INTRODUCTION
Septin 9 (SEPT9) is a cell cycle-related protein[1] and indispensable for coordinating myosin motor proteins during cytokinesis[2]. SEPT filaments and intermediate filaments are non-polar, which distinguishes them from actin filaments and microtubules[1]. SEPT are a family of conserved scaffold proteins with GTP-binding protein activity which interact with each other, lead to the formation of hetero-oligomer complexes, and then polymerize into multi-functional filaments. SEPT interact with microtubule cytoskeletons, actin protein, and phospholipid membranes[3-5] and act as scaffolds for protein-protein interactions or as diffusion barriers for protein compartmentalization during cytoskeleton formation, cell polarization, cytokinesis, cell membrane remodeling, and cell division[6-10]. Different eukaryotes have different numbers of SEPT isoforms[11]. To date, 13 SEPT genes (SEPT1 to SEPT12 and SEPT14) have been identified in mammals and they can be divided into four subgroups (SEPT2, SEPT3, SEPT6, and SEPT7). They form apolar tri-, hexa-, or octameric complexes, consisting of multiple polypeptides, due to alternative promoter usage and splicing[3,12].
SEPT9 stabilizes the polymer and promotes the aggregation of subunits, and plays a key role in the final separation of daughter cells during cell division. Therefore, when the SEPT9 gene is abnormally expressed or deleted, cell division may be seriously affected. The SEPT9 gene is abnormally expressed in many malignant tumors. Tóth et al[13] reported that the SEPT9 mRNA expression decreased during the progression of colorectal adenoma to colorectal cancer (CRC), and SEPT9 protein expression in CRC was significantly lower than that in normal epithelial cells[13]. SEPT9 transcript expression patterns vary in different types of malignant tumor. The expression of SEPT9 v2, v4, v4*, and v5 isoforms was higher in cancer epithelial cells compared to normal cells[14]. The v1 and v4 transcripts showed higher expression in ovarian cancer epithelium compared to benign and borderline tumors[15]. The expression of v1 transcripts was higher in the normal samples than it in cancer cells, whereas SEPT9 transcripts v2, v4, v4*, and v5 overexpression was detected in CRC epithelial cells compared to normal cells[13]. These findings suggest that the SEPT9 gene may play an important role in the development of malignant tumors, and it could be a new target to individualize anti-cancer treatments.
SPECIFIC STRUCTURAL CHARACTERISTICS AND BIOLOGICAL FUNCTIONS OF THE SEPTIN 9 GENE
The SEPT genes are members of a conserved gene family with guanosine triphosphate activity, widely exist in eukaryotes, and were first found in the cell cycle process of budding yeast Saccharomyces cerevisiae[16]. These genes produce diverse splices through variant splicing, resulting in extremely complex family members and gene functions. SEPT consist of a highly variable N-terminal domain, a central GTP-binding domain, and a C-terminal domain[3,12]. GTP-binding domain can form linear filaments using either guanine nucleotide binding site or N- and C-terminal extensions[1]. SEPT can assemble into heteropolymers and form higher order structures[17].
