Published online May 26, 2023. doi: 10.4252/wjsc.v15.i5.281
Peer-review started: December 26, 2022
First decision: February 16, 2023
Revised: March 6, 2023
Accepted: April 17, 2023
Article in press: April 17, 2023
Published online: May 26, 2023
Processing time: 150 Days and 21.1 Hours
Colorectal cancer (CRC) remains the third most prevalent cancer disease and involves a multi-step process in which intestinal cells acquire malignant characte
Core Tip: Colorectal cancer (CRC) represents one of the most prevalent tumors worldwide. The tumor microenvironment (TME) through its proinflammatory role, among others, actively participates in CRC progression and the disturbance of gut microbiota (dysbiosis) can influence this inflammatory process. CRC stem cells (CCSC) are a tumor cell subpopulation that drives CRC initiation, progression and treatment failure. The features and behavior of CCSC are modulated by several factors including TME and gut microbiota. Here, we will give an overview of the synergistic interaction among TME and intestinal microorganisms that condition the CRC environment and shape CCSC characteristics allowing CRC evolution.
- Citation: Novoa Díaz MB, Carriere P, Gentili C. How the interplay among the tumor microenvironment and the gut microbiota influences the stemness of colorectal cancer cells. World J Stem Cells 2023; 15(5): 281-301
- URL: https://www.wjgnet.com/1948-0210/full/v15/i5/281.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v15.i5.281
Colorectal cancer (CRC) is a multi-step process where intestinal cells acquire malignant phenotypic characteristics that allow them to proliferate, migrate, invade and establish in new tissues[1]. In the last decades, screening strategies and treatments have been improved, decreasing the proportion of CRC patients by as much as 65%–88%[2]. However, this disease remains the third most prevalent type of cancer, having an incidence of 10% and ranking second in mortality (9.4% among all cancer deaths) according to global cancer statistics[3]. The leading cause of patient deaths and relapses is the appearance of new CRC subtypes and the acquired resistance to currently used therapies[4]. Moreover, a great number of CRC are diagnosed with distal metastases and these patients have a poor survival rate due to a lack of response to therapy[2]. One of the causes that affect the treatment of this type of tumor by inducing resistance and the appearance of recurrences, is the presence of a small subpopulation of cells called CRC stem cells (CCSC). This small number of cells have mutations in specific oncogenes that allow them to develop the ability to induce tumor initiation, self-renew, differentiate, dedifferentiate, and acquire multidrug resistance[1,5]. The origin of this cell subpopulation is still controversial. They may originate from colorectal normal cells, colorectal normal stem cells, or CRC cells by genetic alterations or by the influence of environmental factors that induce epigenetic changes[5].
It is known that in the tumor niche, different cell types, structures, and biomolecules coexist and interact with cancer cells favoring the growth and development of the tumor. Together these components constitute the tumor microenvironment (TME). In the last decades, several investigations have demonstrated that tumor surrounding ambiance through its proinflammatory role, among others, actively participates in the development, progression and chemoresistance of CRC[1,4].
Researchers have deepened the study of the influence of the complex variety of microorganisms that inhabit the intestinal mucosa, collectively known as the gut microbiota, on this inflammatory microenvironment. Besides contributing to innate and adaptive immune function, it has been observed that the imbalance in the species present in the intestinal microbiota and the consequent variation in microbial products can promote the development of CRC and compromise the efficacy of its therapy[6].
Since all the factors mentioned are involved in the CRC progression and therapy resistance and considering the great influence of CCSC in several events of this disease, this review aims to analyze the available literature that is focused on the interaction of TME and the intestinal microbiota that favors the development and maintenance of CCSC properties.
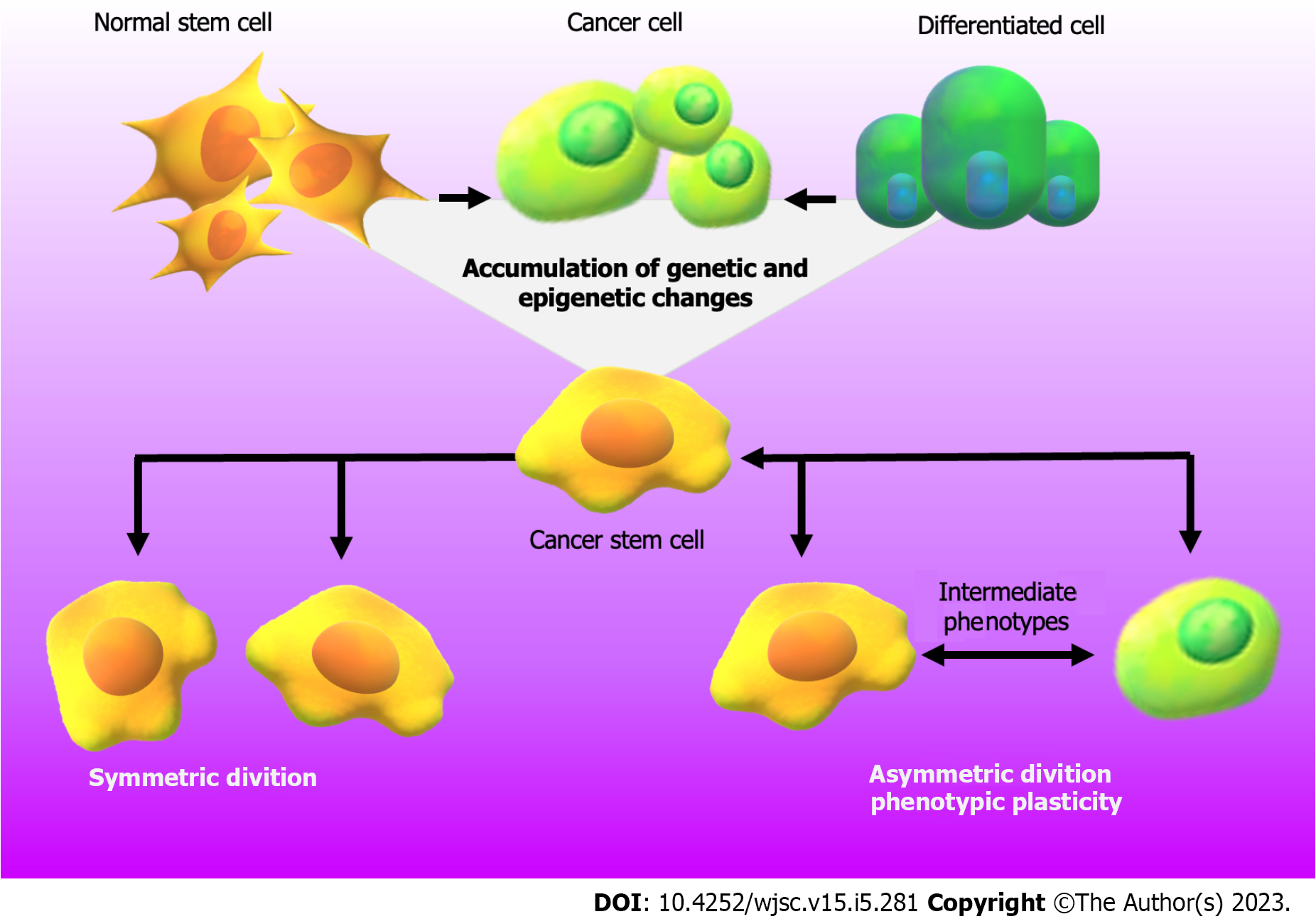
CRC is a heterogeneous pathology that has a variable clinical course and prognosis[7]. The etiology of this disease combines genetic alterations in colorectal epithelial cells with unhealthy lifestyles, such as smoking, alcohol consumption and poor nutritional habits[8,9]. In addition, it has been seen that sex, age, family history of CRC and the persistence of inflammatory processes or infectious agents in the intestinal tract, can be also considered risk factors[5,9-12]. In all these cases, the synergy among genetic mutations, epigenetic alterations and the influence of the TME and gut microorganisms promotes the acquisition of molecular and phenotypic features that allow tumor progression[5,6,11,13,14]. Therefore, within the tumor niche, cells present great heterogeneity but are still strictly organized. In the last 20 years, the focus has been on the study of cancer stem cells (CSC) derived from colorectal tissue (CCSC), a subpopulation of cells that have a substantial tumorigenic capacity and maintain intestinal tumor growth[15]. CSC are responsible for resistance to multiple drugs maintaining a state of undifferentiation and slow cell division and also favoring the efficiency of desoxyribonucleic acid (DNA) damage repair mechanism[16]. Besides, they have similar features to normal stem cells, such as self-renewal, multipotency, cell cycle arrest, quiescence, and reversibility from their resting state[17,18]. As shown in Figure 1, the ability of CSC to maintain their population response to symmetric/asymmetric division, resulted in the first situation in two identical daughter stem cells and, in the second situation in two distinct cells with or without CSC properties[19,20]. In addition to the division theory, CSC undergo a bidirectional conversion process between stem and non-stem phenotype[20]. Although initially a hierarchical model has been established, in which CSC are the initiators of a monoclonal developmental hierarchy, emerging data highlight the concept of phenotypic plasticity of CSC. This new theory is supported by a dynamic state of interconversion between CSC and non-CSC that can be driven by the TME[21-23]. As the reader can see in Figure 1, during this phenomenon, cells can easily exchange their status within the tumor transforming from CSC to intermediate phenotypes to stemless states and vice versa[15,18,22,24]. Therefore, based on the data provided by the literature and shown in Figure 1, it can be concluded that any cell type is capable of initiating and promoting cancer development[24]. This model contributes with new concepts to the classical theory of the origin/behavior of CSC that highlight the importance of taking into account the study of phenotypic plasticity and the reversible state of this type of cells and that support the criterion that cancer cells with or without stem characteristics must be eradicated for successful therapy.
CCSC constitute about 2% of the cell population in the tumor nest and this percentage can be higher with tumor progression, particularly after chemotherapy or radiotherapy treatments[17,18,25]. Since an increase in the proportion of this cell subtype is an indicator of poor prognosis, in the last decades the identification and targeting of CCSC have become one of the key topics of study[26]. The recognition of CCSC is possible by the detection of typical phenotypic characteristics such as the expression of surface markers, membrane transporters and enzymes. Some of them are Prominin-1/cluster of differentiation 133 (CD133), a transmembrane glycoprotein that is associated with metastasis, invasiveness and chemoresistance in CRC[18]; cluster of differentiation 44 (CD44) a receptor of hyaluronic acid in extracellular matrix related to the epithelial to mesenchymal transition (EMT) program and poor survival in CRC patients[5,27]; cluster of differentiation 166 (CD166) and cluster of differentiation 24, both adhesion molecules whose expressions are associated with the aforementioned markers, and that contribute to stratify low, intermediate, and high-risk CRC cases[5,28]; leucine rich repeat containing G-protein coupled receptor 5 (LGR5) a key CCSC biomarker that decreases in advanced stages of CRC[20,29] and aldehyde dehydrogenase (ALDH), an intracellular enzyme found in high concentrations in most of CSC participating in self-renewal, differentiation and self-protection[20,30,31]. In addition, the study of the ATP-binding cassette transporter superfamily through Hoechst 33352 dye efflux is also employed to detect CCSC[15,32]. In experimental models, the identification and characterization of CCSC can also be performed by fluorescence-activated cell sorting, selection by cell culture properties, in vivo transplantation of cells derived from spheroids or organoids, and lineage tracing techniques with labeled CCSC[22]. The above mentioned markers are hallmarks of CCSC and are involved in CRC pharmacotherapy and pathophysiology[33,34], but can also be present in enterocytes and cells of other tissues[20]. Hence, to increase the detection sensitivity and specificity, it is essential to combine the analysis of different biomarkers with CCSC isolation techniques.
Another substantial aspect to consider in the study of CSC is their association with other cellular processes such as EMT, autophagy and the response to cellular stress[15]. In particular, EMT is a physiological process that is also involved in tumor progression. The activation of this program reduces intercellular adhesion and causes epithelial cells to acquire mesenchymal properties that increase the invasiveness and migration of tumor cells[35]. Several studies have reported a link between EMT and the acquisition of CCSC characteristics in both, in vitro and in vivo assays[35-38]. These investigations show that transcription factors and signaling pathways that are altered in the EMT program are also deregulated in CSC, generating this subpopulation to exhibit phenotypes like EMT[39]. However, recent evidence indicates that EMT may not be necessary to acquire CSC properties. Then, although these processes can go along with each other, they can also happen through independent paths[15]. One of the tumor events that is known to be related to EMT and CSC is the high metabolic demand of TME and the existence of a tortuous vasculature that promotes a hypoxic environment. This phenomenon induces the release of factors such as hypoxia-inducible factor 1α (HIF-1α) that promotes not only EMT but also autophagy associated with CSC. In CRC it was demonstrated that blocking this factor with the consequent inhibition of autophagy reduces cell proliferation and the acquisition of stem-like characteri
Another cause that has been reported that promotes a stem-like phenotype on several types of tumor cells is the cellular imbalance derived from oxidative stress[15]. In breast and lung cancer cell lines, studies demonstrate that oxidative stress upregulates the CSC marker SRY-box transcription factor 2 (Sox2) activity, and stem-like properties[41,42]. However, in CRC cells it was shown that the reduction of intracellular reactive oxygen species inhibits the formation of CRC stem-like cells[43]. Since this type of cellular stress is considered potentially cytotoxic, more studies are necessary to know the mechanisms by which it has a positive effect on the development of CCSC[15].
