTB4, BACH2, SLC25A1, KHSRP, NCL, CDK6, TBP, PTBP1, SMARCA4, UBN1, TNPO2 | 17 | 3.02E-07 |
| GO:0005654 | Nucleoplasm | YWHAZ, CCND2, SMG1, PTMA, ZBTB4, BACH2, KHSRP, NCL, CDK6, TBP, PTBP1, SMARCA4, UBN1 | 13 | 5.25E-06 |
| GO:0005737 | Cytoplasm | YWHAZ, MAP1A, CCND2, SMG1, WDR6, TBP, KHSRP, PUM2, CDK6, AKAP11, TNPO2 | 11 | 0.0013 |
| GO:0070062 | Extracellular exosome | YWHAZ, FN1, WDR1, SLC25A1, KHSRP, TPM3, NCL, CLTC, PGAM1, PTBP1, TAGLN2 | 11 | 8.89E-07 |
| GO:0003723 | RNA binding | YWHAZ, WDR6, SMG1, CLTC, KHSRP, PUM2, NCL, MATR3, PTBP1, SMARCA4 | 10 | 1.64E-07 |
| GO:0016020 | Membrane | CLTC, KHSRP, SEC61A1, NCL, MATR3, PTBP1, SMARCA4, AP3D1 | 8 | 0.00032 |
| GO:0042802 | Identical protein binding | YWHAZ, HMGCR, FN1, KHSRP, NCL, MATR3 | 6 | 0.00144 |
| GO:0005730 | Nucleolus | ZNF146, KHSRP, SMARCA4, UBN1, ZBTB4 | 5 | 7.83E-05 |
| GO:0003677 | DNA binding | ZNF146, CCND2, NCL, AKAP11, PTBP1, SMARCA4 | 6 | 0.00369 |
| GO:0000122 | Negative regulation of transcription by RNA polymerase II | BACH2, SMARCA4, CDK6, ZBTB4 | 4 | 0.00569 |
| GO:0019901 | Protein kinase binding | YWHAZ, CCND2, CLTC, ZBTB4 | 4 | 0.00067 |
| GO:0003729 | mRNA binding | PUM2, PTBP1, KHSRP, MATR3 | 4 | 2.90E-05 |
| GO:0032991 | Protein-containing complex | TBP, SMARCA4, CLTC | 3 | 0.01671 |
| GO:0043066 | Negative regulation of apoptotic process | YWHAZ, CCND2, PTMA | 3 | 0.00866 |
| GO:0006468 | Protein phosphorylation | YWHAZ, PHKG2, CDK6 | 3 | 0.00703 |
| GO:0098978 | Glutamatergic synapse | YWHAZ, AP3D1, NPTX1 | 3 | 0.00361 |
| GO:0019899 | Enzyme binding | TBP, PHKG2, FN1 | 3 | 0.00358 |
| GO:0051301 | Cell division | CCND2, CLTC, CDK6 | 3 | 0.00339 |
| GO:0008134 | Transcription factor binding | YWHAZ, TBP, SMARCA4 | 3 | 0.00285 |
| GO:0001525 | Angiogenesis | YWHAZ, NCL, FN1 | 3 | 0.00118 |
| GO:0030054 | Cell junction | KHSRP, SLC7A2, WDR1 | 3 | 0.00062 |
| GO:0005198 | Structural molecule activity | MAP1A, CLTC, MATR3 | 3 | 0.00043 |
| GO:0043488 | Regulation of mRNA stability | YWHAZ, PUM2, KHSRP | 3 | 0.00013 |
| ID | Term | Gene | Count | P value |
| hsa04151 | PI3K-Akt signaling pathway | YWHAZ, CCND2, FN1, CDK6, IGF1, MET, PRLR, MAGI2, CREB5 | 9 | 0.00863 |
| hsa05200 | Pathways in cancer | CCND2, TPM3, FN1, CDK6, IGF2, NOTCH3, MET, CDK6 | 8 | 0.02116 |
| hsa04110 | Cell cycle | YWHAZ, CCND2, CDK6, E2F5, YWHAQ, CCND1, STAG2 | 7 | 0.00751 |
| hsa04115 | p53 signaling pathway | CCND2, CDK6, CCND1, WIG1, CDK6, IGF1 | 6 | 0.03033 |
| hsa04510 | Focal adhesion | CCND2, FN1, ITGB3, IGF1, MET | 5 | 0.02116 |
| hsa05203 | Viral carcinogenesis | YWHAZ, TBP, CCND2, CDK6 | 4 | 0.00190 |
| hsa05165 | Human papillomavirus infection | TBP, CCND2, FN1, CDK6 | 4 | 0.00751 |
| hsa05100 | Bacterial invasion of epithelial cells | CLTC, FN1 | 2 | 0.03033 |
| hsa05222 | Small cell lung cancer | FN1, CDK6 | 2 | 0.03757 |
| hsa04922 | Glucagon signaling pathway | PHKG2, PGAM1 | 2 | 0.03757 |
| hsa04142 | Lysosome | AP3D1, CLTC | 2 | 0.03757 |
| hsa04390 | Hippo signaling pathway | YWHAZ, CCND2 | 2 | 0.03757 |
| hsa04218 | Cellular senescence | CCND2, CDK6 | 2 | 0.03757 |
| hsa05130 | Pathogenic Escherichia coli infection | YWHAZ, NCL | 2 | 0.04294 |