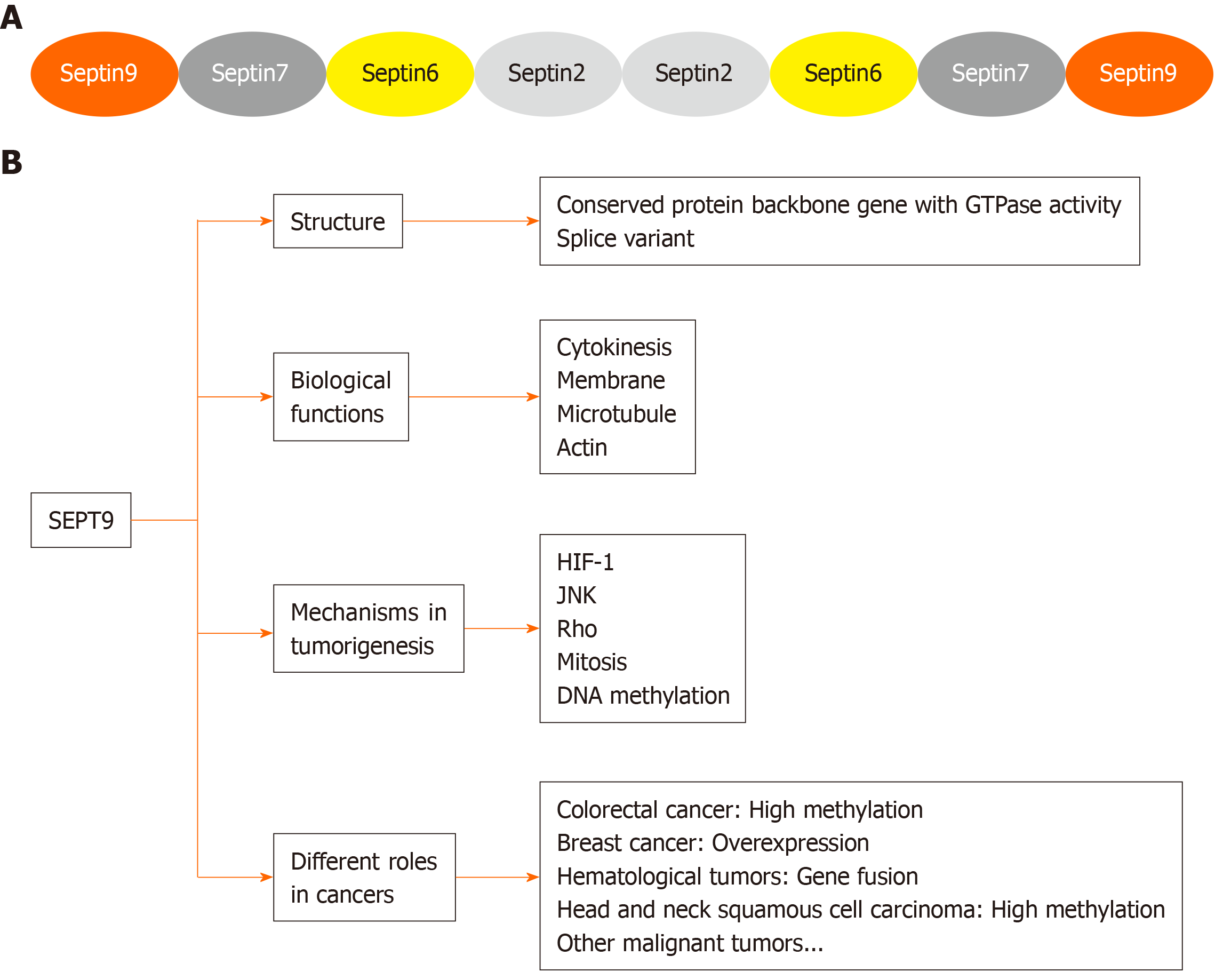
The SEPT9 gene is located on chromosome 17q25.3 and ubiquitous in human cells, with 18 transcript products[18]. Members of the SEPT family have high affinity for each other and form tri-, hexa-, and octameric complexes containing multiple SEPT polypeptides. Mammalian SEPT can form an octameric complex, like SEPT9–SEPT7–SEPT6–SEPT2–SEPT2–SEPT6–SEPT7–SEPT9 (Figure 1A).
Figure 1 Specific structural characteristics and biological functions of Septin 9.
A: Octamers formed by mammalian septin complexes; B: Function and role of Septin 9 in human malignant tumors. SEPT9: Septin 9; HIF-1: Hypoxia inducible factor-1; JNK: Jun N-terminal kinase.
Biological functions
SEPT9 or its complex interact with cellular proteins, like microtubules or actin microfilament, and play different physiological functions during the development of tumor (Figure 1B).
Role in cytokinesis
Cytokinesis is mainly driven by a contractile actomyosin ring (CAR), which controls membrane invagination. The CAR is recruited via a SEPT scaffold at division sites. The mitotic exit network, a Hippo-like kinase cascade, promotes mitotic exit, cytokinesis, and displacement of the SEPT ring from the division site. Mitotic exit network factors localize in the spindle pole body. In budding yeast, SEPT are first assembled as unorganized SEPT patches at the bud neck (constriction between mother and future daughter cells) in the G1 phase of the cell cycle. They are then assembled into the cortical SEPT ring[19]. Microfilaments and microtubules, as the two most important cytoskeleton components during cytokinesis, regulate the assembly of SEPT fibrils. SEPT fibril assembly can be divided into two modes: Actin-dependent and actin-independent. In the former mode, the SEPT fibrils form a SEPT/anillin complex with actin fibrils, through anillin and other adaptor proteins. In this process of complex formation, GTP is not required, but the C-terminal crimp domain is connected to form a 7 nm long filamentous structure. In the latter mode, actin assembly becomes very active under the special circumstances of human cell injury. At this time, the microfilaments of the SEPT-actin complex are separated and curl to form a "C" and "O" ring structure. These two patterns of SEPT assembly play important roles in cell division[3]. The microinjection of an affinity-purified anti-SEPT9 antibody and the use of small interfering RNAs to render cytoplasmic division defective in the early and late stages, eventually leads to the formation of binuclear cells or the failure of daughter cell separation[20].