Furthermore, it is important to note that like all processes and phenomena related to tumorigenesis and malignant progression, CCSC and their features are modulated by the aberrant activation of various signaling pathways. Wnt, NOTCH, hedgehog (HH), and transforming growth factor-β (TGF-β) are important cascades that are usually misregulated in CCSC and play a central role in the therapy resistance of these cells[5,44].
Thus, understanding CCSC features and all the events and factors associated with cell plasticity constitute a fundamental tool for the development of new target therapeutic strategies.
It has been reported that multiple links exist between inflammatory processes and stemness in CRC[2]. In this context, the role of the tumor stroma is crucial. The TME in CRC is a physical shelter for CSC[5] composed of biomolecules from the extracellular matrix, an aberrant vasculature and multiple stromal and immune cell types. These cells include mesenchymal stem cells, cancer-associated fibroblasts (CAFs), endothelial cells (ECs), pericytes, and tumor infiltrating immune cells which comprehend: Macrophages, neutrophils, natural killer cells, Treg cells and cytotoxic T lymphocytes[2,4]. The interaction between CRC cells and the different types of cellular and non-cellular elements of TME involves complex and dynamic crosstalk by paracrine signaling[22]. Therefore, self-renewal, differentiation and properties of CRC cells and CCSC are modified by factors released by the surrounding stroma[1]. These factors are cytokines, growth factors and small nucleic acids, which have different mechanisms of action. Next, we will discuss those derived from TME that modulate CCSC properties and that are summarized in Table 1.
| TME factor | Action | Ref. |
| Growth/inducible factors | ||
| Epidermal growth factor | Regulates and promotes CCSC growth | [50] |
| Insulin-like growth factor | Regulates and promotes CCSC growth | [50] |
| TGF-β | Participates in the initiation of the EMT, invasion, metastasis and initiation of angiogenesis associated to CCSC | [13,29,50] |
| Bone mophogenetic protein 4 | Induces differentiation and decreases the tumorigenic potential of CCSC | [16,60,63] |
| Bone mophogenic protein 2 | Stimulates the differentiation of CCSC inducting autophagic degradation of β-catenin | [44,63] |
| Hepatocyte growth factor | Activates Wnt signaling and the clonogenicity from CCSC | [53,54] |
| Macrophage migration inhibitory factor | Increases CCSC properties | [49] |
| Vascular endothelial growth factor | Promotes growth, epithelial to mesenchymal transition and stemness | [50,51] |
| Platelet derived growth factor | Promotes growth, epithelial to mesenchymal transition and stemness | [50] |
| Osteopontin | Regulates EMT and participates in the activation of the Wnt/β-catenin signaling pathway, promoting stemness | [4,156] |
| HIF-1A | Activates Wnt/β-catenin pathway inducing self-renewal of CCSC. Promotes survival and maintenance of CCSC | [40,157] |
| Citokines/immune associated proteins | ||
| IL-1β | Modulates the expression of CCSC markers | [158] |
| IL-4 | Facilitates the communication of CCSC with stromal cell, maintains their properties and evades the immune system | [5,44] |
| IL-6 | Promotes the expression of the CCSC markers, ALDH1 and LGR5 | [1,47] |
| IL-8 | Induces stemness and EMT | [50,159] |
| IL-17A | Promotes invasiveness and self-renewal and increases CCSC properties | [12] |
| IL-22 | Promotes invasiveness and self-renewal and increases CCSC properties | [12] |
| IL-33 | Induces the expression of core stem cell genes in CRC-derived cells | [160] |
| Chemokine (C-C motif) ligand 2 | Promotes CCSC properties | [4,49] |
| Tumor necrosis factor- α | Modulates CCSC features and induces cell death | [158,161] |
| Parathyroid hormone related-protein | Activates Wnt/β-catenin pathway and promotes events related to stemness | [162-164] |
| Non-coding RNA | ||
| miR-135 a/b and miR-17 | Promote stemness through the activation of Wnt/β-catenin signaling | [157] |
| miR-34 and miR-93 | Inhibit stemness | [157] |
| miR-92a-3p | Promotes Wnt signaling activation and consequently the expression of β-catenin target genes related to stemness, the EMT program, and chemoresistance | [165] |
| miR-20a and miR-106 a/b | Repress TGF-β activity and stemness | [157] |
| miR-146 and Let-7 | Affect stem cell fate or proliferation, activation of several stemness markers in a colon cancer cell line | [157] |
| miR-221/222 and miR-21 | Induce the development and maintenance of CCSC | [157] |
| miR-21 | Promotes the activation of the Wnt/β-catenin signaling pathway and increases the population of CCSC | [157] |
| miR-145 | Represses miR-21 and its expression inversely correlates with that of CCSC markers | [157,166] |
| miR-137 | Suppresses CCSC tumorigenicity | [167] |
| miR-147 | Decreases the expression of CCSC markers | |
| miR-200, miR-203, miR-141 and miR-429 | Regulate CCSC through negative modulation of EMT and self-renewal | [157] |
| lncRNA H19 | Promotes CCSC phenotype and drug resistance | [168] |
| Signaling pathway ligands | ||
| Wnt ligands | Increase CCSC characteristics and enhances tumor-initiating potential | [5,157] |
| Delta like canonical Notch ligand 4 | Participates on CSC maintenance | [44] |
| Jagged1 | Participates on CSC maintenance | [66] |
| SHH | Promotes CCSC survival, self-renewal and drug resistance | [67,68] |
| Enzymes | ||
| Phospholipase D2 | Promotes CRC stemness | [4,49] |
| Extra-cellular matrix components | ||
| Tenascin, fibronectin, collagen type I, secreted protein acidic and rich in cysteine, galectin | Contribute to stemness and CCSC activities | [1] |
Cytokines have been shown to play a key role in CRC stemness. It was reported that TME-derived factors with a pro-inflammatory action such as tumor necrosis factor-α (TNF-α) and interleukin (IL)-1β foster EMT phenotype and stem cell proliferation in human colon cancer cells[45,46]. Besides, it is known that CAFs, one of the most studied cells in the TME, produce IL-6, which promotes the expression of CCSC markers such as ALDH1 and LGR5[1,47].
The acquisition of a stem-like phenotype is also influenced by the expression and secretion of growth factors[48,49]. It was demonstrated that the epidermal growth factor and the insulin-like growth factor regulate and promote CCSC growth[50]. Moreover, Muñoz Galván et al[49] have proved that the treatment of CRC derived cells with hepatocyte growth factor (HGF) and/or macrophage migration inhibitory factor increases the number and size of colonospheras and significantly enhances the expression of putative markers like CD133[49].
Proangiogenic factors like vascular endothelial growth factor (VEGF) and platelet derived growth factor are also implicated in promoting growth and metastasis, both processes directly related to stemness[50]. Furthermore, it was demonstrated that clusters of ECs improve the survival of CCSC and promote their spread[51].
As it is known, all these TME factors modulate the activation of different signaling pathways, altering gene expression and thus modifying the molecular and phenotypic profile of tumor cells[5,44,50]. Wnt signaling is a key stem cell pathway involved in the maintenance of the CCSC and the TME[13,52]. One decade ago, Vermeulen et al[53] observed that high activity of the Wnt signaling was associated with CCSC features. Furthermore, this activity was mainly observed near fibroblasts in the tumor niche. Vermeulen et al[53] then demonstrated that HGF derived from CAFs activates Wnt signaling and the clonogenicity from CCSC[53]. This research had a great impact on the study of CSC and recently, Essex and collaborators replicated these studies and obtained similar results. They found that TME regulates the activation of the Wnt signaling pathway, increases CCSC characteristics and enhances tumor-initiating potential[54]. Regarding this, it is known that several Wnt ligands are secreted mostly by CAFs[53-56]. Moreover, other TME factors participate in the activation of the Wnt/β-catenin pathway (Table 1).
Some ligands from other signaling pathways are also related to stem cell phenotype. TGF-β is a growth factor that belongs to a superfamily of molecules including inhibins and bone morphogenetic proteins (BMP)[13]. It has the ability to promote or suppress tumor development depending on the interactions that take place in the TME[57]. As a pro-tumor factor, TGF-β regulates immune responses and participates in many neoplastic events such as proliferation, EMT and stemness[13]. TGF-β signaling pathway mutations and CCSC are linked[58] and in accordance with this, Zhou et al[29] found an association between TGF-β signaling and the expression of LGR5 biomarker in CRC[29]. Even more, Gu et al[59] have recently demonstrated that the expression of genes related to CCSC features like the carcinoembryonic antigen-related cell adhesion molecule alters TGF-β signaling and promotes CRC[59]. Some other members from the TGF-β family, like bone morphogenetic protein 4 and bone morphogenetic protein 2 (BMP4 and BMP2, respectively), have the capacity to induce CCSC differentiation and increase the response to standard chemotherapy[16,60-62]. Besides, the modulation of the BMP4 pathway by hormones like triiodothyronine was reported in CCSC, decreasing its tumorigenic potential[44,63,64]. This result suggests that CCSC features are modulated not only by local molecules from the TME but also by endocrine factors[44].
Notch signaling is also associated with the expression of CSC features in CRC cells[16,65]. In fact, it was reported that delta like canonical notch ligand 4 and jagged 1, both notch ligands, are overexpressed in this type of tumor providing essential signals for CCSC maintenance[44,66]. Moreover, since HH signaling is implicated in CRC development[20], in the last years several investigations were conducted on the association between this pathway and CCSC properties. Regan et al[67], have shown that the activation of the non-canonical HH pathway is required for CCSC survival and depends on sonic hedgehog protein (SHH) ligand[67]. Recently, it has been also observed that the modulation of HH-related proteins expressions by non-coding ribonucleic acids (ncRNAs) impacts on CCSC self-renewal capacity and drug resistance[68]. In line with this, Skoda and collaborators showed that treatment with HH pathway inhibitors such as vismodegib and sonidegib weakens the ability of CCSC[69]. Since no significant differences have been found in clinical trials[70], more studies are needed to determine the effects of the inhibition of this pathway in CRC patients.
Besides the aberrant activation of several signaling pathways, hypoxia is known as a hallmark of CCSC and TME interaction[5]. This is a condition in the tumor niche whose main cause is the poor vasculature associated with the tumor and the upregulation of HIF-1α, a factor released mainly by ECs[40,71,72]. This condition activates Wnt/β-catenin pathway inducing self-renewal and maintenance of CCSC[50,73]. Also, HIF-1α promotes cancer cell proliferation and CCSC survival[40].
Furthermore, short ncRNAs like microRNAs (miRs) and long ncRNAs are secreted not only by tumor cells but also by stromal cells in the TME[4]. In the last decades, the study of ncRNAs has gained importance in CRC. In the framework of factors and signaling pathways related to CCSC biology, these small nucleic acids have a key role[74]. miRs related to stemness in CRC are exposed in Table 1.
As previously mentioned, the interaction between CRC cells and their TME also involves non-cellular elements. Colonic stromal cells mediate the remodeling of the extra-cellular matrix favoring the healing or progress of the disease[75]. Recently, it has been demonstrated, by lineage tracing, that components of the extra-cellular matrix regulate dormancy in CCSC[76]. Tenascin, fibronectin, collagen type I, secreted protein acidic and rich in cysteine (SPARC), galectin and some other components of the tumor matrix are associated with stemness and CCSC activities[1].
Finally, another important concept to consider in the tumor nest is that CCSC also release various factors and cytokines that enable them to communicate with stromal cells, maintain their properties and evade the immune system, such as IL-4 and the cluster of differentiation 200[5,44].
The aforementioned data (and shown in Table 1) suggest that TME instructs the development, properties, plasticity, maintenance and dissemination of CCSC. In the last decade, the remarkable influence of the stroma on CRC development prompted the postulation of a novel classification of this disease based on its impact on tumor gene expression[5]. This CRC staging contains four consensus molecular subtypes (CMS) plus a group called "unclassified" since their features do not fit into the other CMS. All these subtypes are summarized in Table 2[1,5,77-79]. As the reader can see in this table, the influence of TME determines a low or high degree of immune and inflammatory response depending on the CMS, highlighting the importance of factors from TME in the distinctive characteristics of each CRC subtype. Taking into account that the mentioned inflammatory/immune process (that is relevant for CRC classification) can be influenced by the intestinal microorganisms, next we will discuss the interactions of this microbiota with tumor cells and their microenvironment that modulate the behavior and characteristics of CCSC since it is the focus of this review.
| CMS1-immune (14%) | CMS2-canonical (37%) | CMS3-metabolic (13%) | CMS4-mesenchymal (23%) | Unclassified (13%) | |
| General features | Hypermutated | Epithelial | Epithelial | TGF-β activation. Angiogenesis | Mixed phenotype of multiple CMS |
| Microsatellite unstable | WNT and MYC signaling activation | Metabolic dysregulation | Upregulation of EMT | ||
| Mutations | BRAF, MSH6, RNF43, ATM, TGFBr2, PTEN | APC, KRAS, TP53, PIK3CA | APC, KRAS, TP53, PIK3CA | APC, KRAS, TP53, PIK3CA | |
| TME | Decrease of CAFs | Decrease of CAFs | Decrease of CAFs | Increase of CAFs; Immunosuppressive signature | |
| High immune and inflammatory signature | Low immune and inflammatory signature | Low immune and inflammatory signature |
As we mentioned in this work, the inflammatory microenvironment contributes to promoting CRC initiation and progression. However, the role of the cell types involved in this process, including intestinal microorganisms, has not been completely understood yet.