Currently, we mainly estimate the survival outcome of tumor patients through AJCC stage[31,32]. However, some limitations can be found in our clinical practice when we exclusively use AJCC stage. For instance, some patients with similar AJCC staging and clinical characteristics may differ in response to treatment and survival outcomes. This difference may arise from cancer heterogeneity, which partly root in genetic mutations[33,34]. Therefore, we attempted to establish a comprehensive staging system that combines clinical characteristics with genetic mutations. Moreover, recent studies have demonstrated that microRNAs play have great potential as biomarkers[30,35,36]. For example, low miR-335 expression is an independent prognostic factor and indicates a favorable prognosis in ESCA[37]; miR-1304 can be used as a powerful indicator for the diagnosis and recurrence of ESCA[38]; and miR-21 and miR-93 can be adopted as effective biomarkers for predicting radiotherapy and chemotherapy efficacy in ESCA[39]. Although some research has explored the diagnostic and prognostic roles in colorectal cancer, glioma, and lung cancer, the roles of miR-1269a in ESCA remain to be elucidated. Therefore, it is essential to detect miR-1269a prognosis in ESCA.
Here, we identified miR-1269a expression in ESCA and evaluated its association with clinical variables. Additionally, we performed a Kaplan-Meier analysis assessing its relationship with OS and CSS. ROC curves were drawn for diagnostic value. We estimated independent risk factors for OS and CSS using univariate and multivariate Cox analysis. Ultimately, we successfully confirmed the prognostic ability of miR-1269a for ESCA. Furthermore, a nomogram was created to evaluate the relationship between risk factors and OS and CSS, and a risk model including age, AJCC stage, and miR-1269a expression constructed. Kaplan-Meier analysis and ROC curves indicated that our risk model has substantial predictive value. Moreover, some advantages can be found in our nomogram such as simplicity, accuracy, easy to use and understand. These features enable the surgeon to quickly assess and make treatment decisions, which indicated that our nomogram was an effective tool for clinical applications.
In addition, the result from functional analysis revealed that miR-1269a might play important roles in cancer pathways including cell cycle pathway, pathway in cancer and PI3K/AKT pathway. Moreover, some studies also supported our prediction. For instance, miR-1269a overexpression in gastric cancer cell could stimulate the activation of PI3K/AKT pathway, thereby inducing cell proliferation and invasion; Meanwhile, the overexpression of miR-1269a inhibited RASSF9 expression, thereby impeding the cell apoptosis of AGS/MKN-45 cells through Bax/Bcl-2 signaling pathway[40]. miR-1269a contributes to down-regulation of FOXO1 and affects the dysregulation of cyclin D1, CDK2 and Bcl-2, thereby inducing cell proliferation, migration, and invasion[41]. The low expression of ATRX can be controlled by miR-1269a, also promoting proliferation and progression in vitro[18]. And this suppressive effects can be rescue by regulating ATRX overexpression in glioma cells. Those studies indicated that miR-1269a could serve as a potential therapeutic target for cancer.