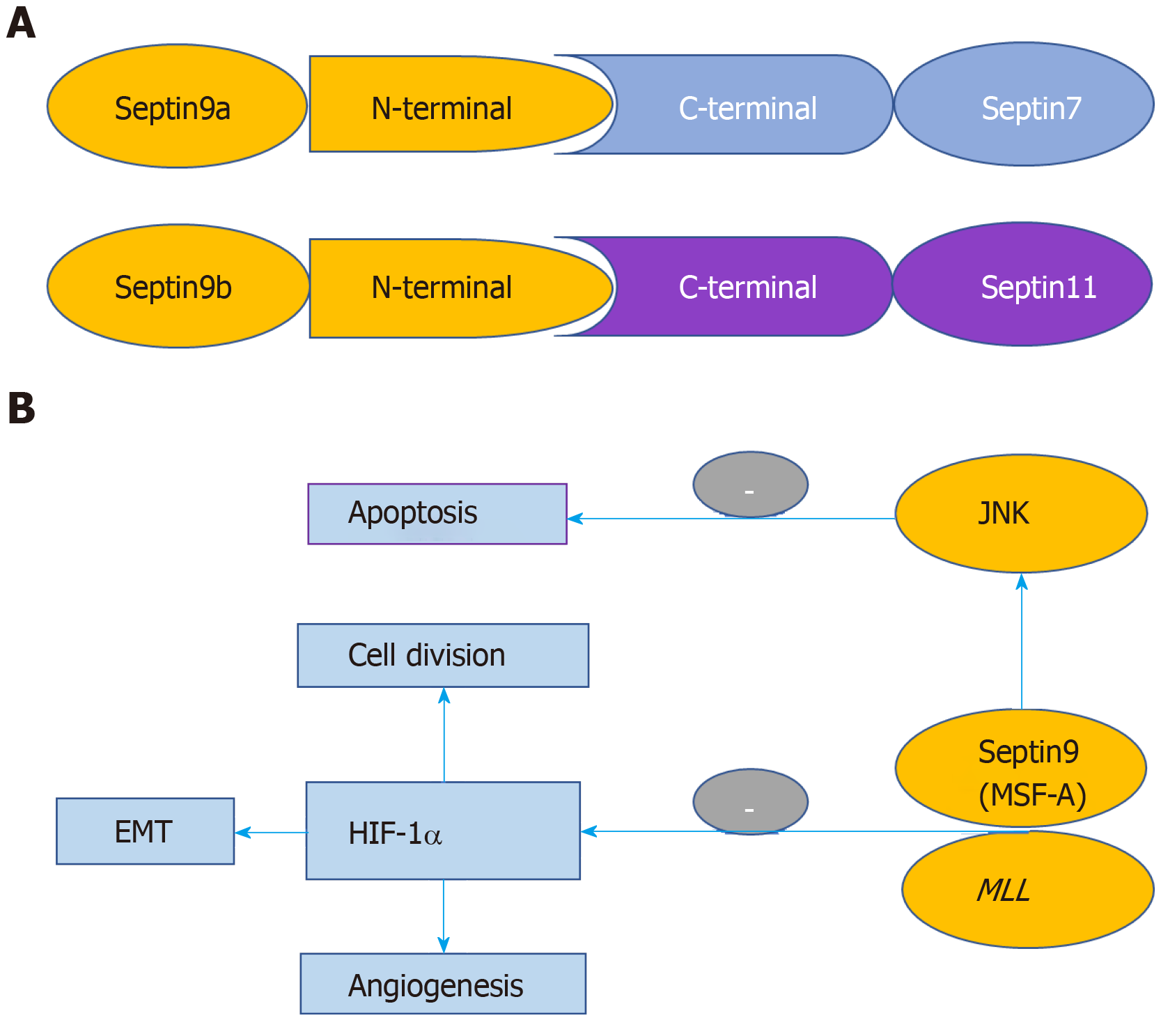
SEPT9 occupies the ends of mammalian octameric SEPT complex. The SEPT9 has a stable multimeric structure, promotes the polymerization of each subunit, and plays a key role in the final separation of cells during cell division[21]. SEPT 9b binds to C-termini of both SEPT 7 and SEPT 11 through its long N-terminal extension[22] (Figure 2A).
Figure 2 Structure of the filaments containing Septins 7/9b/11 and Jun N-terminal kinase signaling pathway.
A: Structure of the filaments containing Septins 7/9b/11; B: Regulatory mechanism of Septin 9 in the development and progression of malignant tumors. HIF-1: Hypoxia inducible factor-1; JNK: Jun N-terminal kinase; EMT: Epithelial-mesenchymal transformation; MLL: Mixed-lineage leukemia.
Role in membrane
SEPT, when self-assembled into filaments and higher-order structures, acts as a scaffold for diverse cellular functions. This is achieved by diffusion-driven annealing to the plasma membrane[23], which alters the cortical rigidity of migrating cells by applying membrane curvature and inducing tumor metastasis[24]. SEPT9 levels correlate with the cell surface levels of epidermal growth factor receptors (EGFRs). SEPT9 induces tumor progression by disordering EGFRs. Plasma membrane-related SEPT9 prevents the binding of the adaptor protein, CIN85 (SH3KBP1), with the ubiquitin ligase, Cbl, thereby reducing the degradation of ubiquitin-independent EGFR[25].
Role in microtubule
Intracellular transport is an important means of substance exchange between eukaryotic cells and their internal environment. It is also a necessary condition to maintain the physiological activities of organisms. The realization of this process requires the participation and regulation of many substances, among which, the most important regulatory factor is the major component of cytoskeleton-tubulin. SEPT filaments specifically localize with microtubules and the formation of tubulin is closely related to SEPT[26,27]. SEPT compete with the microtubule-associated protein, MAP4, to associate with the microtubule lattice and reduce its stability[28,29]. Microtubule-targeting agents are often used in cancer chemotherapy. SEPT9_V1 overexpression or dysregulation is associated with a poor prognosis and resistance to microtubule-targeting agents. SEPT9_V1, associated with microtubules, is strongly correlated with the susceptibility of cancer cells to 2-methoxyestradiol and paclitaxel[30]. Some studies have shown that SEPT9 protein directly co-localizes with microtubules and spindles in HeLa cells. After treatment with the specific inhibitor of microtubule polymerization, colchicine, the structure of SEPT9 is affected and it is distributed in the cytoplasm as dots[11].