The human microbiome, a concept that is mentioned throughout this section, represents microorganisms with their genetic elements and the interactions arising with the environment in which they are found[80]. Advances in the characterization of this human microbiome have led to the consideration that the role of the microbiota in metabolic functions and maintenance of homeostasis is more important than previously believed. Currently, the human is considered as a holobiont organism inhabited by millions of microorganisms including bacteria, archaea and fungi[81]. The gut microbiota is a complex ecosystem that contains more than 500 bacteria species involved in physiological processes like immune regulation and maintenance of human health[6] and its composition relies fundamentally on diet and lifestyle[74].
In physiological conditions, stromal and immune cells from the gut mucosa interact with this ecosystem to maintain intestinal equilibrium[82]. Cells from the immune system recognize antigens from foreign cells and generate memory and effector cells, which control or avoid the generation of diseases[82].
It has been observed that sustained shifts in this ecosystem, known as intestinal dysbiosis, have unfavorable repercussions on health[74,83]. In this sense, the presence of harmful microorganisms (“drivers”) could induce changes in the intestinal mucosa and favor the colonization by opportunistic bacteria (“passengers”)[84]. This model is known as driver-passenger[84] and could involve changes in the immune system allowing the advance of the damage in the intestinal epithelium tissue[85,86]. This imbalance of the local microbiota promotes the restructuring of the intestinal environment and alters the immune status of the host contributing to the appearance of malignant cells and a favorable niche for tumor development, invasion and metastasis[85,87,88]. The mechanisms of these microorganisms that influence directly the immune system are different and involve the synthesis of immunomodulatory compounds and metabolites, like short-chain fatty acids (SCFAs), polyamines and other fermentation products[89,90]. Moreover, it is known that the intratumoral composition of microorganisms affects T-cell-mediated cytotoxicity and anti-tumor immune surveillance[91]. The unfavorable changes in the intestinal microbiota can promote a pro-inflammatory environment and impair anti-cancer immunity[91]. In this context, cells from TME secrete factors like interferon-γ, TGF-β, IL-6, IL-8, CXCL1 and
The increased expression of PD-L1 in CRC cells both in vitro and in vivo is a mechanism involved in the influence of certain pathogenic bacteria associated with an immunosuppressive TME[96]. In contrast, bacteria associated with healthy microbiota improve the efficacy of anti-PD-L1 therapy by enhancing the accumulation of cytotoxic T cells in the TME[97]. This suggests that TME reprogramming through manipulation of the microbiota can modulate the response to immunotherapies in CRC[98]. Concerning all this information, CRC could be considered as a bacterial-induced disease and disturbance in microbiota could be potentially useful as diagnostic biomarker, indicator of risk and predictor of response to therapies for this type of cancer[74,88].
On the other hand, CRC modifies the local metabolic environment[99]. In this context, it is important to mention that metabolites and factors derived from CRC cells and TME cells such as spermidine, L-valine, L-lysine or stearic acid confer an advantage for the growth and development of certain bacterial species, conditioning changes in the intestinal microbiota[99]. Although different factors produce changes in gut microbiota, recently it has been seen that the shift in the metabolome of tumor cells and TME cells is a key aspect in this event[86,99,100]. Thus, TME can be the consequence or the cause of intestinal dysbiosis.
The gut microorganisms cited below in this section are described in the available literature due to their role in CCSC development and maintenance. They are also summarized in Table 3.
| Microorganism | Action | Ref. |
| Bacterioides dorei, Bacterioides vulgatum, Parabacterioides distasonis, Lachnoclostridium sp., and Mordavella sp | Inhibit the action of factors related to CCSC phenotype. Inhibit CRC development and progression | [59,114] |
| Bacterioides fragilis | Releases an enterotoxin that promotes immune TME cells activation with secretion of factors related to CCSC | [118] |
| Citrobacter rodentium | Protects the inflammatory CCSC niche | [121] |
| Clostridium septicum | Contributes to CRC development and to the activation of signaling pathways associated with CCSC | [59] |
| Enterococcus faecalis | Induces the expression of TGF-β, thereby activating signaling pathways associated with CCSC. Activates Wnt/β-catenin signaling and pluripotent transcription factors associated with CCSC | [113,115] |
| Escherichia coli | Upregulates the expression of CCSC-associated genes. Releases genotoxin colibactin which induces the production of growth factors related to CCSC | [112,117,79] |
| Fusobacterium nucleatum | Stimulates the secretion of immune factors related to CCSC | [79] |
| Helicobacter pylori | Promotes the expression of markers associated with stemness | [101,102] |
| Lactobacillus acidophilus | Promotes proliferation or death in CCSC depending on dose | [104] |
| Porphyromonas gingivalis | Promotes the expression of markers associated with stemness | [101,102] |
| Shigella, and Citrobacter | Upregulate the expression of CCSC-associated genes | [112] |
Regarding CSC properties, some pathogenic bacteria such as Helicobacter pylori and Porphyromonas gingivalis can promote the expression of markers associated with stemness such as CD44 and CD133 in gastrointestinal tumors[101,102]. This association between the presence of certain bacteria genera in the gut and the expression of CSC markers has led to the study of the effects of microorganisms shifts and bacterial metabolites on CRC. Several models of tumorigenesis induced by bacteria have been proposed, suggesting how the interactions of host-microorganism promote the development and progression of this type of cancer[101]. In fact, it is known that the metabolites from the intestinal microbiota have the potential to act as tumorigenic factors. However, others can act as anti-tumorigenic factors since many of these microbiota-derived products are capable of inhibiting CRC progression[103]. Kim et al[43] have demonstrated that ursodeoxycholic acid, a secondary bile acid produced by Clostridium species, including Clostridium absonum and Clostridium baratii, regulates the oxidative stress suppressing CCSC growth and CRC cells proliferation[43]. Moreover, it has been observed that niacin, a product of the metabolism of some intestinal bacteria, such as Lactobacillus acidophilus, has different effects on CCSC. Depending on the dose, this vitamin can promote proliferation or death in this cell subtype[104]. Additionally, bowel microorganisms produce SCFAs such as butyrate, propionate and acetate[92]. It has been reported that these SCFAs favor beneficial bacteria proliferation and stimulate regulatory T cells to reduce inflammatory mediators, regulating immune response[105]. Butyrate participates in epithelial integrity maintenance and has antitumor effects. Several investigations show that in CRC, this product inhibits events associated with CSC such as invasion and proliferation[6]. Interestingly, butyrate inhibits cell proliferation to a greater extent in CRC derived cells than in non-cancerous cells[92]. Although butyrate was reported as an anti-tumor and chemopreventive agent[92,106], other studies have shown that it has variable outcomes on CCSC[92]. So, more investigations are necessary to determine the mechanistic action of this type of fatty acid. Experiments with other SCFAs like acetate and propionate with similar results demonstrated that these acids have opposing effects[6,107]. Besides that, a large number of microbial products such as deoxycholic acid, lithocholic acid chenodeoxycholic acid, taurochenodeoxycholic acid and others, are associated with the promotion of gastrointestinal tumors including CRC[6,108]. Recent studies have found that in CRC patients the microbial composition of the colonic crypt is different from that of the intestinal lumen. In the environment of the crypt of the colorectal tumor, groups such as Proteobacteria and anaerobes, such as Acinetobacter, Stenotrophomonas and Delftia were found[109]. Therefore, specific microorganisms could have a role in the maintenance of CCSC, located in the crypt, through the production of specific metabolites[110]. However, more studies are needed in this field since the molecular mechanisms underlying the effects of intestinal microbial products on CCSC have not yet been fully elucidated.
Currently, the study of mechanisms involved in the communication between the microbiota, the tumor cells and their microenvironment has gained impact on CRC. One reported mechanism for this interaction is through pattern recognition receptors located on intestinal epithelial cells that have the ability to detect distinctive microbial macromolecular ligands called pathogen-associated molecular patterns such as lipopolysaccharides and peptidoglycans[111]. Congruently, a recent work documented an altered function of CSC in a CRC murine model due to intruding bacteria like Escherichia, Shigella, and Citrobacter. This effect results in the activation of a toll-like receptor (TLR), a class of pattern recognition receptors, and the consequent upregulation of stem cell-associated genes such as Cd44v6 and Lgr5[110,112]. In line with this, the microorganisms are capable of activating several signaling pathways in tumor cells and/or TME cells inducing the secretion of factors associated with CCSC features. In this context, it has been observed in a murine model that microorganisms such as Enterococcus faecalis cause colitis after infection and induce expression of TGF-β, thereby activating the Smad signaling pathway[113]. A recent study has demonstrated an inverse correlation between the expression of molecules associated with TGF-β signaling pathway and stem cells- related genes in CRC. Moreover, the authors of this work have compared feces from mice with defects in TGF-β signaling with feces from wild-type (WT) mice, and have shown that the first ones had increased bacterial species associated with the development and progression of CRC, such as Clostridium septicum, and diminished amounts of favorable microorganisms including Bacteroides vulgatus and Parabacteroides distasonis[59]. Similar results were obtained by Wang and collaborators who showed that the amounts of beneficial species (Bacterioides dorei, Lachnoclostridium sp., and Mordavella sp.) are recovered in WT mice but not in those with mutated TGF-β signaling after chemotherapy treatment[114]. These investigations demonstrate the close relationship between the microbiota, the production and release of TGF-β and CCSC in the tumor.
Concerning other signaling pathways, Wang et al[115] have shown that Enterococcus faecalis are capable of polarizing macrophages by activating Wnt/β-catenin signaling and pluripotent transcription factors associated with the dedifferentiation, reprogramming and development of CCSC such as cellular myelocytomatosis oncogene, Kruppel-like factor 4, octamer-binding transcription factor 4 (Oct4), and Sox2[115]. These events respond to the microbiota-induced bystander effect theory based on the fact that macrophages induce genetic mutations and chromosomal instability in intestinal cells[116].
Other signaling pathways associated with pro-inflammatory and growth factors can be activated in response to bacterial products. For instance, the unbalance in the amount of the gut bacteria Escherichia coli, correlates with CRC progression by producing the genotoxin colibactin[79]. This toxin accelerates tumor progression and involves the production of growth factors related to CCSC, such as the HGF and the consequent activation of its signaling pathway[79,117]. Also, the enterotoxin produced by Bacteroides fragilis promotes immune TME cells activation with the secretion of IL-17 which favors CCSC properties[118]. Furthermore, as we have previously mentioned, gut microorganisms shape the immune environment promoting tumor evolution and CCSC features. For example, Fusobacterium nucleatum stimulates IL-8 secretion by TME cells and the inhibition of T and NK cell functions[79]. This bacteria has been deeply studied, since clinical analysis of specimens from CRC patients showed that the levels of F. nucleatum are significantly higher in neoplastic tissues than in adjacent normal tissues, and correlate with tumor invasion and metastasis[119]. These results support the role of F. nucleatum in the regulation of CCSC plasticity and EMT[101]. Also, it is known that F. nucleatum and other microorganisms like Epstein–Barr virus are capable of incorporating human ncRNAs favoring microbial growth[74]. In this regard, Tarallo et al[120] found a human and microbial ncRNA signature in CRC in which many miRs associated with CSC features, are overexpressed including miR-21 and miR-200[74,120]. A recent study conducted by Wang showed that Citrobacter rodentium infection induces the inhibition of miR-34a, which protects the inflammatory CCSC niche[121]. These investigations suggest a close relationship between the intestinal microbiota and the regulation of ncRNAs involved in CCSC properties.
Finally, not only the shift in the number of microorganisms is responsible for stemness and CRC progression, but the interaction and collaboration between several types of bacteria in biofilm communities also participate in bowel inflammation and CRC. It was demonstrated that biofilms correlate with an increase in IL-6 secretion by TME cells playing a key role in proliferation, cell transformation and stemness[79].
The data in this section demonstrate a close interrelationship between the gut microbiota, the TME, and CCSC. This information highlights the relevance of further investigating the intestinal microbiota switch in patients with CRC and the associated mechanisms that lead to TME changes and promote stemness.
Standard chemotherapeutic approaches for CRC are based on attacking the replicative mechanisms of tumor cells to induce tumor regression. However, considering CSC properties, this subpopulation usually results unharmed by the treatment because they present a low division rate as well as a great capacity to correct DNA defects[122]. This entails therapy resistance of CSC and the subsequent treatment failure and disease progression. It is interesting to note that in CRC, CSC represent around 2.5% of neoplastic cells but due to their phenotypic plasticity, they constitute a dynamic population[123,124]. This fact, together with the lack of response to therapies, highlights the need of new clinical strategies targeting CCSC[125].