Although our miR-1269a-related nomogram suggested a good performance in prognostic prediction of ESCA, there are some limitations. First, we only compared miR-1269a expression in tissue from ESCA and normal subjects. Further investigations are needed to detect the plasma difference of miR-1269a in ESCA. Additionally, retrospective study might weaken the validity of our research. Randomized controlled trials with larger populations are required to confirm our results. Lastly, the relevant mechanisms of miR-1269a in vivo remain to be elucidated. Therefore, we hope to carry out basic experiments in the future that could provide more evidence for the clinical application of miR-1269a in ESCA.
In conclusion, our findings suggest that miR-1269a expression could serve as a prognostic indicator for ESCA. Moreover, the simplicity and accuracy of our nomo
Esophageal cancer (ESCA) is a heterogeneous cancer with variable outcomes that are challenging to predict. MicroRNA (miR)-1269a is a newly discovered non-coding RNA that shows promising prognostic prediction in other cancers, but its clinical value in ESCA remains unclear.
This study established a comprehensive staging system that combines clinical characteristics with genetic mutations in ESCA.
This study aimed to determine the prognostic value of miR-1269a, and to develop a nomogram to succinctly predict the prognosis in esophageal carcinoma.
miR-1269a expression in ESCA were detected using quantitative real-time polymerase chain reaction. Then we determined its prognostic value with clinical variables through multivariate Cox analysis. A nomogram based on miR-1269a expression using age and American Joint Committee on Cancer (AJCC) stage was developed and assessed its prognostic performance. Finally, we predicted the target genes of miR-1269a and analyzed their potential function in caner development using Gene Onto
High expression of miR-1269a in ESCA showed poor prognosis in overall survival (OS) and cancer-specific survival (CSS), suffered increased rates of low differentiation and metastasis, and exhibited tumor stage T3 + T4, positive lymph stage, and AJCC stage III + IV. The area under the receiver operating characteristic curve of miR-1269a was 0.716 for OS and 0.764 for CSS. Multivariate Cox analysis revealed that AJCC stage and miR-1269a were independent factors for OS and CSS. Combing with age, we constructed a nomogram for prognostic prediction, which showed excellent perfor
miR-1269a can be used as a potential indicator for the prognosis of esophageal cancer. We developed an easy-to-use nomogram with excellent prognostic prediction for clinical use.
In further research, the molecular mechanism of miR-1304 in esophageal cancer will be elucidated.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Faloppi L, Koshy A, Sharma M S-Editor: Wang JL L-Editor: Filipodia P-Editor: Yuan YY
| 1. | Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL, Siegel RL. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69:363-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2417] [Cited by in RCA: 3074] [Article Influence: 512.3] [Reference Citation Analysis (0)] |
| 2. | Wei W, Zeng H, Zheng R, Zhang S, An L, Chen R, Wang S, Sun K, Matsuda T, Bray F, He J. Cancer registration in China and its role in cancer prevention and control. Lancet Oncol. 2020;21:e342-e349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 300] [Article Influence: 60.0] [Reference Citation Analysis (0)] |
| 3. | Yang F, Wang S, Guo J, Liu X, Ge N, Wang G, Sun S. EUS-guided fine-needle technique facilitates the establishment of organoid biobanks. Endosc Ultrasound. 2020;9:355-360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 4. | Tang C, Zhu J, Hu D, Chen G. Dysphagia caused by a rare esophageal external compression lesion (with video). Endosc Ultrasound. 2020;9:205-206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 5. | Rabinowitz SS, Grossman E, Gress F. Potential pitfalls in diagnostic EUS of the esophagus. Endosc Ultrasound. 2020;9:272-273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 6. | Pesenti C, Bories E, Caillol F, Ratone JP, Godat S, Monges G, Poizat F, Raoul JL, Ries P, Giovannini M. Characterization of subepithelial lesions of the stomach and esophagus by contrast-enhanced EUS: A retrospective study. Endosc Ultrasound. 2019;8:43-49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 7. | Zhang Y, Zhang B, Wang Y, Zhang J, Wu Y, Xiao T, Liao Y, Bao Y, Qiu H, Sun S, Guo J. Advances in the Prevention and Treatment of Esophageal Stricture after Endoscopic Submucosal Dissection of Early Esophageal Cancer. J Transl Int Med. 2020;8:135-145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 8. | Rubenstein JH, Shaheen NJ. Epidemiology, Diagnosis, and Management of Esophageal Adenocarcinoma. Gastroenterology. 2015;149:302-17.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 262] [Article Influence: 26.2] [Reference Citation Analysis (1)] |