Role in actin
SEPT control actin remodeling during cancer cell migration and invasion, prevent the depolymerization of actin by myosin and cofilin, and protect the integrity of actin filaments[31]. Epithelial-mesenchymal transformation (EMT) is a specific biological process in which epithelial cells are transformed into cells with a mesenchymal phenotype. It plays an important role in cancer metastasis. The main characteristics of EMT are a decrease in the expression of cell adhesion molecules (such as E-cadherin), the transformation of the cytokeratin cytoskeleton into a Vimentin-based cytoskeleton, and the induction of a mesenchymal cell morphology. Through EMT, epithelial cells lose their polarity, their connection with the basement membrane, and other epithelial phenotypes and they obtain greater invasion, migration, and anti-apoptosis abilities; become capable of degrading the extracellular matrix; and assume other interstitial phenotypes.
Knockdown of SEPT9 in metastatic cells inhibits actin cytoskeleton dynamics, induces EMT[32], increases the activation of the Src/Vav2/RhoA pathway, and induces the proliferation of endothelial cells[33]. Moreover, some studies have shown that the structure of filaments containing SEPTs 7/9b/11 depends on the integrity of actin filaments in REF52 cells[22]. Cell division may be severely affected when SEPT9 gene expression is abnormal or absent. Mutations in the GTP domain of the SEPT9 gene can destabilize the cell genome and overexpression of this domain can affect SEPT9 polymer formation, ultimately leading to tumor formation[34].
EXPRESSION OF THE SEPTIN 9 GENE IN TUMORIGENESIS AND DEVELOPMENT OF DIFFERENT MALIGNANT TUMORS
Hypoxia inducible factor-1 signaling pathway
Hypoxia inducible factor-1 (HIF-1) is a transcription factor regulating tumor cells in hypoxic environment, and consists of HIF-1-alpha and HIF-1-beta subunits. The expression of HIF-1-alpha is related to tumor invasion, metastasis, drug resistance, and prognosis. Amir et al[35] reported that the mammalian SEPT family member MSF-A (a homologue of SEPT9) and HIF-1-alpha can interact with each other. SEPT9 overexpression can inhibit ubiquitination and degradation of HIF-1-alpha, which leads to the enhancement of the transcriptional activity of HIF-1. This will in turn increase the expression of downstream genes, such as vascular endothelial growth factor, and ultimately promote tumor metastasis and invasion. High expression of vascular endothelial growth factor can promote angiogenesis of tumor tissue, which provides the basis of the proliferation and infiltration of tumors[35].
Jun N-terminal kinase signaling pathway
Jun N-terminal kinase (JNK) is a signal transduction protein of mitogen-activated protein kinase, which is important for cell growth, division, transformation of cancer cells, and regulation of cell cycle (Figure 2B). SEPT9_v1 can maintain the activity of JNK by preventing the degradation of JNK. The retention of JNK activity can inhibit cell apoptosis and promote cell proliferation[36].
Rho signaling pathway
SEPT protein assembly is regulated by the small GTPase superfamily. The activity of this family is regulated by GTPase-activating proteins and guanine exchange factors. Rho protein is a small protein with GTPase activity. It can regulate the expression of downstream effector molecules, thus affecting the formation of the cytoskeleton, mediating cell-cell and cell-matrix adhesion, and affecting the metastasis and invasion of tumor cells[37]. SEPT9 is a negative upstream effector for RhoA. It is found at significantly higher levels in endothelial cells on hydrogels with low stiffness than those on hydrogels with high stiffness. Moreover, low-stiffness hydrogels can inhibit endothelial cell proliferation by inactivating integrin, resulting in an increase in SEPT9 expression and thus inhibiting the activation of the Src/Vav2/RhoA pathway[33]. Rho activity is regulated by Rho-guanine nucleotide exchange factor (Rho GEF). SEPT-associated RhoGEF (SA-RhoGEF) can directly combine with SEPT9b and distribute along actin stress fibers. Transient transfection of SEPT9b can inhibit SA-RhoGEF-dependent Rho activity in COS7 and HeLa cells, while SA-RhoGEF transfection can alter the structure of SEPT9b fibrils[38]. Rhetokin is the downstream effector of Rho. It can block the formation of SEPT9 fibers, thus affecting normal physiological activities, such as cell division. The conformational change in the SEPT protein induced by rhetokin is similar to the change seen during Rho activation. Thus, rhetokin may be the node connecting the SEPT protein and the Rho signaling pathway[39].