As we explain throughout this review, the influence of the TME and the intestinal microorganisms on CSC properties makes these factors a promising tool in therapy. Many therapeutic agents are capable of inhibiting those events associated with the maintenance of CCSC. For instance, Apatinib napabucasin, Bigelovin, Wogonin and Metformin are drugs whose mechanisms are associated with the inhibition of EMT or angiogenesis in CRC[1]. Moreover, it has been demonstrated that therapeutic agents such as Genistein cause the inhibition of CSC characteristics by glioma-associated oncogene1 signaling pathway[126]. Targeting the activation of those signaling pathways associated with CCSC can also be considered as a mechanism to reduce stemness in CRC tumors and thus improve the response to the therapy. LGK974, Foxy-5, PRI-724[127] and DKN-01[126] are agents that act targeting the Wnt/β-catenin pathway. However, the clinical application of most of these drugs is still under study.
The tumor protective niche also could be modified to eradicate CCSC and overcome chemoresistance. As we have mentioned in previous sections, in the TME, immune cells modulate cancer development and progression. For that reason, in the last decade the treatment of patients with immune checkpoint inhibitors such as CTLA-4 and PD-1/PD-L1 has been studied. Even though employing these drugs leads to various systemic and organic complications, immunotherapy may be promising in sorting these obstacles and could ameliorate the response of CRC patients to the treatment[1,128]. In fact, coadjuvant therapy with FOLFOX (a combination of leucovorin, fluorouracil and oxaliplatin which are first-line chemotherapeutic drugs)[129], and PD-1/PD-L1 inhibitors had an objective response rate of 50% in clinical trials[130]. In addition, in a phase II trial in CRC metastatic patients, immune checkpoints inhibitors like nivolumab and nivolumab-ipilimumab show improvement in patients survival rate[130]. Moreover, monoclonal antibodies against CAFs and antifibrotic drugs were also tested in clinical studies[5]. Another type of antitumor therapy was accomplished through the production of a cell-based vaccine with specific antigens of CCSC[5].
In addition, plenty of compounds were designed in the last decade to target CCSC signaling pathways[5]. These strategies include the inhibition of HH signaling components, NOTCH pathway inhibitors, anti-angiogenic agents and Wnt ligand blockers. All these drugs are undergoing clinical trials[129]. Despite being an encouraging strategy, it still has limitations like the inhibition of signaling pathways involved in physiologic processes.
In the last years, the particularities exhibited by extracellular vesicles (EVs) have led researchers to consider them as a therapeutic delivery strategy of great value in CRC and other types of tumors. Within the different types of EVs are the exosomes, which are secreted by a variety of cells. These vesicles carry out the molecular content of donor cells and enable cellular communication over short and long distances. These EVs are loaded with coding nucleic acids, ncARNs and bioactive proteins which determine their functions. Exosomes can target a specific tissue and internalize in a cell type by the recognition of surface ligands/receptors[131]. In this regard, Han et al[132] investigated the delivery of human cord blood-derived MSC exosomes loaded with miRs as CRC targeted therapy. The results showed an inhibition of tumor growth in vitro and in vivo, as well as a selective increase of these ncRNAs in CRC cells[132]. The relation between miRs and CCSC was mentioned in previous sections so their delivery may be strong weapons to confront drug resistance and CCSC maintenance[5]. Circular RNAs are ncRNAs that exhibit cell-type and tissue-specific signatures. There has recently been considerable attention on these ncRNAs as they modulate miRs expression[129]. In CRC, recent studies have focused on their study as biomarkers. However, they have not been applied in patients’ therapy yet[133,134]. Moreover, the importance that these small molecules could have in CRC is unknown[129].
Foods containing biologically active ingredients are termed functional foods or nutraceuticals[135,136]. In the past years, the influence of diet on CRC development and evolution was demonstrated. A diet with natural products like phytochemicals and nutritional herbs has shown protective effects in overcoming CRC associated dysbiosis[137,138]. Diets enriched in dairy are a major source of products that are known to have a protective effect on CRC development such as, calcium, vitamin D and folate[138]. Sulforaphane, a sulfur-rich compound found in cruciferous vegetables like broccoli, has been documented to diminish CSC markers and improve the chemotherapeutic efficacy of drugs commonly used in CRC treatment such as cisplatin, doxorubicin and fluorouracil[137]. It has been observed that dietary polyphenols like quercetin have similar effects[137,139]. Other polyphenols or flavonoids are known to target ABCG-2 transporters and miRs strictly associated with CCSC[139]. Curcumin is one of several substances present in turmeric plants. It has been demonstrated that this bioactive agent inhibits the activation of several signaling pathways related to CSC characteristics. The treatment with this natural product on a CSC model diminished the expression of CD44 and CD133 markers[137]. Moreover, some other natural products have been observed that interfere with intrinsic CSC pathways, like epigallocatechin-3-gallate (EGCG), resveratrol and genistein[140].
Diet can also manipulate the gut microbiota. Indeed, this is achieved by the administration of probiotics in the diet. As probiotics and their active metabolites can exert immunomodulatory and anti-tumorigenic effects[135], the study of them and their metabolites has gained ground in recent decades. Probiotics are live microorganisms, normally lactic acid bacteria, recognized as safe by the United States Food and Drug Administration[135]. Defined as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host”[141], they can improve health by administration along vegetable fibers and other prebiotics stimulating beneficial bacterial growth in the intestine[142].
Probiotics administration can be done by different routes, commonly through functional foods, but also by commercial supplements or vaccines[135,138]. It is known that probiotic oral vaccines promote mucosal immunity that prevents enteric infections and could complement the standard therapy in the patient[143]. Microorganisms administration including probiotics and synbiotics (pharmaceutical preparation that contains probiotics and prebiotics that implies a synergy between both) are a potential resource for prophylaxis and therapy in CRC[138,144]. In addition, the luminal cocktail of microorganisms in the bowel can be modified not only by dietary approaches but also with the use of antibiotics or fecal microbiota transplantation (FMT)[145,146]. In particular, FMT has gained considerable interest in recent years as a strategy to treat different gastrointestinal disorders[147-149]. It consists of introducing a healthy microbial population from a disease-free host into a diseased host that has a dysbiotic community to restore microbial homeostasis[150]. Although there are limited data on the use of FMT in the treatment of CRC, several studies are under development to answer relevant questions such as if CRC can be detected, treated or prevented with this method. Rosshart and collaborators observed that mice treated with this method improved their resistance against colorectal tumorigenesis induced by azoximetane[151]. Besides, it has been seen that FMT in Balb-c mice prevents intestinal damage, and chemotherapy-induced toxicity[152]. Interestingly, the fecal microbiota from CRC patients has been shown to cause tumors in healthy and germen-free Apcmin/+ mice through the activation of the Wnt signaling pathway. In these mice, the intestinal barrier is also altered and the presence of pro-inflammatory cytokines is increased[153]. These data reveal that the composition of the microbiota may play a determinant role in TME conditions during tumorigenesis. Nevertheless, the subjacent mechanisms of all these treatments or how they ameliorate the side effects of chemotherapy is not clear yet.
In summary, we need a favorable and efficient clearance of tumor cells, all tumorigenic cells including CCSC and a restructuring of the TME for the complete eradication of CRC. Based on everything described in this review, a specific combination of techniques and therapies for each tumor and patient would be necessary to achieve this goal.
According to the information stated in the previous sections, in CRC occurs an alliance between the TME, intestinal microorganisms and CCSC that favors tumor progression. In this scenario, it is emerging a new query regarding the direct effects of CCSC on gut microbiota. Perhaps the appearance of CCSC by spontaneous mutations favors (through paracrine signals and the release of specific factors) a dysbiotic and pro-inflammatory environment but in this regard, new investigations are necessary to evaluate the regulation of CCSC on CRC microbiota. So, there is great potential in the study of the interrelationship between these three components in the tumor niche, mostly for the development of new therapies aimed at the eradication of CCSC and non-stem cells, the restructuring of the TME and the growth induction of microorganisms that are beneficial to the intestinal mucosa.
Many of the therapies currently in use or under clinical evaluation are associated with systemic toxicity since they do not act on a well-defined target[137]. Therefore, the combination of radiotherapy and chemotherapy has still remained the strategy of choice in CRC[145] and not much attention is paid to nutritional accompaniment. Since the gut microbiota seems to be a pivotal factor in inflammatory disease and CRC development, overcoming therapy resistance could also improve with changes in diet. For this purpose, is crucial the development of foods containing compounds with anti-CCSC activity such as flavonoids but with better bioaccessibility and bioavailability[154]. Moreover, bacteriotherapy is a great opportunity to customize CRC treatment and the following tools that we will mention could be useful in this type of therapy. The modification of patient microbiome tending to resolve dysbiosis through the administration of beneficial bacteria could significantly improve conventional treatment[93]. Even more, considering that some microbial species exhibit tumor targeting specificity, this strategy could ameliorate cytotoxicity in non-tumor cells. Regarding bacterial products, given their low molecular weight and hydrophobicity, they can easily enter tumor tissues and exert their action[155]. These features result in the use of microorganisms with potential preventive or palliative action in CRC currently receiving special attention. In fact, microbe-based therapies, and bacteria-mediated modulatory strategies are studied to be used for the delivery of drugs to the tumor site and to produce anti-cancer vaccines[145,155]. However, the information about the toxins, metabolism of microbial-derived agents and complications from bacteriotherapy is still limited155]. Thus, placing emphasis on clinical research that allows the use of these new therapies, overcoming the obstacles related to it, will be essential in the coming years.
In addition, as we discussed in the previous section, it is also necessary to focus on the restructuring of the TME in favor of improving conventional CRC treatment. Restructuring the extracellular matrix, modulating the immune response with vaccines, antibodies, or inhibitory drugs, employing drugs that induce changes in the secretion profile of TME cells, switching macrophages polarization and inhibiting CAFs and processes like fibrosis and inflammation are some of the potential effective techniques under investigation[1,5,116,128].
The development of vaccines containing CSC-specific antigens is also under investigation[5]. However, since many of the antigens present in this cell subtype are also found in differentiated cells or normal stem cells, this is a challenge to overcome for successful therapy.
So, the combination of conventional therapies with new targeted inhibitors (e.g. inhibitors of signaling pathways or molecules derived from TME) plus an appropriate diet that favors beneficial colonic microbiota, as well as the use of targeting methods such as charged nanoparticles or specific bacterial species, could constitute a reliable alternative to fight with CRC chemoresistance and relapses. The use of different in vitro and in vivo preclinical models of CCSC such as colonospheras, organoids and xenografts, is essential to achieve this goal and bring it to clinical research.
In the near future, the challenge will be the development of selective and combined therapies to promote: (1) CSC eradication; (2) Eradication of cancer cells, owing to their phenotypic plasticity, even in the absence of CSC features; and (3) Reduction of the damage to cells outside the tumor bulk.
In any case, it is clear that the standardization of treatment protocols is not always effective for this disease. It is advisable to resort to a combined and personalized therapy that considers the needs and responses of each patient.