| 9. | Rustgi AK, El-Serag HB. Esophageal carcinoma. N Engl J Med. 2014;371:2499-2509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 817] [Cited by in RCA: 998] [Article Influence: 90.7] [Reference Citation Analysis (0)] |
| 10. | Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1289] [Cited by in RCA: 1426] [Article Influence: 203.7] [Reference Citation Analysis (0)] |
| 11. | Feng Q, Zhang H, Yao D, Chen WD, Wang YD. Emerging Role of Non-Coding RNAs in Esophageal Squamous Cell Carcinoma. Int J Mol Sci. 2019;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 60] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 12. | Liang J, Chen W, Lin J. LncRNA: An All-rounder in Rheumatoid Arthritis. J Transl Int Med. 2019;7:3-9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 13. | Saliminejad K, KhorramKhorshid HR, SoleymaniFard S, Ghaffari SH. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234:5451-5465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1431] [Cited by in RCA: 1335] [Article Influence: 222.5] [Reference Citation Analysis (0)] |
| 14. | Sharma P, Sharma R. miRNA-mRNA crosstalk in esophageal cancer: From diagnosis to therapy. Crit Rev Oncol Hematol. 2015;96:449-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 15. | Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460-469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1310] [Cited by in RCA: 1551] [Article Influence: 141.0] [Reference Citation Analysis (0)] |
| 16. | Huang J, Zhang SY, Gao YM, Liu YF, Liu YB, Zhao ZG, Yang K. MicroRNAs as oncogenes or tumour suppressors in oesophageal cancer: potential biomarkers and therapeutic targets. Cell Prolif. 2014;47:277-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 80] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 17. | Bu P, Wang L, Chen KY, Rakhilin N, Sun J, Closa A, Tung KL, King S, Kristine Varanko A, Xu Y, Huan Chen J, Zessin AS, Shealy J, Cummings B, Hsu D, Lipkin SM, Moreno V, Gümüş ZH, Shen X. miR-1269 promotes metastasis and forms a positive feedback loop with TGF-β. Nat Commun. 2015;6:6879. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 106] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 18. | Zhang Y, Wang Q, Luo N, Liu J, Ren H, Shao X, Zhang L, Yu Y. MicroRNA-1269a Promotes Proliferation and Arrest of Apoptosis of Glioma Cells by Directly Targeting ATRX. Front Oncol. 2020;10:563901. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 19. | Wang X, Jiang X, Li J, Wang J, Binang H, Shi S, Duan W, Zhao Y, Zhang Y. Serum exosomal miR-1269a serves as a diagnostic marker and plays an oncogenic role in non-small cell lung cancer. Thorac Cancer. 2020;11:3436-3447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 20. | Jang HJ, Lee HS, Burt BM, Lee GK, Yoon KA, Park YY, Sohn BH, Kim SB, Kim MS, Lee JM, Joo J, Kim SC, Yun JS, Na KJ, Choi YL, Park JL, Kim SY, Lee YS, Han L, Liang H, Mak D, Burks JK, Zo JI, Sugarbaker DJ, Shim YM, Lee JS. Integrated genomic analysis of recurrence-associated small non-coding RNAs in oesophageal cancer. Gut. 2017;66:215-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 21. | Rice TW, Blackstone EH, Rusch VW. 7th edition of the AJCC Cancer Staging Manual: esophagus and esophagogastric junction. Ann Surg Oncol. 2010;17:1721-1724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 571] [Cited by in RCA: 639] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 22. | Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, Chien CH, Wu MC, Huang CY, Tsou AP, Huang HD. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39:D163-D169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 879] [Cited by in RCA: 1021] [Article Influence: 68.1] [Reference Citation Analysis (0)] |
| 23. | Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou D, Kavakiotis I, Maniou S, Skoufos G, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018;46:D239-D245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 507] [Cited by in RCA: 811] [Article Influence: 135.2] [Reference Citation Analysis (0)] |
| 24. | Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4013] [Cited by in RCA: 5416] [Article Influence: 541.6] [Reference Citation Analysis (0)] |
| 25. | Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24735] [Cited by in RCA: 27977] [Article Influence: 1748.6] [Reference Citation Analysis (0)] |