Mitosis
During mitosis, the filaments formed by the polymerization of SEPT9 protein in interphase are dispersed in the cytoplasm in prophase and then aggregate in the cleavage groove in anaphase. High SEPT9 expression promotes genetic instability, causes mitotic spindle defects, hinders the cytoplasmic division, and leads to chromosomal segregation disorders. If SEPT9 gene expression is downregulated or absent in actively mitotic cells, cell division is impaired and cannot be completed and the spindle cannot be correctly attached, resulting in polyploidy and aneuploidy[40-42].
DNA methylation
DNA methylation is one of the important epigenetic modifications. It refers to the transfer of a methyl group to a base by DNA methyltransferase, with S-adenosylmethionine as the methyl donor. In mammals, the methyl group is transferred to the 5th carbon atom of the cytosine in a cytosine-guanine dinucleotide (CpG) to form 5-methylcytosine[43]. CpG islands are the main sites of DNA methylation, which is closely related to the occurrence and development of tumors. Many abnormal gene methylation processes are closely related to the pathogenesis, prognosis, and chemotherapy response of cancer. The main manifestations of these abnormalities are low total DNA methylation levels in nonpromoter regions and high methylation levels in gene promoter regions[44]. Hypomethylation is related to transcriptional activation, resulting in increased expression of oncogenes, loss of gene imprinting, and increased chromosomal instability. By contrast, hypermethylation is related to gene expression silencing and genomic instability; reduced expression of tumor suppressor genes and DNA repair genes; and effects on normal cellular functions, such as apoptosis, DNA repair, and cell cycle regulation. SEPT9 is a tumor suppressor gene and it has been shown to be methylated in a variety of tumors. DNA methylation is one of the most thoroughly studied epigenetic markers in eukaryotes, with studies focusing on hypomethylation of the whole genome and hypermethylation of specific repair genes and tumor suppressor genes[45]. In vertebrates, approximately half of all gene promoters have CpG islands and abnormal hypermethylation often occurs in these regions. Abnormal methylation of the SEPT9 gene causes abnormal gene expression and reduced gene transcription activity, leading to abnormal physiological function and finally, the development of cancer.
SEPT9 AND COLORECTAL CANCER
It is reported that hypermethylation of the CpG island in the promoter region of the SEPT9 gene can inhibit its expression, abrogating its tumor suppressor functions, and ultimately results in the malignant tumor formation[13]. Upon detecting the methylation status of the SEPT9 gene in different types of colon lesions and their adjacent sites, Wasserkort et al[46] proved that hypermethylation of the SEPT9 gene occurred in CPG island 3 of V2 transcript in CRC and adenoma epithelial cells[46]. Hypermethylation extends from the core region to the 5' terminus in CRC tissues, but there is no hypermethylation at the 5' terminus of CPG island 3 in adenomas, which suggests that the expansion of hypermethylation may be a late event in the progression of adenoma to CRC. This characteristic of hypermethylation may be the reason for the lower sensitivity of the SEPT9 gene methylation detection assay in adenoma than in CRC. Abnormal methylation of the SEPT9 gene not only occurs in CRC, but also in other malignant tumors. Grützmann et al[47] compared the methylation status of the SEPT9 gene in the plasma of patients with various malignant tumors. They showed that the positive rate of methylation in CRC patients was 72% (90/125), whereas it was only 11.5% (11/96) in non-CRC patients, which confirmed that SEPT9 gene methylation may act as a specific diagnostic marker of CRC[47]. Now, the peripheral blood mSEPT9 detection has been widely used in the screening, diagnosis, prognosis, and recurrence monitoring of CRC in China because of its high specificity and sensitivity and low price.
Pathogenic Mechanism of the Septin 9 Gene in Colorectal Cancer
SEPT9 protein, being a component of the fourth cytoskeleton, can aggregate and form fibrillary microfilaments and other complexes during active cell division. It also recruits target proteins and stabilizes cytokinesis. SEPTs—Cdc3, Cdc10, Cdc11, Cdc12, and Shs1 form the bud neck ring, and induce the separation of the mother and daughter cells during cell division. The phosphorylation of SEPT9 by cyclin-dependent kinase 1 plays an important role in the disjunction of daughter cells by interacting with proline isomerase. It also affects chromosome congression and segregation during anaphase of cell division. Thus, phosphorylated SEPT9 regulates mitosis and promotes cell proliferation and survival[48]. SEPT9 mislocalization can mediate cytokinesis failure and lead to aneuploidy, centrosome amplification, and multipolar mitoses[40]. Elongated N-terminal tail of SEPT9 is enriched with proline residues, and interacts with actin microfilaments and microtubule[31]. SEPT9-Spr3 and Spr28 also induce the formation of prospore membrane during sporulation[49]. When the SEPT9 gene in active cells was knocked out, the cells did not divide smoothly to form multinuclei, which resulted in abnormal spindle-related functions, disordered chromosome segregation, and unstable cell genome, ultimately leading to canceration of tissues[42].