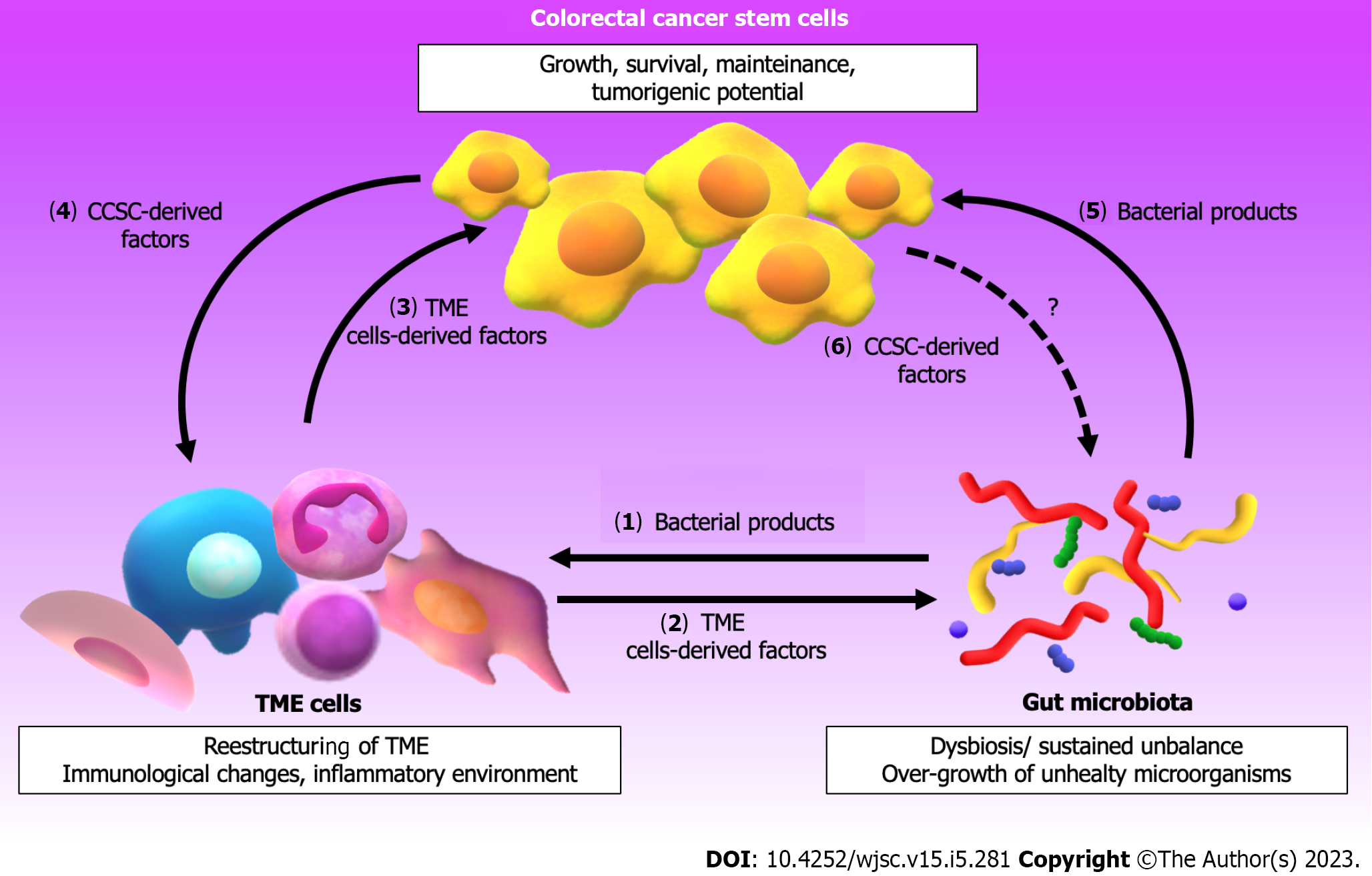
Figure 2 shows the interplay between the TME and the gut microbiota that influences the properties/behavior of CCSC. Besides, the reader can appreciate that CCSC influence on cells from TME favoring CRC progression but probably also on gut microbiota. The knowledge described in the present review provides data that may promote future research aimed at addressing the complexity of the components in the CRC-associated microenvironment and microbiota. Compounding such complexity, CRC is not an isolated neoplasm, but it’s rather emerging as a dynamic pathology whose actors are capable, regrettably, of contributing to evasion mechanisms of the current therapeutic strategies. Although the incidence and mortality from CRC have decreased in recent years, a large number of patients still suffer from relapses due to resistance to treatment. The development of metastases and chemoresistance is undoubtedly one of the greatest challenges in CRC therapy. As we have seen in this work, the properties of the CCSC make this cell subtype have the main responsibility for the recurrences. The shift in the tumor niche and the intestinal microbiota favors the acquisition of CSC characteristics, promoting a worse prognosis of CRC. Although much is currently known about the interrelationship between components of the TME, the microorganisms present in the intestinal mucosa and CCSC, there is still much to be discovered in this field.
The authors would like to thank Antonela Rossi Bertone for her helpful advice. Being a teacher of English graduated from Instituto Superior Juan XXIII (from Bahia Blanca city, Argentina) and currently working at San Cayetano school (in the same city), she revised the paper and made useful comments as regards the spelling, grammar and punctuation of this manuscript.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Cell and tissue engineering
Country/Territory of origin: Argentina
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gao W, China; Liu W, China; Peng XC, China; Ventura C, Italy S-Editor: Li L L-Editor: A P-Editor: Zhang XD
| 1. | Islas JF, Quiroz-Reyes AG, Delgado-Gonzalez P, Franco-Villarreal H, Delgado-Gallegos JL, Garza-Treviño EN, Gonzalez-Villarreal CA. Cancer Stem Cells in Tumor Microenvironment of Adenocarcinoma of the Stomach, Colon, and Rectum. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Reference Citation Analysis (0)] |
| 2. | Briede I, Balodis D, Gardovskis J, Strumfa I. Stemness, Inflammation and Epithelial-Mesenchymal Transition in Colorectal Carcinoma: The Intricate Network. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 3. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64722] [Article Influence: 16180.5] [Reference Citation Analysis (177)] |
| 4. | Novoa Díaz MB, Martín MJ, Gentili C. Tumor microenvironment involvement in colorectal cancer progression via Wnt/β-catenin pathway: Providing understanding of the complex mechanisms of chemoresistance. World J Gastroenterol. 2022;28:3027-3046. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 4] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 5. | Jahanafrooz Z, Mosafer J, Akbari M, Hashemzaei M, Mokhtarzadeh A, Baradaran B. Colon cancer therapy by focusing on colon cancer stem cells and their tumor microenvironment. J Cell Physiol. 2020;235:4153-4166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 97] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 6. | Fang Y, Yan C, Zhao Q, Xu J, Liu Z, Gao J, Zhu H, Dai Z, Wang D, Tang D. The roles of microbial products in the development of colorectal cancer: a review. Bioengineered. 2021;12:720-735. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 79] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 7. | Yadav VK, Huang YJ, George TA, Wei PL, Sumitra MR, Ho CL, Chang TH, Wu ATH, Huang HS. Preclinical Evaluation of the Novel Small-Molecule MSI-N1014 for Treating Drug-Resistant Colon Cancer via the LGR5/β-catenin/miR-142-3p Network and Reducing Cancer-Associated Fibroblast Transformation. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 8. | Weinberg B, Marshall J. L. Colon Cancer in Young Adults: Trends and Their Implications. Curr Oncol Rep. 18 ;21:3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 87] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 9. | Wang S, Miao Z, Yang Q, Wang Y, Zhang J. The Dynamic Roles of Mesenchymal Stem Cells in Colon Cancer. Can J Gastroenterol Hepatol. 2018;2018:7628763. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 10. | Stastna M, Janeckova L, Hrckulak D, Kriz V, Korinek V. Human Colorectal Cancer from the Perspective of Mouse Models. Genes (Basel). 2019;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 11. | Koliaraki V, Pallangyo CK, Greten FR, Kollias G. Mesenchymal Cells in Colon Cancer. Gastroenterology. 2017;152:964-979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 152] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 12. | Tauriello DV, Calon A, Lonardo E, Batlle E. Determinants of metastatic competency in colorectal cancer. Mol Oncol. 2017;11:97-119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 183] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 13. | Chruścik A, Gopalan V, Lam AK. The clinical and biological roles of transforming growth factor beta in colon cancer stem cells: A systematic review. Eur J Cell Biol. 2018;97:15-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 14. | Robinson BD, Sica GL, Liu YF, Rohan TE, Gertler FB, Condeelis JS, Jones JG. Tumor microenvironment of metastasis in human breast carcinoma: a potential prognostic marker linked to hematogenous dissemination. Clin Cancer Res. 2009;15:2433-2441. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 284] [Cited by in RCA: 262] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 15. | Hung KF, Yang T, Kao SY. Cancer stem cell theory: Are we moving past the mist? J Chin Med Assoc. 2019;82:814-818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 16. | Dzobo K, Senthebane DA, Ganz C, Thomford NE, Wonkam A, Dandara C. Advances in Therapeutic Targeting of Cancer Stem Cells within the Tumor Microenvironment: An Updated Review. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 75] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 17. | Najafi M, Mortezaee K, Majidpoor J. Cancer stem cell (CSC) resistance drivers. Life Sci. 2019;234:116781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 295] [Article Influence: 49.2] [Reference Citation Analysis (0)] |
| 18. | Vincent A, Ouelkdite-Oumouchal A, Souidi M, Leclerc J, Neve B, Van Seuningen I. Colon cancer stemness as a reversible epigenetic state: Implications for anticancer therapies. World J Stem Cells. 2019;11:920-936. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (1)] |
| 19. | López-Lázaro M. The stem cell division theory of cancer. Crit Rev Oncol Hematol. 2018;123:95-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 20. | Silva VR, Santos LS, Dias RB, Quadros CA, Bezerra DP. Emerging agents that target signaling pathways to eradicate colorectal cancer stem cells. Cancer Commun (Lond). 2021;41:1275-1313. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 70] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 21. | Chen X, Wang Y, Feng T, Yi M, Zhang X, Zhou D. The overshoot and phenotypic equilibrium in characterizing cancer dynamics of reversible phenotypic plasticity. J Theor Biol. 2016;390:40-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 22. | De Angelis ML, Francescangeli F, Zeuner A, Baiocchi M. Colorectal Cancer Stem Cells: An Overview of Evolving Methods and Concepts. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 23. | Eun K, Ham SW, Kim H. Cancer stem cell heterogeneity: origin and new perspectives on CSC targeting. BMB Rep. 2017;50:117-125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 177] [Cited by in RCA: 255] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 24. | Vaiopoulos AG, Kostakis ID, Koutsilieris M, Papavassiliou AG. Colorectal cancer stem cells. Stem Cells. 2012;30:363-371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 180] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 25. | Merlos-Suárez A, Barriga FM, Jung P, Iglesias M, Céspedes MV, Rossell D, Sevillano M, Hernando-Momblona X, da Silva-Diz V, Muñoz P, Clevers H, Sancho E, Mangues R, Batlle E. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 666] [Cited by in RCA: 742] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 26. | Roudi R, Barodabi M, Madjd Z, Roviello G, Corona SP, Panahei M. Expression patterns and clinical significance of the potential cancer stem cell markers OCT4 and NANOG in colorectal cancer patients. Mol Cell Oncol. 2020;7:1788366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 27. | Leng Z, Xia Q, Chen J, Li Y, Xu J, Zhao E, Zheng H, Ai W, Dong J. Lgr5+CD44+EpCAM+ Strictly Defines Cancer Stem Cells in Human Colorectal Cancer. Cell Physiol Biochem. 2018;46:860-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 57] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 28. | Shafaei S, Sharbatdaran M, Kamrani G, Khafri S. The association between CD166 detection rate and clinicopathologic parameters of patients with colorectal cancer. Caspian J Intern Med. 2013;4:768-772. [PubMed] |
| 29. | Zhou X, Geng L, Wang D, Yi H, Talmon G, Wang J. R-Spondin1/LGR5 Activates TGFβ Signaling and Suppresses Colon Cancer Metastasis. Cancer Res. 2017;77:6589-6602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 30. | Wang Y, Chen Y, Garcia-Milian R, Golla JP, Charkoftaki G, Lam TT, Thompson DC, Vasiliou V. Proteomic profiling reveals an association between ALDH and oxidative phosphorylation and DNA damage repair pathways in human colon adenocarcinoma stem cells. Chem Biol Interact. 2022;368:110175. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 31. | Tomita H, Tanaka K, Tanaka T, Hara A. Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget. 2016;7:11018-11032. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 321] [Cited by in RCA: 436] [Article Influence: 54.5] [Reference Citation Analysis (0)] |
| 32. | Guo Q, Grimmig T, Gonzalez G, Giobbie-Hurder A, Berg G, Carr N, Wilson BJ, Banerjee P, Ma J, Gold JS, Nandi B, Huang Q, Waaga-Gasser AM, Lian CG, Murphy GF, Frank MH, Gasser M, Frank NY. ATP-binding cassette member B5 (ABCB5) promotes tumor cell invasiveness in human colorectal cancer. J Biol Chem. 2018;293:11166-11178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 33. | Alfaro Alfaro ÁE, Murillo Castillo B, Cordero García E, Tascón J, Morales AI. Colon Cancer Pharmacogenetics: A Narrative Review. Pharmacy (Basel). 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 34. | Jelic MD, Mandic AD, Maricic SM, Srdjenovic BU. Oxidative stress and its role in cancer. J Cancer Res Ther. 2021;17:22-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 352] [Article Influence: 88.0] [Reference Citation Analysis (0)] |
| 35. | Ning X, Wang C, Zhang M, Wang K. Ectopic Expression of miR-147 Inhibits Stem Cell Marker and Epithelial-Mesenchymal Transition (EMT)-Related Protein Expression in Colon Cancer Cells. Oncol Res. 2019;27:399-406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 36. | Takigawa H, Kitadai Y, Shinagawa K, Yuge R, Higashi Y, Tanaka S, Yasui W, Chayama K. Mesenchymal Stem Cells Induce Epithelial to Mesenchymal Transition in Colon Cancer Cells through Direct Cell-to-Cell Contact. Neoplasia. 2017;19:429-438. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 65] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 37. | Li W, Cho MY, Lee S, Jang M, Park J, Park R. CRISPR-Cas9 mediated CD133 knockout inhibits colon cancer invasion through reduced epithelial-mesenchymal transition. PLoS One. 2019;14:e0220860. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 38. | Pouyafar A, Rezabakhsh A, Rahbarghazi R, Heydarabad MZ, Shokrollahi E, Sokullu E, Khaksar M, Nourazarian A, Avci ÇB. Treatment of cancer stem cells from human colon adenocarcinoma cell line HT-29 with resveratrol and sulindac induced mesenchymal-endothelial transition rate. Cell Tissue Res. 2019;376:377-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 39. | Tanabe S, Quader S, Cabral H, Ono R. Interplay of EMT and CSC in Cancer and the Potential Therapeutic Strategies. Front Pharmacol. 2020;11:904. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 112] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 40. | Zakaria S, Elsebaey S, Allam S, El-Sisi A. Modulating the Siah2-PHD3-HIF1α axis and/or autophagy potentially retard colon cancer proliferation possibly, due to the damping of colon cancer stem cells. Biomed Pharmacother. 2022;154:113562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 41. | Gopal K, Gupta N, Zhang H, Alshareef A, Alqahtani H, Bigras G, Lewis J, Douglas D, Kneteman N, Lavasanifar A, Lai R. Oxidative stress induces the acquisition of cancer stem-like phenotype in breast cancer detectable by using a Sox2 regulatory region-2 (SRR2) reporter. Oncotarget. 2016;7:3111-3127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 42. | Saijo H, Hirohashi Y, Torigoe T, Horibe R, Takaya A, Murai A, Kubo T, Kajiwara T, Tanaka T, Shionoya Y, Yamamoto E, Maruyama R, Nakatsugawa M, Kanaseki T, Tsukahara T, Tamura Y, Sasaki Y, Tokino T, Suzuki H, Kondo T, Takahashi H, Sato N. Plasticity of lung cancer stem-like cells is regulated by the transcription factor HOXA5 that is induced by oxidative stress. Oncotarget. 2016;7:50043-50056. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 43. | Kim EK, Cho JH, Kim E, Kim YJ. Ursodeoxycholic acid inhibits the proliferation of colon cancer cells by regulating oxidative stress and cancer stem-like cell growth. PLoS One. 2017;12:e0181183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 44. | Hirata A, Hatano Y, Niwa M, Hara A, Tomita H. Heterogeneity of Colon Cancer Stem Cells. Adv Exp Med Biol. 2019;1139:115-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 45. | Goodla L, Xue X. The Role of Inflammatory Mediators in Colorectal Cancer Hepatic Metastasis. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 46. | Borowczak J, Szczerbowski K, Maniewski M, Kowalewski A, Janiczek-Polewska M, Szylberg A, Marszałek A, Szylberg Ł. The Role of Inflammatory Cytokines in the Pathogenesis of Colorectal Carcinoma-Recent Findings and Review. Biomedicines. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 31] [Reference Citation Analysis (0)] |