| 26. | Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316-W322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2658] [Cited by in RCA: 3476] [Article Influence: 248.3] [Reference Citation Analysis (0)] |
| 27. | Ravani P, Parfrey P, MacRae J, James M, Quinn R, Malberti F, Brunori G, Mandolfo S, Tonelli M, Hemmelgarn B, Manns B, Barrett B. Modeling survival of arteriovenous accesses for hemodialysis: semiparametric vs parametric methods. Clin J Am Soc Nephrol. 2010;5:1243-1248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 28. | Simon N, Friedman J, Hastie T, Tibshirani R. Regularization Paths for Cox's Proportional Hazards Model via Coordinate Descent. J Stat Softw. 2011;39:1-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1263] [Cited by in RCA: 1357] [Article Influence: 96.9] [Reference Citation Analysis (0)] |
| 29. | Li J, Ma S. Time-dependent ROC analysis under diverse censoring patterns. Stat Med. 2011;30:1266-1277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 30. | Mishra PJ. MicroRNAs as promising biomarkers in cancer diagnostics. Biomark Res. 2014;2:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 72] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 31. | Rice TW, Gress DM, Patil DT, Hofstetter WL, Kelsen DP, Blackstone EH. Cancer of the esophagus and esophagogastric junction-Major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017;67:304-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 209] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 32. | Rice TW, Ishwaran H, Ferguson MK, Blackstone EH, Goldstraw P. Cancer of the Esophagus and Esophagogastric Junction: An Eighth Edition Staging Primer. J Thorac Oncol. 2017;12:36-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 490] [Article Influence: 54.4] [Reference Citation Analysis (0)] |
| 33. | Lin DC, Wang MR, Koeffler HP. Genomic and Epigenomic Aberrations in Esophageal Squamous Cell Carcinoma and Implications for Patients. Gastroenterology. 2018;154:374-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 185] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 34. | Yan T, Cui H, Zhou Y, Yang B, Kong P, Zhang Y, Liu Y, Wang B, Cheng Y, Li J, Guo S, Xu E, Liu H, Cheng C, Zhang L, Chen L, Zhuang X, Qian Y, Yang J, Ma Y, Li H, Wang F, Liu J, Liu X, Su D, Wang Y, Sun R, Li Y, Cheng X, Liu Z, Zhan Q, Cui Y. Multi-region sequencing unveils novel actionable targets and spatial heterogeneity in esophageal squamous cell carcinoma. Nat Commun. 2019;10:1670. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 108] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 35. | Klingenberg M, Matsuda A, Diederichs S, Patel T. Non-coding RNA in hepatocellular carcinoma: Mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67:603-618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 279] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 36. | Zheng D, Ding Y, Ma Q, Zhao L, Guo X, Shen Y, He Y, Wei W, Liu F. Identification of Serum MicroRNAs as Novel Biomarkers in Esophageal Squamous Cell Carcinoma Using Feature Selection Algorithms. Front Oncol. 2018;8:674. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 37. | Zhang BJ, Gong HY, Zheng F, Liu DJ, Liu HX. Up-regulation of miR-335 predicts a favorable prognosis in esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:6213-6218. [PubMed] |
| 38. | Luo YG, Duan LW, Ji X, Jia WY, Liu Y, Sun ML, Liu GM. Expression of miR-1304 in patients with esophageal carcinoma and risk factors for recurrence. World J Gastroenterol. 2020;26:670-685. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 5] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 39. | Wang WT, Guo CQ, Cui GH, Zhao S. Correlation of plasma miR-21 and miR-93 with radiotherapy and chemotherapy efficacy and prognosis in patients with esophageal squamous cell carcinoma. World J Gastroenterol. 2019;25:5604-5618. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 11] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 40. | Liu WL, Wang HX, Shi CX, Shi FY, Zhao LY, Zhao W, Wang GH. MicroRNA-1269 promotes cell proliferation via the AKT signaling pathway by targeting RASSF9 in human gastric cancer. Cancer Cell Int. 2019;19:308. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 41. | Yang XW, Shen GZ, Cao LQ, Jiang XF, Peng HP, Shen G, Chen D, Xue P. MicroRNA-1269 promotes proliferation in human hepatocellular carcinoma via downregulation of FOXO1. BMC Cancer. 2014;14:909. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 67] [Article Influence: 6.1] [Reference Citation Analysis (0)] |