The high methylation of CpG loci of the SEPT9 gene will block its gene expression and result in a loss of its anti-cancer function. This DNA methylation inhibits gene expression in two ways and promotes tumorigenesis. On one hand, DNA methylation changes its conformation, making it unconducive to bind transcription factors, and ultimately inhibits gene expression[46]. On the other hand, methylated DNA molecules recruit and bind methylated CpG binding domain proteins. As these methylated CpG binding domain proteins continue to bind other proteins (such as histone deacetylase), eventually it forms a compact and inactive abnormal chromatin, which inhibits gene expression.
Autophagy, a regulatory cellular mechanism, prevents malignant transformation of cells by reducing oxidative stress, eliminating oncogene proteins, and maintaining genomic stability, thereby exerting its anti-cancer potential[50]. Studies have shown that cell autophagy inhibits the occurrence of CRC[51]. SEPT protein complex participates in cell autophagy[52,53]. Methylation of the SEPT9 gene inhibits the expression of SEPT9 protein, decreasing cell autophagy function, and thereby, promotes the occurrence and development of CRC. For example, in microsatellite-unstable CRC, frameshift mutation of UVRAG counteracts its anti-tumor function of inducing autophagy and DNA repair, which in turn promotes tumorigenesis, epithelial-mesenchymal transformation, and metastasis[27,54,55].
Application of Septin 9 in Early Screening, Diagnosis, and Prognosis of Colorectal Cancer
Since 2008, different research groups have published a series of clinical data on screening early stage CRC by detecting SEPT9 gene methylation. Since the second generation detection kit EpiproColon 2.0 (German Epigenomics AG) was used, its sensitivity has increased to 79.3%-95.6% and specificity has reached 84.8%-98.9%[56]. In the CRC screening process, the high specificity of screening methods is conducive for excluding non-cancer samples to avoid unnecessary colonoscopy, and the high-risk population who may miss diagnosis can be compensated by regular fecal occult blood test review[57]. For screening of precancerous lesions by SEPT9 methylation detection, the sensitivity is 11%-19% and specificity is 85%-94% in adenomas. For polyps, the sensitivity is 3%-8% and specificity is 90%-97%. The sensitivity is low both in adenomas and polyps, suggesting that detection of SEPT9 gene methylation has limited diagnostic value for precancerous lesions of CRC[58]. The positive rates of SEPT9 gene methylation in peripheral blood of patients with stages I, II, III, and IV CRC were 45%, 70%, 76%, and 79%, respectively, which indicated that SEPT9 gene methylation detection still had appreciated value for detecting early CRC, and can provide a basis for diagnosis[59]. The positive rate of methylation increased with the progression of CRC, which may be related to the increase of necrosis and apoptosis of primary tumor cells and the increase of methylated SEPT9 gene DNA released into peripheral blood. A study observed the changes in plasma SEPT9 gene methylation levels in CRC patients before and after radical surgery. During the follow-up period of approximately 4 mo, 8 out of 9 patients with positive preoperative methylation turned negative (88.9%). Survival analysis showed that there was a significant negative correlation between SEPT9 methylation levels and disease-free survival after CRC surgery[60]. Although the sample size of that study was small, it indicated that SEPT9 gene methylation in peripheral blood can be used as a monitoring index for recurrence after CRC surgery. Whether this index can be used as an evaluation index for the efficacy of CRC radiotherapy and chemotherapy remains to be further explored.
Studies have compared the diagnostic efficacy of SEPT9 gene methylation detection and fecal immunoassay test (FIT) alone or combined in CRC screening. The results showed that the sensitivity and specificity of SEPT9 and FIT alone were 73.3%-81.5% and 53.8%-88.9%, and 81.5%-95.3% and 54.1%-97.4%, respectively. The sensitivity and specificity of the combined diagnosis of SEPT9 and fecal occult blood test were 88.7%-97.8% and 52.9%-78.8%, respectively. SEPT9 methylation was superior to FIT in sensitivity, and joint detection was helpful to further improve sensitivity[61]. Glycoprotein markers for CRC screening include CEA, CA19-9, CA125, CA724, and CA242. The sensitivity and specificity of the diagnostic panel consisting of CEA, CA19-9, CA242, CA211, and CA724 were 83% and 95%, respectively, which were similar to those of detection of SEPT9 gene methylation in peripheral blood. The sensitivity and specificity of the most commonly used single serum CEA were lower than those of SEPT9 gene methylation[62-64]. Colonoscopy biopsy combined with pathological diagnosis is the golden standard for the diagnosis of CRC. In recent years, endoscopic treatment of colorectal tumors has become more developed. The application of endoscopic surgery such as endoscopic mucosal resection and endoscopic submucosal dissection in the treatment of precancerous lesions has gradually increased. However, colonoscopy has few limitations. Large-scale screening by colonoscopy can consume a lot of medical resources. It requires thorough bowel preparation to ensure a clear visual field. Moreover, contraindications and complications risk can occur during colonoscopy. All these limitations may lead to poor patient compliance. Therefore, the consensus of early CRC screening process is recommended by the clinical experts in China in order to stratify the risk of screening population. FIT and/or SEPT9 gene methylation detection can be used for low-risk population, and colonoscopy can be used for high-risk population[65].