| 47. | Huynh PT, Beswick EJ, Coronado YA, Johnson P, O'Connell MR, Watts T, Singh P, Qiu S, Morris K, Powell DW, Pinchuk IV. CD90(+) stromal cells are the major source of IL-6, which supports cancer stem-like cells and inflammation in colorectal cancer. Int J Cancer. 2016;138:1971-1981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 79] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 48. | Todaro M, Gaggianesi M, Catalano V, Benfante A, Iovino F, Biffoni M, Apuzzo T, Sperduti I, Volpe S, Cocorullo G, Gulotta G, Dieli F, De Maria R, Stassi G. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell. 2014;14:342-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 476] [Cited by in RCA: 579] [Article Influence: 52.6] [Reference Citation Analysis (0)] |
| 49. | Muñoz-Galván S, Lucena-Cacace A, Perez M, Otero-Albiol D, Gomez-Cambronero J, Carnero A. Tumor cell-secreted PLD increases tumor stemness by senescence-mediated communication with microenvironment. Oncogene. 2019;38:1309-1323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 50. | Quiroz-Reyes AG, Islas JF, Delgado-Gonzalez P, Franco-Villarreal H, Garza-Treviño EN. Therapeutic Approaches for Metastases from Colorectal Cancer and Pancreatic Ductal Carcinoma. Pharmaceutics. 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 51. | Cima I, Kong SL, Sengupta D, Tan IB, Phyo WM, Lee D, Hu M, Iliescu C, Alexander I, Goh WL, Rahmani M, Suhaimi NA, Vo JH, Tai JA, Tan JH, Chua C, Ten R, Lim WJ, Chew MH, Hauser CA, van Dam RM, Lim WY, Prabhakar S, Lim B, Koh PK, Robson P, Ying JY, Hillmer AM, Tan MH. Tumor-derived circulating endothelial cell clusters in colorectal cancer. Sci Transl Med. 2016;8:345ra89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 94] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 52. | Basu S, Haase G, Ben-Ze'ev A. Wnt signaling in cancer stem cells and colon cancer metastasis. F1000Res. 2016;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 137] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 53. | Vermeulen L, De Sousa E Melo F, van der Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M, Merz C, Rodermond H, Sprick MR, Kemper K, Richel DJ, Stassi G, Medema JP. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12:468-476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1301] [Cited by in RCA: 1447] [Article Influence: 96.5] [Reference Citation Analysis (1)] |
| 54. | Essex A, Pineda J, Acharya G, Xin H, Evans J; Reproducibility Project: Cancer Biology. Replication Study: Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Elife. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 55. | Kamal Y, Schmit SL, Frost HR, Amos CI. The tumor microenvironment of colorectal cancer metastases: opportunities in cancer immunotherapy. Immunotherapy. 2020;12:1083-1100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 56. | Unterleuthner D, Neuhold P, Schwarz K, Janker L, Neuditschko B, Nivarthi H, Crncec I, Kramer N, Unger C, Hengstschläger M, Eferl R, Moriggl R, Sommergruber W, Gerner C, Dolznig H. Cancer-associated fibroblast-derived WNT2 increases tumor angiogenesis in colon cancer. Angiogenesis. 2020;23:159-177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 197] [Cited by in RCA: 216] [Article Influence: 43.2] [Reference Citation Analysis (0)] |
| 57. | Villalba M, Evans SR, Vidal-Vanaclocha F, Calvo A. Role of TGF-β in metastatic colon cancer: it is finally time for targeted therapy. Cell Tissue Res. 2017;370:29-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 83] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 58. | Bellam N, Pasche B. Tgf-beta signaling alterations and colon cancer. Cancer Treat Res. 2010;155:85-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 119] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 59. | Gu S, Zaidi S, Hassan MI, Mohammad T, Malta TM, Noushmehr H, Nguyen B, Crandall KA, Srivastav J, Obias V, Lin P, Nguyen BN, Yao M, Yao R, King CH, Mazumder R, Mishra B, Rao S, Mishra L. Mutated CEACAMs Disrupt Transforming Growth Factor Beta Signaling and Alter the Intestinal Microbiome to Promote Colorectal Carcinogenesis. Gastroenterology. 2020;158:238-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 60. | Lombardo Y, Scopelliti A, Cammareri P, Todaro M, Iovino F, Ricci-Vitiani L, Gulotta G, Dieli F, de Maria R, Stassi G. Bone morphogenic protein 4 induces differentiation of colorectal cancer stem cells and increases their response to chemotherapy in mice. Gastroenterology. 2011;140:297-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 179] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 61. | Ouahoud S, Voorneveld PW, van der Burg LRA, de Jonge-Muller ESM, Schoonderwoerd MJA, Paauwe M, de Vos T, de Wit S, van Pelt GW, Mesker WE, Hawinkels LJAC, Hardwick JCH. Bidirectional tumor/stroma crosstalk promotes metastasis in mesenchymal colorectal cancer. Oncogene. 2020;39:2453-2466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 62. | Panda PK, Naik PP, Praharaj PP, Meher BR, Gupta PK, Verma RS, Maiti TK, Shanmugam MK, Chinnathambi A, Alharbi SA, Sethi G, Agarwal R, Bhutia SK. Abrus agglutinin stimulates BMP-2-dependent differentiation through autophagic degradation of β-catenin in colon cancer stem cells. Mol Carcinog. 2018;57:664-677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 63. | Catalano V, Dentice M, Ambrosio R, Luongo C, Carollo R, Benfante A, Todaro M, Stassi G, Salvatore D. Activated Thyroid Hormone Promotes Differentiation and Chemotherapeutic Sensitization of Colorectal Cancer Stem Cells by Regulating Wnt and BMP4 Signaling. Cancer Res. 2016;76:1237-1244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 65] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 64. | Qian Y, Wu X, Yokoyama Y, Okuzaki D, Taguchi M, Hirose H, Wang J, Hata T, Inoue A, Hiraki M, Ohtsuka M, Takahashi H, Haraguchi N, Mizushima T, Tanaka S, Mori M, Yamamoto H. E-cadherin-Fc chimera protein matrix enhances cancer stem-like properties and induces mesenchymal features in colon cancer cells. Cancer Sci. 2019;110:3520-3532. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 65. | Fender AW, Nutter JM, Fitzgerald TL, Bertrand FE, Sigounas G. Notch-1 promotes stemness and epithelial to mesenchymal transition in colorectal cancer. J Cell Biochem. 2015;116:2517-2527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 120] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 66. | Kawai S, Yamazaki M, Shibuya K, Fujii E, Nakano K, Suzuki M. Three-dimensional culture models mimic colon cancer heterogeneity induced by different microenvironments. Sci Rep. 2020;10:3156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 67. | Regan JL, Schumacher D, Staudte S, Steffen A, Haybaeck J, Keilholz U, Schweiger C, Golob-Schwarzl N, Mumberg D, Henderson D, Lehrach H, Regenbrecht CRA, Schäfer R, Lange M. Non-Canonical Hedgehog Signaling Is a Positive Regulator of the WNT Pathway and Is Required for the Survival of Colon Cancer Stem Cells. Cell Rep. 2017;21:2813-2828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 106] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 68. | Zhou H, Xiong Y, Peng L, Wang R, Zhang H, Fu Z. LncRNA-cCSC1 modulates cancer stem cell properties in colorectal cancer via activation of the Hedgehog signaling pathway. J Cell Biochem. 2020;121:2510-2524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 69. | Skoda AM, Simovic D, Karin V, Kardum V, Vranic S, Serman L. The role of the Hedgehog signaling pathway in cancer: A comprehensive review. Bosn J Basic Med Sci. 2018;18:8-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 507] [Article Influence: 72.4] [Reference Citation Analysis (0)] |
| 70. | Geyer N, Gerling M. Hedgehog Signaling in Colorectal Cancer: All in the Stroma? Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 71. | Calvo N, Carriere P, Martín MJ, Gigola G, Gentili C. PTHrP treatment of colon cancer cells promotes tumor associated-angiogenesis by the effect of VEGF. Mol Cell Endocrinol. 2019;483:50-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 72. | Dong HJ, Jang GB, Lee HY, Park SR, Kim JY, Nam JS, Hong IS. The Wnt/β-catenin signaling/Id2 cascade mediates the effects of hypoxia on the hierarchy of colorectal-cancer stem cells. Sci Rep. 2016;6:22966. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 64] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 73. | Kalra H, Gangoda L, Fonseka P, Chitti SV, Liem M, Keerthikumar S, Samuel M, Boukouris S, Al Saffar H, Collins C, Adda CG, Ang CS, Mathivanan S. Extracellular vesicles containing oncogenic mutant β-catenin activate Wnt signalling pathway in the recipient cells. J Extracell Vesicles. 2019;8:1690217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 67] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 74. | Zygulska AL, Pierzchalski P. Novel Diagnostic Biomarkers in Colorectal Cancer. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 135] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 75. | Jasso GJ, Jaiswal A, Varma M, Laszewski T, Grauel A, Omar A, Silva N, Dranoff G, Porter JA, Mansfield K, Cremasco V, Regev A, Xavier RJ, Graham DB. Colon stroma mediates an inflammation-driven fibroblastic response controlling matrix remodeling and healing. PLoS Biol. 2022;20:e3001532. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 66] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 76. | Ohta Y, Fujii M, Takahashi S, Takano A, Nanki K, Matano M, Hanyu H, Saito M, Shimokawa M, Nishikori S, Hatano Y, Ishii R, Sawada K, Machinaga A, Ikeda W, Imamura T, Sato T. Cell-matrix interface regulates dormancy in human colon cancer stem cells. Nature. 2022;608:784-794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 112] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 77. | Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, Bot BM, Morris JS, Simon IM, Gerster S, Fessler E, De Sousa E Melo F, Missiaglia E, Ramay H, Barras D, Homicsko K, Maru D, Manyam GC, Broom B, Boige V, Perez-Villamil B, Laderas T, Salazar R, Gray JW, Hanahan D, Tabernero J, Bernards R, Friend SH, Laurent-Puig P, Medema JP, Sadanandam A, Wessels L, Delorenzi M, Kopetz S, Vermeulen L, Tejpar S. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350-1356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3408] [Cited by in RCA: 3562] [Article Influence: 356.2] [Reference Citation Analysis (0)] |
| 78. | Becht E, de Reyniès A, Giraldo NA, Pilati C, Buttard B, Lacroix L, Selves J, Sautès-Fridman C, Laurent-Puig P, Fridman WH. Immune and Stromal Classification of Colorectal Cancer Is Associated with Molecular Subtypes and Relevant for Precision Immunotherapy. Clin Cancer Res. 2016;22:4057-4066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 409] [Article Influence: 45.4] [Reference Citation Analysis (0)] |
| 79. | Fidelle M, Yonekura S, Picard M, Cogdill A, Hollebecque A, Roberti MP, Zitvogel L. Resolving the Paradox of Colon Cancer Through the Integration of Genetics, Immunology, and the Microbiota. Front Immunol. 2020;11:600886. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 80. | Berg G, Rybakova D, Fischer D, Cernava T, Vergès MC, Charles T, Chen X, Cocolin L, Eversole K, Corral GH, Kazou M, Kinkel L, Lange L, Lima N, Loy A, Macklin JA, Maguin E, Mauchline T, McClure R, Mitter B, Ryan M, Sarand I, Smidt H, Schelkle B, Roume H, Kiran GS, Selvin J, Souza RSC, van Overbeek L, Singh BK, Wagner M, Walsh A, Sessitsch A, Schloter M. Microbiome definition re-visited: old concepts and new challenges. Microbiome. 2020;8:103. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 716] [Cited by in RCA: 981] [Article Influence: 196.2] [Reference Citation Analysis (0)] |
| 81. | Postler TS, Ghosh S. Understanding the Holobiont: How Microbial Metabolites Affect Human Health and Shape the Immune System. Cell Metab. 2017;26:110-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 456] [Cited by in RCA: 581] [Article Influence: 72.6] [Reference Citation Analysis (0)] |
| 82. | Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3003] [Cited by in RCA: 2993] [Article Influence: 230.2] [Reference Citation Analysis (0)] |
| 83. | Weiss GA, Hennet T. Mechanisms and consequences of intestinal dysbiosis. Cell Mol Life Sci. 2017;74:2959-2977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 393] [Cited by in RCA: 437] [Article Influence: 54.6] [Reference Citation Analysis (0)] |
| 84. | Avril M, DePaolo RW. "Driver-passenger" bacteria and their metabolites in the pathogenesis of colorectal cancer. Gut Microbes. 2021;13:1941710. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 85. | Hanus M, Parada-Venegas D, Landskron G, Wielandt AM, Hurtado C, Alvarez K, Hermoso MA, López-Köstner F, De la Fuente M. Immune System, Microbiota, and Microbial Metabolites: The Unresolved Triad in Colorectal Cancer Microenvironment. Front Immunol. 2021;12:612826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 126] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 86. | Seely KD, Morgan AD, Hagenstein LD, Florey GM, Small JM. Bacterial Involvement in Progression and Metastasis of Colorectal Neoplasia. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 87. | Roberti MP, Yonekura S, Duong CPM, Picard M, Ferrere G, Tidjani Alou M, Rauber C, Iebba V, Lehmann CHK, Amon L, Dudziak D, Derosa L, Routy B, Flament C, Richard C, Daillère R, Fluckiger A, Van Seuningen I, Chamaillard M, Vincent A, Kourula S, Opolon P, Ly P, Pizzato E, Becharef S, Paillet J, Klein C, Marliot F, Pietrantonio F, Benoist S, Scoazec JY, Dartigues P, Hollebecque A, Malka D, Pagès F, Galon J, Gomperts Boneca I, Lepage P, Ryffel B, Raoult D, Eggermont A, Vanden Berghe T, Ghiringhelli F, Vandenabeele P, Kroemer G, Zitvogel L. Chemotherapy-induced ileal crypt apoptosis and the ileal microbiome shape immunosurveillance and prognosis of proximal colon cancer. Nat Med. 2020;26:919-931. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 145] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 88. | Lin Y, Kong DX, Zhang YN. Does the Microbiota Composition Influence the Efficacy of Colorectal Cancer Immunotherapy? Front Oncol. 2022;12:852194. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 89. | Rezasoltani S, Yadegar A, Asadzadeh Aghdaei H, Reza Zali M. Modulatory effects of gut microbiome in cancer immunotherapy: A novel paradigm for blockade of immune checkpoint inhibitors. Cancer Med. 2021;10:1141-1154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 90. | Qiu Q, Lin Y, Ma Y, Li X, Liang J, Chen Z, Liu K, Huang Y, Luo H, Huang R, Luo L. Exploring the Emerging Role of the Gut Microbiota and Tumor Microenvironment in Cancer Immunotherapy. Front Immunol. 2020;11:612202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 85] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 91. | Aghamajidi A, Maleki Vareki S. The Effect of the Gut Microbiota on Systemic and Anti-Tumor Immunity and Response to Systemic Therapy against Cancer. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 48] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 92. | Zeng H, Taussig DP, Cheng WH, Johnson LK, Hakkak R. Butyrate Inhibits Cancerous HCT116 Colon Cell Proliferation but to a Lesser Extent in Noncancerous NCM460 Colon Cells. Nutrients. 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 93. | Lu Y, Yuan X, Wang M, He Z, Li H, Wang J, Li Q. Gut microbiota influence immunotherapy responses: mechanisms and therapeutic strategies. J Hematol Oncol. 2022;15:47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 278] [Article Influence: 92.7] [Reference Citation Analysis (0)] |
| 94. | Ren Y, Qian Y, Ai L, Xie Y, Gao Y, Zhuang Z, Chen J, Chen YX, Fang JY. TRAPPC4 regulates the intracellular trafficking of PD-L1 and antitumor immunity. Nat Commun. 2021;12:5405. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 46] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 95. | Li C, Chi H, Deng S, Wang H, Yao H, Wang Y, Chen D, Guo X, Fang JY, He F, Xu J. THADA drives Golgi residency and upregulation of PD-L1 in cancer cells and provides promising target for immunotherapy. J Immunother Cancer. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 96. | Gao Y, Zou T, Xu P, Wang Y, Jiang Y, Chen YX, Chen H, Hong J, Fang JY. Fusobacterium nucleatum stimulates cell proliferation and promotes PD-L1 expression via IFIT1-related signal in colorectal cancer. Neoplasia. 2023;35:100850. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 35] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 97. | Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML, Chang EB, Gajewski TF. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084-1089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1979] [Cited by in RCA: 2850] [Article Influence: 285.0] [Reference Citation Analysis (2)] |
| 98. | Canale FP, Basso C, Antonini G, Perotti M, Li N, Sokolovska A, Neumann J, James MJ, Geiger S, Jin W, Theurillat JP, West KA, Leventhal DS, Lora JM, Sallusto F, Geiger R. Metabolic modulation of tumours with engineered bacteria for immunotherapy. Nature. 2021;598:662-666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 334] [Article Influence: 83.5] [Reference Citation Analysis (0)] |
| 99. | Garza DR, Taddese R, Wirbel J, Zeller G, Boleij A, Huynen MA, Dutilh BE. Metabolic models predict bacterial passengers in colorectal cancer. Cancer Metab. 2020;8:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 100. | Gao R, Wu C, Zhu Y, Kong C, Gao Y, Zhang X, Yang R, Zhong H, Xiong X, Chen C, Xu Q, Qin H. Integrated Analysis of Colorectal Cancer Reveals Cross-Cohort Gut Microbial Signatures and Associated Serum Metabolites. Gastroenterology. 2022;163:1024-1037.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 61] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 101. | Vergara D, Simeone P, Damato M, Maffia M, Lanuti P, Trerotola M. The Cancer Microbiota: EMT and Inflammation as Shared Molecular Mechanisms Associated with Plasticity and Progression. J Oncol. 2019;2019:1253727. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 102. | Ha NH, Woo BH, Kim DJ, Ha ES, Choi JI, Kim SJ, Park BS, Lee JH, Park HR. Prolonged and repetitive exposure to Porphyromonas gingivalis increases aggressiveness of oral cancer cells by promoting acquisition of cancer stem cell properties. Tumour Biol. 2015;36:9947-9960. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 110] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 103. | Ganesan K, Jayachandran M, Xu B. Diet-Derived Phytochemicals Targeting Colon Cancer Stem Cells and Microbiota in Colorectal Cancer. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 104. | Sen U, Shenoy P S, Bose B. Opposing effects of low versus high concentrations of water soluble vitamins/dietary ingredients Vitamin C and niacin on colon cancer stem cells (CSCs). Cell Biol Int. 2017;41:1127-1145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 105. | Quaglio AEV, Grillo TG, De Oliveira ECS, Di Stasi LC, Sassaki LY. Gut microbiota, inflammatory bowel disease and colorectal cancer. World J Gastroenterol. 2022;28:4053-4060. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 219] [Cited by in RCA: 222] [Article Influence: 74.0] [Reference Citation Analysis (115)] |
| 106. | Rangan P, Mondino A. Microbial short-chain fatty acids: a strategy to tune adoptive T cell therapy. J Immunother Cancer. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 52] [Reference Citation Analysis (0)] |
| 107. | Hou H, Chen D, Zhang K, Zhang W, Liu T, Wang S, Dai X, Wang B, Zhong W, Cao H. Gut microbiota-derived short-chain fatty acids and colorectal cancer: Ready for clinical translation? Cancer Lett. 2022;526:225-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 135] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 108. | Chattopadhyay I, Gundamaraju R, Jha NK, Gupta PK, Dey A, Mandal CC, Ford BM. Interplay between Dysbiosis of Gut Microbiome, Lipid Metabolism, and Tumorigenesis: Can Gut Dysbiosis Stand as a Prognostic Marker in Cancer? Dis Markers. 2022;2022:2941248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 33] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 109. | Saffarian A, Mulet C, Regnault B, Amiot A, Tran-Van-Nhieu J, Ravel J, Sobhani I, Sansonetti PJ, Pédron T. Crypt- and Mucosa-Associated Core Microbiotas in Humans and Their Alteration in Colon Cancer Patients. mBio. 2019;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 105] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 110. | Marzano M, Fosso B, Piancone E, Defazio G, Pesole G, De Robertis M. Stem Cell Impairment at the Host-Microbiota Interface in Colorectal Cancer. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 111. | Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65-80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1071] [Cited by in RCA: 921] [Article Influence: 57.6] [Reference Citation Analysis (0)] |
| 112. | Peuker K, Muff S, Wang J, Künzel S, Bosse E, Zeissig Y, Luzzi G, Basic M, Strigli A, Ulbricht A, Kaser A, Arlt A, Chavakis T, van den Brink GR, Schafmayer C, Egberts JH, Becker T, Bianchi ME, Bleich A, Röcken C, Hampe J, Schreiber S, Baines JF, Blumberg RS, Zeissig S. Epithelial calcineurin controls microbiota-dependent intestinal tumor development. Nat Med. 2016;22:506-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 91] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 113. | Ruiz PA, Shkoda A, Kim SC, Sartor RB, Haller D. IL-10 gene-deficient mice lack TGF-beta/Smad-mediated TLR2 degradation and fail to inhibit proinflammatory gene expression in intestinal epithelial cells under conditions of chronic inflammation. Ann N Y Acad Sci. 2006;1072:389-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 35] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 114. | Wang Z, Hopson LM, Singleton SS, Yang X, Jogunoori W, Mazumder R, Obias V, Lin P, Nguyen BN, Yao M, Miller L, White J, Rao S, Mishra L. Mice with dysfunctional TGF-β signaling develop altered intestinal microbiome and colorectal cancer resistant to 5FU. Biochim Biophys Acta Mol Basis Dis. 2021;1867:166179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 115. | Wang X, Yang Y, Huycke MM. Commensal-infected macrophages induce dedifferentiation and reprogramming of epithelial cells during colorectal carcinogenesis. Oncotarget. 2017;8:102176-102190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 116. | Wang X, Undi RB, Ali N, Huycke MM. It takes a village: microbiota, parainflammation, paligenosis and bystander effects in colorectal cancer initiation. Dis Model Mech. 2021;14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 117. | Dalmasso G, Cougnoux A, Delmas J, Darfeuille-Michaud A, Bonnet R. The bacterial genotoxin colibactin promotes colon tumor growth by modifying the tumor microenvironment. Gut Microbes. 2014;5:675-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 209] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 118. | Chung L, Thiele Orberg E, Geis AL, Chan JL, Fu K, DeStefano Shields CE, Dejea CM, Fathi P, Chen J, Finard BB, Tam AJ, McAllister F, Fan H, Wu X, Ganguly S, Lebid A, Metz P, Van Meerbeke SW, Huso DL, Wick EC, Pardoll DM, Wan F, Wu S, Sears CL, Housseau F. Bacteroides fragilis Toxin Coordinates a Pro-carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe. 2018;23:203-214.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 345] [Cited by in RCA: 379] [Article Influence: 54.1] [Reference Citation Analysis (0)] |