A large number of clinical data confirmed that the detection of SEPT9 gene methylation in peripheral blood has a high sensitivity and specificity for the diagnosis of CRC. It can be used for early CRC screening. The results are not affected by sex and tumor location, and can guide the selection of follow-up screening methods and improve the compliance of the population to colonoscopy. Our previous study showed that methylation of SEPT9 is useful for the screening, diagnosis, and recurrence monitoring of CRC, and methylation of SEPT9 is associated with TNM category, Dukes stage, gross tumor volume, and mismatch repair-deficiency status[63]. Another study also showed that patients with high levels of methylation of SEPT9 had a tendency toward lower overall survival, which demonstrated that methylation of SEPT9 may be a useful biomarker for the prognosis of CRC[66].
SEPTIN 9 AND BREAST CANCER
The SEPT9 gene is also closely related to the occurrence and development of breast cancer. The expression level of the SEPT9 gene had significantly increased in mice with breast cancer induced by transgenic technology. Thsp1 and Bax-mediated apoptosis decreased in human breast cancer and breast cancer cell lines with high SEPT9 expression. Silencing SEPT9 expression reversed this effect[67], suggesting that the inhibitory effect of SEPT9 on apoptosis could promote the progression of breast cancer. The expression of SEPT9 in human breast cancer cell lines MCF10A and MCF-7 and breast cancer tissues was high. Overexpression of SEPT9 could accelerate cell proliferation, increase invasiveness of cancer tissues, and cause chromosomal abnormalities[68]. In addition, the expression of SEPT9 isoforms in different tumors was also different. The main isoform closely related to breast cancer is SEPT9-v1. Overexpression of SEPT9-v1 promoted the transformation of breast epithelial cells to breast stromal cells, thus promoting the development of breast cancer. Few studies reported that SEPT9-v3 was closely related to breast cancer and its expression was regulated by methylation[14]. Methylation can decrease the expression of SEPT9-v3, while demethylation can increase the levels of SEPT9-v3 protein and mRNA. However, a study found that SEPT9_v2 was also related to breast cancer, and the SEPT9_v2 promoter hypermethylation was found in 67% of breast cancer cell lines and 53% of breast cancer tissue, but not in normal breast tissue[69]. Other studies showed increased transcription of four variants of the SEPT9 gene, v1, v3, v6, and v7, in breast cancer tissues[70]. In addition, a study also found that SEPT9 and SEPT2, SEPT8, and SEPT11 were involved in the mechanism of paclitaxel resistance in human breast cancer cell lines by regulating cellular microenvironment[71]. The overexpression of the SEPT9-i1 isoform enhanced cell resistance to paclitaxel, which was further enhanced by microtubule polyglutamylation[72]. So far, several studies have confirmed the close relationship between SEPT9 and breast cancer. This provides a new idea for searching tumor markers for early screening and diagnosis of breast cancer, as well as clues for finding new therapeutic targets for breast cancer.
SEPTIN 9 AND HEMATOLOGICAL TUMORS
In a large number of studies on hematological malignancy, gene fusion was often found in leukemia patients. Mixed-lineage leukemia (MLL) gene rearrangement is very common in leukemia[73]. The N-terminal base sequence of the MLL gene and SEPT9 gene can fuse in frame and produce chimeric protein[74]. SEPT family genes such as SEPT2, SEPT5, SEPT6, SEPT9, and SEPT11 can also fuse with MLL. Diseases associated with the fusion of the MLL and SEPT9 genes include acute myeloid leukemia, acute lymphoblastic leukemia, and myelodysplastic syndrome, but their mechanism of action is not yet clear. In 2007, Kreuziger et al[75] reported the fusion of the MLL gene and SEPT9 gene in a patient with myelodysplastic syndrome[75]. In one case study of acute monocytic leukemia, t(11;17)(q23;q25) appeared, accompanied by fusion of the SEPT9 gene and MLL gene[76]. Another study found that exon 8 at the 3' end of the MLL gene was fused with exon 2 and exon 3 at the 5' end of the SEPT9 gene in patients with acute myeloid leukemia[77]. In addition, AML1 gene amplification and 17q25 deletion were also found in leukemia patients[78]. In a study involving 45 patients with leukemia, MLL gene rearrangements were found to be associated with SEPT9 gene[79]. A study also showed that patients with acute megakaryoblastic leukemia and t(11;17)(q23;25) had a KMT2A–SEPT9 fusion[80]. Overall, these studies have shown that SEPT9 gene is closely related to the occurrence and development of leukemia, and its fusion with the MLL gene may be one of the pathogeneses of leukemia. And in other hematological tumors, like T-cell lymphomas, study found that murine retrovirus SL3-3 was integrated into the SEPT9 gene locus[81].
SEPTIN 9 AND HEAD AND NECK SQUAMOUS CELL CARCINOMA
In a study, five highly methylated genes, including SEPT9, were detected in head and neck squamous cell carcinomas by restriction marker genome scanning[82]. This suggests that the SEPT9 gene is associated with head and neck tumors. It was also found that the expression of SEPT9-v1 was higher in head and neck squamous cell carcinoma than in normal tissues. The higher the expression of SEPT9-v1, the higher the stage of head and neck squamous cell carcinoma[83]. A cohort study by Schrock et al[84] showed that detection of plasma methylated SEPT9 is a valuable method for early diagnosis and monitoring of head and neck squamous cell carcinoma[84]. Hence, these studies suggest that SEPT9 may also be a marker for head and neck squamous cell carcinoma.
OTHER MALIGNANT TUMORS
Studies have confirmed that SEPT9 is also associated with various kinds of tumors. The SEPT9 gene is closely related to ovarian cancer[15]. Detection of SEPT9 expression in benign, borderline, and malignant serous and mucinous ovarian tumors by fluorescent quantitative PCR showed that SEPT-9-v1 and SEPT-9-v4 were significantly increased in borderline ovarian tumors, but not in benign ovarian tumors and ovarian cancer. At the same time, the high expression of SEPT9-v1 transcript coincided with the high expression of SEPT9-v4 transcrips, and the ratio between them was related to the phenotype of borderline and malignant tumors. These results suggested that the high expression of SEPT9-v1 and SEPT9-v4 may be associated with the occurrence of borderline ovarian tumors. The mean level of plasma SEPT9 in epithelial ovarian carcinoma patients was significantly higher than that in healthy women. The mean level of plasma SEPT9 in epithelial ovarian cancer patients with a tumor family history or distant metastases was significantly higher than that of other patients[85].
The methylation of SEPT9 promoter region was found in lung cancer patients, which resulted in a decrease in the expression of SEPT9 protein[86]. This was similar to the increased methylation of the SEPT9 gene in patients with colorectal cancer. A study involving 70 lung cancer patients and 100 healthy people explored the relationship between lung cancer and SEPT9. The results showed that the positive rate of methylated SEPT9 gene in peripheral blood of the lung cancer group was much higher than that of the control group. This study suggests that detection of methylated SEPT9 gene level in peripheral blood may have some clinical significance in early diagnosis of lung cancer[87].
Li et al[88] analyzed the expression level of SEPT9 in 102 cases of gastric cancer and their adjacent tissues by tissue microarray and immunohistochemical staining, and studied the relationship between SEPT9 and the degree of differentiation of cancer. The results showed that the expression of SEPT9 was higher in gastric cancer tissues than in the adjacent tissues and related to the degree of differentiation of cancer[88].
AREAS FOR FUTURE RESEARCH
DNA methylation may be one of the mechanisms regulating the differential expression of SEPT9 transcripts. DNA methylation is an important epigenetic modification, and CpG islands are the main sites of DNA methylation. Abnormal gene methylations have been observed in different kinds of malignant tumor. DNA methylation is closely related to the development, progression, occurrence, prognosis, and chemotherapy response of malignant tumors[43,44]. DNA methylation includes hypermethylation and hypomethylation. It is believed that hypermethylation leads to gene silencing and decreased protein expression, while hypomethylation leads to the activation of genes and increased protein expression. Detection of methylation level of the SEPT9 gene in peripheral blood has a high sensitivity and specificity for the diagnosis of some malignant tumors, especially CRC, and the detection result is not affected by the patient's gender and the location of the tumor. Furthermore, detection of methylation level of the SEPT9 gene in peripheral blood by PCR can be used to assess the risk of some malignant tumor initiation and relapse, and can be widely used in clinic because of the use of non-fasting, easy-to-obtain blood samples, noninvasiveness, and good patient compliance[56,89].
CONCLUSION
It has been more than 40 years since the initial discovery of the SEPT gene in yeast. During this period, there has been much in-depth research on SEPTs. According to their different tissue expression patterns and different proteins they interact with, members of the SEPT gene family can perform different cellular functions. However, due to the nature of the SEPT family structure and the complexity of its members, the detailed physiological functions of SEPTs require further study. SEPT gene expression is strictly controlled to maintain the correct assembly of filaments and normal cellular functions. However, if gene mutations result in changes to SEPT proteins or their expression levels, this may lead to the development of disease.
As reviewed above, a large number of studies have confirmed the correlation between the SEPT9 gene and a diverse range of tumor types. The expression levels, mutations, and functions of the SEPT9 gene are different in different tumors. Genetic and epigenetic changes in the SEPT9 gene are linked to proliferation, autophagy, angiogenesis, cell invasion, chromosomal instability, microsatellite instability, resistance to anti-cancer drugs, and the induction of tumor growth by the cancer-associated microenvironment. Several clinical trials have confirmed that methylation of the SEPT9 gene is a specific biological marker for some tumors. However, further studies are required to determine the specific mechanism whereby the regulation of SEPT9 affects tumorigenesis and tumor progression and to determine the correlation between SEPT9 and other pathogenic mechanisms. In the future, with ongoing research, a novel role for SEPT9 may be identified in the early screening, diagnosis, targeted therapy, prognosis, and recurrence monitoring of different cancers.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Gazouli M, Mohamed SY, Sung WW S-Editor: Zhang L L-Editor: Wang TQ E-Editor: Qi LL