| 119. | Yan X, Liu L, Li H, Qin H, Sun Z. Clinical significance of Fusobacterium nucleatum, epithelial-mesenchymal transition, and cancer stem cell markers in stage III/IV colorectal cancer patients. Onco Targets Ther. 2017;10:5031-5046. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 86] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 120. | Tarallo S, Ferrero G, Gallo G, Francavilla A, Clerico G, Realis Luc A, Manghi P, Thomas AM, Vineis P, Segata N, Pardini B, Naccarati A, Cordero F. Altered Fecal Small RNA Profiles in Colorectal Cancer Reflect Gut Microbiome Composition in Stool Samples. mSystems. 2019;4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 71] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 121. | Wang L, Wang E, Wang Y, Mines R, Xiang K, Sun Z, Zhou G, Chen KY, Rakhilin N, Chao S, Ye G, Wu Z, Yan H, Shen H, Everitt J, Bu P, Shen X. miR-34a is a microRNA safeguard for Citrobacter-induced inflammatory colon oncogenesis. Elife. 2018;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 122. | Voutsadakis I. The pluripotency network in colorectal cancer pathogenesis and prognosis: an update. Biomark Med. 2018;12:653-665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 123. | Shibata M, Hoque MO. Targeting Cancer Stem Cells: A Strategy for Effective Eradication of Cancer. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 129] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 124. | da Silva-Diz V, Lorenzo-Sanz L, Bernat-Peguera A, Lopez-Cerda M, Muñoz P. Cancer cell plasticity: Impact on tumor progression and therapy response. Semin Cancer Biol. 2018;53:48-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 142] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 125. | Zalewski A, Snook AE, Waldman SA. Stem cells as therapeutic targets in colorectal cancer. Per Med. 2021;18:171-183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 126. | Pádua D, Figueira P, Ribeiro I, Almeida R, Mesquita P. The Relevance of Transcription Factors in Gastric and Colorectal Cancer Stem Cells Identification and Eradication. Front Cell Dev Biol. 2020;8:442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 127. | Kim MJ, Huang Y, Park JI. Targeting Wnt Signaling for Gastrointestinal Cancer Therapy: Present and Evolving Views. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 128. | Wojtukiewicz MZ, Rek MM, Karpowicz K, Górska M, Polityńska B, Wojtukiewicz AM, Moniuszko M, Radziwon P, Tucker SC, Honn KV. Inhibitors of immune checkpoints-PD-1, PD-L1, CTLA-4-new opportunities for cancer patients and a new challenge for internists and general practitioners. Cancer Metastasis Rev. 2021;40:949-982. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 165] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 129. | Garza Treviño EN, González PD, Valencia Salgado CI, Martinez Garza A. Effects of pericytes and colon cancer stem cells in the tumor microenvironment. Cancer Cell Int. 2019;19:173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 130. | Wang DK, Zuo Q, He QY, Li B. Targeted Immunotherapies in Gastrointestinal Cancer: From Molecular Mechanisms to Implications. Front Immunol. 2021;12:705999. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 131. | Guo G, Tan Z, Liu Y, Shi F, She J. The therapeutic potential of stem cell-derived exosomes in the ulcerative colitis and colorectal cancer. Stem Cell Res Ther. 2022;13:138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 78] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 132. | Han S, Li G, Jia M, Zhao Y, He C, Huang M, Jiang L, Wu M, Yang J, Ji X, Liu X, Chen C, Chu X. Delivery of Anti-miRNA-221 for Colorectal Carcinoma Therapy Using Modified Cord Blood Mesenchymal Stem Cells-Derived Exosomes. Front Mol Biosci. 2021;8:743013. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 133. | Zhang M, Xin Y. Circular RNAs: a new frontier for cancer diagnosis and therapy. J Hematol Oncol. 2018;11:21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 159] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 134. | Xiong W, Ai YQ, Li YF, Ye Q, Chen ZT, Qin JY, Liu QY, Wang H, Ju YH, Li WH. Microarray Analysis of Circular RNA Expression Profile Associated with 5-Fluorouracil-Based Chemoradiation Resistance in Colorectal Cancer Cells. Biomed Res Int. 2017;2017:8421614. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 68] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 135. | Dahiya D, Nigam PS. The Gut Microbiota Influenced by the Intake of Probiotics and Functional Foods with Prebiotics Can Sustain Wellness and Alleviate Certain Ailments like Gut-Inflammation and Colon-Cancer. Microorganisms. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 57] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 136. | Damián MR, Cortes-Perez NG, Quintana ET, Ortiz-Moreno A, Garfias Noguez C, Cruceño-Casarrubias CE, Sánchez Pardo ME, Bermúdez-Humarán LG. Functional Foods, Nutraceuticals and Probiotics: A Focus on Human Health. Microorganisms. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 60] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 137. | Telang N. Drug-Resistant Stem Cells: Novel Approach for Colon Cancer Therapy. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 138. | Gomes S, Teixeira-Guedes C, Silva E, Baltazar F, Preto A. Colon microbiota modulation by dairy-derived diet: new strategy for prevention and treatment of colorectal cancer. Food Funct. 2022;13:9183-9194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 139. | Meerson A, Khatib S, Mahajna J. Natural Products Targeting Cancer Stem Cells for Augmenting Cancer Therapeutics. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 140. | Naujokat C, McKee DL. The "Big Five" Phytochemicals Targeting Cancer Stem Cells: Curcumin, EGCG, Sulforaphane, Resveratrol and Genistein. Curr Med Chem. 2021;28:4321-4342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 67] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 141. | Salminen S, Collado MC, Endo A, Hill C, Lebeer S, Quigley EMM, Sanders ME, Shamir R, Swann JR, Szajewska H, Vinderola G. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat Rev Gastroenterol Hepatol. 2021;18:649-667. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 304] [Cited by in RCA: 996] [Article Influence: 249.0] [Reference Citation Analysis (1)] |
| 142. | Markowiak P, Śliżewska K. Effects of Probiotics, Prebiotics, and Synbiotics on Human Health. Nutrients. 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1200] [Cited by in RCA: 1241] [Article Influence: 155.1] [Reference Citation Analysis (1)] |
| 143. | Singh S, Singh M, Gaur S. Probiotics as multifaceted oral vaccines against colon cancer: A review. Front Immunol. 2022;13:1002674. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 144. | Alam Z, Shang X, Effat K, Kanwal F, He X, Li Y, Xu C, Niu W, War AR, Zhang Y. The potential role of prebiotics, probiotics, and synbiotics in adjuvant cancer therapy especially colorectal cancer. J Food Biochem. 2022;46:e14302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 23] [Reference Citation Analysis (0)] |
| 145. | Saeed M, Shoaib A, Kandimalla R, Javed S, Almatroudi A, Gupta R, Aqil F. Microbe-based therapies for colorectal cancer: Advantages and limitations. Semin Cancer Biol. 2022;86:652-665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 32] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 146. | Chattopadhyay I, Dhar R, Pethusamy K, Seethy A, Srivastava T, Sah R, Sharma J, Karmakar S. Exploring the Role of Gut Microbiome in Colon Cancer. Appl Biochem Biotechnol. 2021;193:1780-1799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 79] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 147. | Konturek PC, Koziel J, Dieterich W, Haziri D, Wirtz S, Glowczyk I, Konturek K, Neurath MF, Zopf Y. Successful therapy of Clostridium difficile infection with fecal microbiota transplantation. J Physiol Pharmacol. 2016;67:859-866. [PubMed] |
| 148. | Myneedu K, Deoker A, Schmulson MJ, Bashashati M. Fecal microbiota transplantation in irritable bowel syndrome: A systematic review and meta-analysis. United European Gastroenterol J. 2019;7:1033-1041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 149. | Sokol H, Landman C, Seksik P, Berard L, Montil M, Nion-Larmurier I, Bourrier A, Le Gall G, Lalande V, De Rougemont A, Kirchgesner J, Daguenel A, Cachanado M, Rousseau A, Drouet É, Rosenzwajg M, Hagege H, Dray X, Klatzman D, Marteau P; Saint-Antoine IBD Network, Beaugerie L, Simon T. Fecal microbiota transplantation to maintain remission in Crohn's disease: a pilot randomized controlled study. Microbiome. 2020;8:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 249] [Article Influence: 49.8] [Reference Citation Analysis (1)] |
| 150. | Fong W, Li Q, Yu J. Gut microbiota modulation: a novel strategy for prevention and treatment of colorectal cancer. Oncogene. 2020;39:4925-4943. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 382] [Article Influence: 76.4] [Reference Citation Analysis (0)] |
| 151. | Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, Hickman HD, McCulloch JA, Badger JH, Ajami NJ, Trinchieri G, Pardo-Manuel de Villena F, Yewdell JW, Rehermann B. Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell. 2017;171:1015-1028.e13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 421] [Cited by in RCA: 578] [Article Influence: 72.3] [Reference Citation Analysis (0)] |
| 152. | Chang CW, Lee HC, Li LH, Chiang Chiau JS, Wang TE, Chuang WH, Chen MJ, Wang HY, Shih SC, Liu CY, Tsai TH, Chen YJ. Fecal Microbiota Transplantation Prevents Intestinal Injury, Upregulation of Toll-Like Receptors, and 5-Fluorouracil/Oxaliplatin-Induced Toxicity in Colorectal Cancer. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 145] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 153. | Min Ho PY, Hu W, Lee YY, Gao C, Tan YZ, Cheen HH, Wee HL, Lim TG, Ong WC. Health-related quality of life of patients with inflammatory bowel disease in Singapore. Intest Res. 2019;17:107-118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 154. | Marzagalli M, Fontana F, Raimondi M, Limonta P. Cancer Stem Cells-Key Players in Tumor Relapse. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 91] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 155. | Mueller AL, Brockmueller A, Fahimi N, Ghotbi T, Hashemi S, Sadri S, Khorshidi N, Kunnumakkara AB, Shakibaei M. Bacteria-Mediated Modulatory Strategies for Colorectal Cancer Treatment. Biomedicines. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 156. | Ishigamori R, Komiya M, Takasu S, Mutoh M, Imai T, Takahashi M. Osteopontin Deficiency Suppresses Intestinal Tumor Development in Apc-Deficient Min Mice. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 157. | De Robertis M, Poeta ML, Signori E, Fazio VM. Current understanding and clinical utility of miRNAs regulation of colon cancer stem cells. Semin Cancer Biol. 2018;53:232-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 158. | Watanabe S, Hibiya S, Katsukura N, Kitagawa S, Sato A, Okamoto R, Watanabe M, Tsuchiya K. Influence of chronic inflammation on the malignant phenotypes and the plasticity of colorectal cancer cells. Biochem Biophys Rep. 2021;26:101031. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 159. | Ma X, Liu J, Yang X, Fang K, Zheng P, Liang X. Mesenchymal stem cells maintain the stemness of colon cancer stem cells via interleukin-8/mitogen-activated protein kinase signaling pathway. Exp Biol Med (Maywood). 2020;245:562-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 160. | Fang M, Li Y, Huang K, Qi S, Zhang J, Zgodzinski W, Majewski M, Wallner G, Gozdz S, Macek P, Kowalik A, Pasiarski M, Grywalska E, Vatan L, Nagarsheth N, Li W, Zhao L, Kryczek I, Wang G, Wang Z, Zou W, Wang L. IL33 Promotes Colon Cancer Cell Stemness via JNK Activation and Macrophage Recruitment. Cancer Res. 2017;77:2735-2745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 145] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 161. | Lee SH, Kim MJ, Kim DW, Kang CD, Kim SH. Amurensin G enhances the susceptibility to tumor necrosis factor-related apoptosis-inducing ligand-mediated cytotoxicity of cancer stem-like cells of HCT-15 cells. Cancer Sci. 2013;104:1632-1639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 162. | Martín MJ, Gigola G, Zwenger A, Carriquiriborde M, Gentil F, Gentili C. Potential therapeutic targets for growth arrest of colorectal cancer cells exposed to PTHrP. Mol Cell Endocrinol. 2018;478:32-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 163. | Carriere P, Calvo N, Novoa Díaz MB, Lopez-Moncada F, Herrera A, Torres MJ, Alonso E, Gandini NA, Gigola G, Contreras HR, Gentili C. Role of SPARC in the epithelial-mesenchymal transition induced by PTHrP in human colon cancer cells. Mol Cell Endocrinol. 2021;530:111253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 164. | Novoa Díaz MB, Carriere PM, Martín MJ, Calvo N, Gentili C. Involvement of parathyroid hormone-related peptide in the aggressive phenotype of colorectal cancer cells. World J Gastroenterol. 2021;27:7025-7040. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 3] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 165. | Hu JL, Wang W, Lan XL, Zeng ZC, Liang YS, Yan YR, Song FY, Wang FF, Zhu XH, Liao WJ, Liao WT, Ding YQ, Liang L. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol Cancer. 2019;18:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 240] [Cited by in RCA: 559] [Article Influence: 93.2] [Reference Citation Analysis (0)] |
| 166. | Zhu Y, Wang C, Becker SA, Hurst K, Nogueira LM, Findlay VJ, Camp ER. miR-145 Antagonizes SNAI1-Mediated Stemness and Radiation Resistance in Colorectal Cancer. Mol Ther. 2018;26:744-754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 99] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 167. | Sakaguchi M, Hisamori S, Oshima N, Sato F, Shimono Y, Sakai Y. miR-137 Regulates the Tumorigenicity of Colon Cancer Stem Cells through the Inhibition of DCLK1. Mol Cancer Res. 2016;14:354-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 69] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 168. | Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen S, Wang Y, Wang T, Hou Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8:3932-3948. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 296] [Cited by in RCA: 558] [Article Influence: 79.7] [Reference Citation Analysis (0)] |
| 169. | Trinh A, Lädrach C, Dawson HE, Ten Hoorn S, Kuppen PJK, Reimers MS, Koopman M, Punt CJA, Lugli A, Vermeulen L, Zlobec I. Tumour budding is associated with the mesenchymal colon cancer subtype and RAS/RAF mutations: a study of 1320 colorectal cancers with Consensus Molecular Subgroup (CMS) data. Br J Cancer. 2018;119:1244-1251. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |