Published online Jul 7, 2022. doi: 10.3748/wjg.v28.i25.2900
Peer-review started: February 21, 2022
First decision: April 5, 2022
Revised: April 8, 2022
Accepted: May 28, 2022
Article in press: May 28, 2022
Published online: July 7, 2022
Processing time: 133 Days and 5.2 Hours
Gastric cancer (GC) is the fourth leading cause of cancer-related death. The occurrence and development of GC is a complex process involving multiple biological mechanisms. Although traditional regulation modulates molecular functions related to the occurrence and development of GC, the comprehensive mechanisms remain unclear. Ultraconserved region (UCR) refers to a genome sequence that is completely conserved in the homologous regions of the human, rat and mouse genomes, with 100% identity, without any insertions or deletions, and often located in fragile sites and tumour-related genes. The transcribed UCR (T-UCR) is transcribed from the UCR and is a new type of long noncoding RNA. Recent studies have found that the expression level of T-UCRs changes during the occurrence and development of GC, revealing a new mechanism underlying GC. Therefore, this article aims to review the relevant research on T-UCRs in GC, as well as the function of T-UCRs and their regulatory role in the occurrence and development of GC, to provide new strategies for GC diagnosis and treatment.
Core Tip: Transcribed ultraconserved region (T-UCR) is abnormally expressed in gastric cancer (GC) cells and tumors. It has been found that a variety of T-UCR affects downstream genes and related pathways, and plays a regulatory role in the proliferation, migration and invasion of GC. However, there are few relevant reviews, and this paper aims to review the related studies of T-UCR in GC. And the function of T-UCR and its regulatory role in the occurrence and development of GC, thus providing a new strategy for the diagnosis and treatment of GC.
- Citation: Gao SS, Zhang ZK, Wang XB, Ma Y, Yin GQ, Guo XB. Role of transcribed ultraconserved regions in gastric cancer and therapeutic perspectives. World J Gastroenterol 2022; 28(25): 2900-2909
- URL: https://www.wjgnet.com/1007-9327/full/v28/i25/2900.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i25.2900
Gastric cancer (GC) is one of the most common malignant tumours in the world. Although the incidence of GC has decreased, GC ranks fourth in cancer-related deaths due to its high lethality[1,2]. Due to the limited specific biomarkers of GC, patients usually have advanced GC when they are diagnosed. The tumourigenesis and development of GC is a multistep process involving countless signal transduction pathways and gene regulation. Among them, oncogenic and tumour suppressor factors, such as the transcribed ultraconserved region (T-UCR), play a key role[3,4].
In the human genome, 93% of genes can be transcribed to produce RNA, but only 2% are translated to protein. This type of RNA that lacks the potential for translation into protein is called noncoding RNA[5-9]. According to the number of bases it contains, noncoding RNA is divided into long noncoding RNAs (lncRNAs, > 200 bp) and small noncoding RNAs (< 200 bp, including rRNA, miRNA, snRNA, snoRNA, siRNA and pi-RNA)[10-13]. T-UCRs are lncRNAs, transcribed from UCRs. Recent studies have found that the expression level of T-UCRs is altered and abnormally expressed in human GC[14-17], which reveals a new mechanism for the occurrence and development of GC. Therefore, this article aims to review the relevant research on T-UCRs in GC, to further understand the specific mechanism of T-UCRs in GC cells, which will facilitate preventive measures against GC, early diagnoses, and new treatments.
In 2004, Bejerano et al[18] compared the genomes of humans, mice, and rats. They found highly conserved DNA sequences, which were subsequently renamed ultraconserved regions (UCRs)[19]. These regions were absolutely conserved in the three species (100% identical, no insertions or deletions) and were often located in fragile sites and genomic regions of tumour-related genes. UCRs are highly conserved in the evolutionary process, due to the long-distance enhancers and ultraconserved exons that originated from the short interspersed repetitive element retroposon family 400 million years ago. At present, such extreme conservation is still active in the Indonesian "coelacanth"[20].
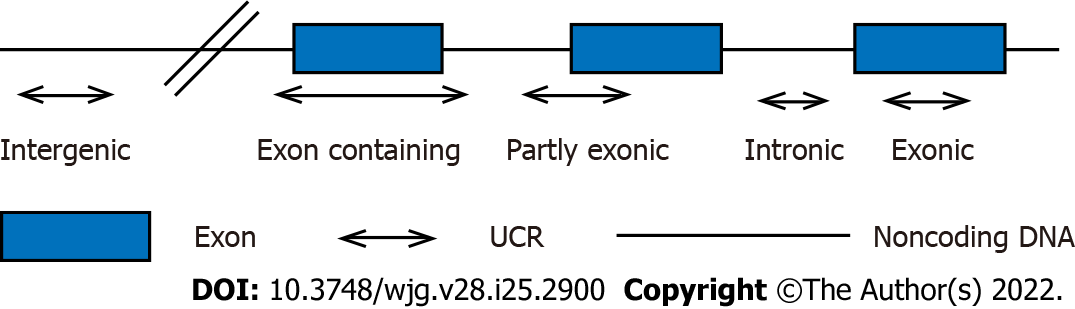
UCRs represent a small part of the human genome, forming a subset of conserved sequences in intragenic and intergenic regions. UCRs are functional but do not encode proteins[21]. The length of UCRs is between 200 and 799 bp. To date, humans have found at least 481 UCR regions[18,22]. It is known that a large part of these regions can actively transcribe RNA, some of them overlap with known protein-coding sequences, and more than half are predicted to not contain any protein-coding sequences[23]. Based on their overlap with known protein-coding genes, these 481 ultraconserved elements were initially divided into three categories: Nonexons, exons and possibly exons[18,19]. However, based on the positional relationship between UCRs and genes, Mestdagh et al[24] reclassified them into 5 categories: Exon containing (4.2%), exonic (5%), partly exonic (5%), intergenic (38.7%) and intronic (42.6%) (Figure 1). The sense/antisense strand of each region produces two transcripts, resulting in a total of 962 possible transcripts: One corresponding to the sense genome sequence (named “+”) and the other corresponding to the antisense sequence (named “+ A”)[25].
T-UCRs are the transcripts of UCRs[26]. Due to their high degree of conservation, T-UCRs may have fundamental functional importance for the ontogeny and phylogeny of mammals and other vertebrates. Recent studies have shown that a T-UCR acts as a regulator in a variety of pathways (such as pri-miRNA processing, transcription regulation, translation and chromatin modification)[27-29]. It is speculated that T-UCRs may be candidate genes for cancer susceptibility because the transcription level of some UCRs is dysregulated in cancer[30,31].
Although transcribed T-UCRs are a type of lncRNA, transcribed by UCRs, they are completely different from lncRNA. T-UCRs are absolutely conserved, while lncRNAs are the least conserved among noncoding RNA[32]. The amazing evolutionary retention of T-UCRs strongly suggests their profound biological role in various physiological responses. As a type of evolutionarily conserved ncRNA, T-UCRs are regarded as essential for life by acting as an antisense transcription inhibitors for nearby protein coding RNA and other ncRNA. T-UCRs are thought to be involved in RNA processing or transcriptional regulation[18]. It has previously been demonstrated that T-UCRs act as regulators of gene expression[20,33]. In recent decades, increasing evidence has shown that T-UCRs are involved in carcinogenesis[23,30,31,34]. Recent studies have identified changes in T-UCR expression patterns associated with specific tumour phenotypes, such as hepatocellular carcinoma[35-37], pancreatic cancer[38,39], bladder cancer[40,41], colorectal cancer[22,26,42,43], prostate cancer[16,25,44,45], cervical cancer[46], neuroblastoma[24], breast cancer[47], lung cancer[22,48-50] and leukaemia[51], indicating a mechanism by which T-UCRs are involved in cancer development. Related studies in GC have also described changes in the expression pattern of T-UCRs[3,4,14-16], indicating that T-UCRs also play a regulatory role in the occurrence and development of GC.
Uc.160+ is transcribed from the UCR on the 5q14.1 chromosome band. Honma et al[15] used real-time fluorescent quantitative polymerase chain reaction and in situ hybridization to detect Uc.160+. Compared with nontumour tissues, Uc.160+ expression is downregulated in GC and adenoma tissues. To further understand the biological mechanism of Uc.160+ in GC, the Uc.160+ overexpression vector was used to transfect GC cell lines MKN-1 and MKN-45, and then Western blotting was performed to detect the involvement of mitogens. Compared with the control group, the expression of phos
Sakamoto et al[14] analysed the expression and distribution of Uc.63+ by using qRT-PCR and in situ hybridization, and they found that all GC tissues showed high Uc.63+ expression compared with normal tissues. The expression of Uc.63+ was also elevated in GC cell lines. After overexpression of Uc.63+ was induced by transfection of Uc.63+ expression vector, GC cell proliferation was significantly enhanced. After the expression of Uc.63+ was inhibited by siRNA, the proliferation of GC cells was inhibited. These results supported the possibility that Uc.63+ had a carcinogenic effect in GC. Additionally, they found that Uc.63+ had no effect on the survival rate of cancer patients, but Uc.63+ overexpression was associated with advanced cancer and GC classification. Uc.63+ was preferentially overexpressed in diffuse GC, but not in intestinal GC. In conclusion, Uc.63+ plays a key role in the classification and progression of GC.
Goto et al[16] found that UC.416+A is overexpressed in GC compared with normal tissues by in situ hybridization. The growth of GC cells was significantly inhibited after Uc.416+A was downregulated by siRNA. These results suggest that UC.416+A, as an oncogene, plays an important role in promoting the proliferation of GC cells. Global genetic analysis using Affymetrix GeneChips showed that the most upregulated gene was IGFBP1 (insulin-like growth factor binding protein 1) when Uc.416+A was overexpressed. Compared with the corresponding nontumour gastric mucosa, IGFBP6 was significantly downregulated in GC tissue, indicating that Uc.416+A might promote the proliferation of GC cells by inhibiting IGFBP6.
In addition, the expression levels of Uc.118, Uc.158, Uc.241 and Uc.346 were found to be significantly downregulated in GC; Uc.244, Uc.249, Uc.252, Uc.261, Uc.282, Uc.283 and Uc.359 Levels were undetermined (Table 1).
| T-UCR name | Location | Orientation | Expression in gastric cancer | Biological or molecular functions in gastric cancer | Ref. |
| Uc.63+ | Chr.2 | Sense | Downregulated | Associated with the classification and progression of gastric cancer | [14] |
| Uc.118 | Chr.3 | Antisense | Downregulated | - | [16] |
| Uc.158 | Chr.5 | Antisense | Downregulated | - | [16] |
| uc.160 | Chr.5 | Sense | Downregulated | Inhibits cell proliferation and promotes apoptosis in AGS and SGC-7901 cell lines, promoting gastric cancer tumourigenesis | [15,17] |
| Uc.241 | - | - | Downregulated | - | [16] |
| Uc.244 | Chr.8 | Sense | Down/upregulated | - | [16,23] |
| Uc.249 | Chr.9 | Sense | Down/upregulated | - | [16,23] |
| Uc.252 | Chr.9 | Antisense | Down/upregulated | - | [16,23] |
| Uc.261 | Chr.9 | Sense | Down/upregulated | - | [16,23] |
| Uc.282 | Chr.9 | Sense | Down/upregulated | - | [16,23] |
| Uc.283 | - | - | - | - | [16] |
| Uc.346 | Chr.12 | - | Downregulated | - | [23] |
| Uc.359 | Chr.14 | Antisense | Down/upregulated | - | [16,23] |
| Uc.416 | - | Antisense | Upregulated | Promotes proliferation in MKN-74 cell line, promoting gastric cancer tumourigenesis | [16] |
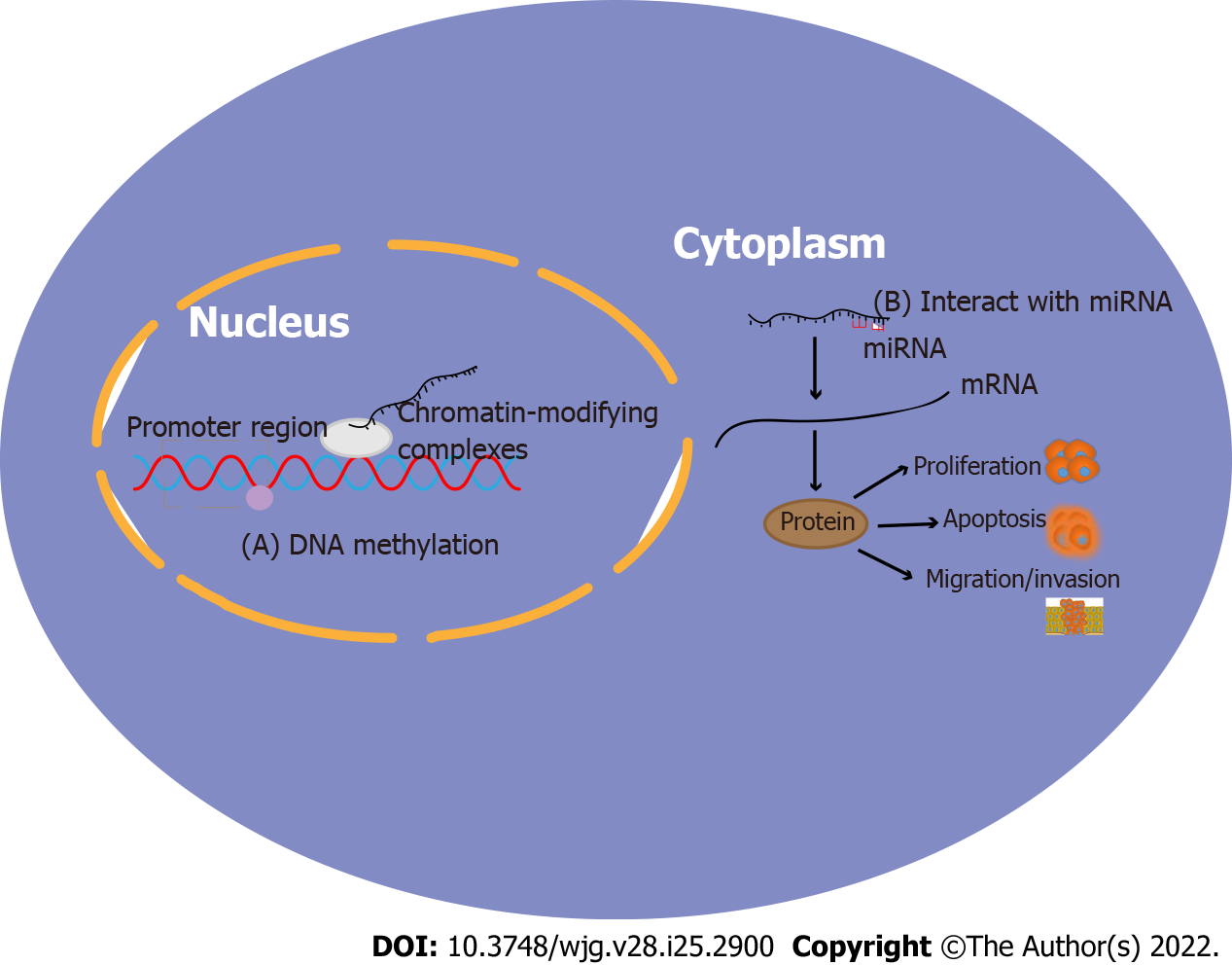
The regulatory mechanism of T-UCRs in tumourigenesis and the development of cancer is largely unclear, but current studies have found that T-UCR are involved in three different regulatory mechanisms in tumours: CpG island methylation[52], interaction with miRNAs[53-55] and direct binding to the target mRNAs[27,43]. In GC related research, it was only found that T-UCRs participate in the occurrence and development of GC through the first two pathways (Figure 2).
The field of epigenetics describes the transmission of information through heritable changes in phenotypes that do not involve changes in DNA sequences during cell division[56]. CpG island methylation, histone modification and chromatin delivery structure are the potential mechanisms of epigenetic transmission, and CpG island methylation is a key component of the changes in gene expression associated with human cancer, especially the expression of GC related genes[57-60]. GC is the most susceptible of all cancers to epigenetic changes without any changes in DNA[59,61-63]. CpG islands are DNA fragments of at least 0.5 kb, that are rich in G:C and CpG content, and are present in approximately 70% of human gene promoters. In other words, CpG islands are tandem repeats of cytosine (C) and guanidine (G), where p is the phosphoric acid between C and G. In brief, the methylation of CpG occurs under the action of methylase whereby the hydrogen on the cytosine (C) 5 carbon atom is replaced by methyl (CH3)[60,64-66]. Demethylation is the opposite process. It has been found that more than 50% of human genes are regulated by promoters including CpG islands[67]. In normal cells, except for genes with inactive X chromosomes or genes related to imprinted genes, promoter CpG islands are usually unmethylated[68]. Although the aetiology is still unclear, promoter CpG island methylation may be related to cancer development and ageing[69]. It is a common feature of human cancers that low expression of tumour-suppressive ncRNA causes CpG island methylation and then affects epigenetic silencing. Promoter CpG island methylation has been found in almost all human cancer tissue types, and it is an important mechanism for the inactivation of tumour suppressor genes and tumour-related genes[70]. GC is the most common human cancer caused by methylation of the promoter CpG island. Interestingly, T-UCRs undergo DNA methylation-related silencing in cancer cells, and promoter CpG island methylation is now considered to be important for inactivating tumour suppressor genes or tumour-related genes. Previous studies have shown that T-UCR regulation in the occurrence and development of GC is closely related to the methylation of CpG islands of the host gene promoter[16].
Goto et al[16] discovered that there are CpG islands approximately 500bp upstream of the Uc.158+A transcription gene. Bisulfite genomic sequencing of GC cell lines and GC tissue samples showed specific DNA methylation of GC, which contained UCRs. The luciferase vector of the CpG island upstream of the Uc.158+A transcription gene proved that when the upstream sequence of the Uc.158+A transcription gene was methylated, reporter activity was significantly inhibited. Thus, this suggests that Uc.158+A expression is silenced by DNA methylation in the promoter region upstream of its transcription gene.
Previous studies have shown that miRNAs affect the regulation of gene expression at both the transcriptional and posttranscriptional levels[71-73]. MiRNAs are also almost completely conserved[74,75]. Do T-UCRs interact with miRNAs to regulate the occurrence and development of cancer? In 2007, Calin and his collaborators demonstrated for the first time that certain T-UCRs are altered due to the direct regulation of high-level miRNAs in chronic lymphocytic leukaemia, and the expression of each T-UCR is negatively correlated with the corresponding microRNA level[19]. Recent studies have found that a T-UCR interacts with miR-596 and synergistically promotes the development of bladder cancer[28]. Terreri et al[55] found that the formation of T-UCR::miRNA pairs may have different effects, either targeting T-UCR or forming sponges that capture miRNAs. They also reported that the interaction between miRNAs and T-UCRs can act as a network to regulate the availability of certain lncRNAs in bladder urothelial carcinoma cells. These results indicated that miRNAs are involved in the process of T-UCR-mediated tumour regulation. In GC, we found that T-UCRs represent the possible targets of miRNAs[14-16], and these interactions may have biological and prognostic significance for cancer patients.
Goto et al[16] determined that the expression of miR-153 in GC cell lines with higher expression of Uc.416+A was significantly reduced, suggesting an inverse correlation between Uc.416+A and miR-153. The direct interaction between Uc.416+A and miR-153 was confirmed by a luciferase activity assay. This result showed that the overexpression of Uc.416+A is related to the downregulation of miR-153 in GC. Overexpression of UC.416+A caused changes in downstream related genes. The expression of insulin-like growth factor binding protein 6 (IGFBP6) was upregulated, and alcohol dehydrogenase 1C (ADH1C), homeobox B5 (HOXB5) and homeobox B6 (HOXB6) levels were downregulated. The expression of IGFBP6, HOXB5 and HOXB6 in GC and adjacent tissues showed statistically significant differences, indicating that the overexpression of UC.416+A is regulated by miR-153, which then affects the gene changes of downstream coding proteins and plays a promoting role in the growth of GC.
Honma et al[15] found that the low expression of Uc.160+ in GC was caused by methylation of its upstream promoter region, and the low expression of Uc.160+ caused a decrease in PTEN expression, an increase in phosphatase activity and activation of the MAPK pathway. In addition, Pang et al[17]'s study found that UC.160 was also regulated by miR-155, which then regulated PTEN and affected the MAPK pathway. In other words, the low expression of UC.160 in GC is regulated by miR-155 and affected by methylation in the upstream promoter region of its transcriptional genes.
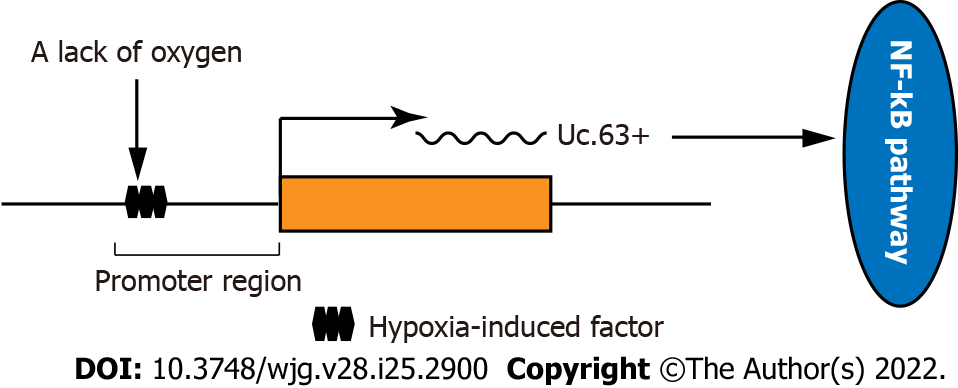
Sakamoto et al[14] found that the expression of UC.63+ is regulated by hypoxia. The promoter region located upstream of the Uc.63+ transcription gene is considered to have hypoxia-induced binding sites. Hypoxia induces overexpression of UC.63+, and the overexpression of UC.63+ upregulates its downstream target P65 and activates the NF-KB signalling pathway to promote the occurrence of GC (Figure 3).
In GC, T-UCRs change at the transcriptional level, and the abnormal expression of T-UCRs can lead to the occurrence and development of GC. Compared with normal cells, T-UCRs in GC cells have a unique expression profile, which indicates that changes in T-UCRs are involved in this malignant process. This study may provide new ideas and directions for GC diagnosis and prognosis. Compared with coding RNA, T-UCRs have incomparable advantages. Because T-UCRs do not code for proteins, they are relatively less regulated and more accurate. In addition, some T-UCR expression levels are completely different in different stages of cancer, and can also be used as one of the criteria for judging prognosis.
Despite the fact that this new type of dysregulated molecule seems to be useful in future clinical applications, further research is needed before it can be used as a valuable clinical biomarker for GC. First, although approximately 98% of the entire genome contains nonprotein coding genes, the proportion of T-UCRs is currently unclear. Second, current screening methods for T-UCRs (RT-qPCR, T-UCR chip) are useful, but simpler and more practical techniques will help in the identification and screening of disease-related T-UCRs. Third, it has been proven that T-UCRs are involved in the occurrence and development of GC, but their biological mechanism has not been fully elucidated. In addition, more powerful strategies are needed to clarify the regulatory role of T-UCR by constructing an interaction network, or to evaluate its function in typical signalling pathways. Since the knowledge of T-UCR is still in its infancy-especially when compared to other ncRNAs, further research is needed to convincingly incorporate these ncRNAs into the growing field of cancer therapeutics.
At present, there are few studies on T-UCR therapy, so there is still much room to explore treatment measures aimed at T-UCRs. Perhaps we can achieve a treatment purpose by changing the expression of T-UCRs. In the reference cell experiment, restoring the downregulated levels of T-UCRs or suppressing the overexpressed levels of T-UCRs via overexpression vectors or small interfering RNA methods can reverse the tumour phenotype. In clinical treatment, we can use the negative regulation between T-UCRs and miRNAs to inhibit the expression of T-UCRs, and methylation inhibitors can be used to restore the expression of T-UCRs with downregulated levels in tumours, thereby delaying or even reversing tumour progression.
Although these hypotheses have not been confirmed, we believe that as research on T-UCRs continues, the insight gained will definitely provide a novel strategy for the diagnosis, prognosis and treatment of tumours.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kamran M, Pakistan; Senchukova M, Russia A-Editor: Liu X, China S-Editor: Yan JP L-Editor: A P-Editor: Yan JP
| 1. | Thrift AP, El-Serag HB. Burden of Gastric Cancer. Clin Gastroenterol Hepatol. 2020;18:534-542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 432] [Cited by in RCA: 952] [Article Influence: 190.4] [Reference Citation Analysis (1)] |
| 2. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 63856] [Article Influence: 15964.0] [Reference Citation Analysis (174)] |
| 3. | Ghafouri-Fard S, Taheri M. Long non-coding RNA signature in gastric cancer. Exp Mol Pathol. 2020;113:104365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 4. | Yu H, Rong L. Emerging role of long non-coding RNA in the development of gastric cancer. World J Gastrointest Oncol. 2018;10:260-270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 19] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 5. | ENCODE Project Consortium. . An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57-74. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14554] [Cited by in RCA: 12864] [Article Influence: 989.5] [Reference Citation Analysis (0)] |
| 6. | Oe S, Kimura T, Yamada H. Regulatory non-coding RNAs in nervous system development and disease. Front Biosci (Landmark Ed). 2019;24:1203-1240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 7. | Tsagakis I, Douka K, Birds I, Aspden JL. Long non-coding RNAs in development and disease: conservation to mechanisms. J Pathol. 2020;250:480-495. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 141] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 8. | Rederstorff M, Hüttenhofer A. Small non-coding RNAs in disease development and host-pathogen interactions. Curr Opin Mol Ther. 2010;12:684-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 9. | Dey BK, Mueller AC, Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 2014;5:e944014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 273] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 10. | Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol Rev. 2016;96:1297-1325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1378] [Cited by in RCA: 1324] [Article Influence: 147.1] [Reference Citation Analysis (1)] |
| 11. | Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol. 2017;14:1705-1714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 365] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 12. | Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2018;19:143-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 600] [Cited by in RCA: 974] [Article Influence: 121.8] [Reference Citation Analysis (0)] |
| 13. | Romano G, Veneziano D, Acunzo M, Croce CM. Small non-coding RNA and cancer. Carcinogenesis. 2017;38:485-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 321] [Article Influence: 40.1] [Reference Citation Analysis (0)] |
| 14. | Sakamoto N, Sekino Y, Fukada K, Pham QT, Honma R, Taniyama D, Ukai S, Takashima T, Hattori T, Naka K, Tanabe K, Ohdan H, Yasui W. Uc.63+ contributes to gastric cancer progression through regulation of NF-kB signaling. Gastric Cancer. 2020;23:863-873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 15. | Honma R, Goto K, Sakamoto N, Sekino Y, Sentani K, Oue N, Yasui W. Expression and function of Uc.160+, a transcribed ultraconserved region, in gastric cancer. Gastric Cancer. 2017;20:960-969. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 16. | Goto K, Ishikawa S, Honma R, Tanimoto K, Sakamoto N, Sentani K, Oue N, Teishima J, Matsubara A, Yasui W. The transcribed-ultraconserved regions in prostate and gastric cancer: DNA hypermethylation and microRNA-associated regulation. Oncogene. 2016;35:3598-3606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 17. | Pang L, Li Q, Zhang Y, Deng B, Wu F, Wang J, Wu K, Ding Y, Yu D. Transcribed ultraconserved noncoding RNA uc.160 acts as a negative regulator in gastric cancer. Am J Transl Res. 2018;10:2822-2833. [PubMed] |
| 18. | Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321-1325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1212] [Cited by in RCA: 1223] [Article Influence: 58.2] [Reference Citation Analysis (0)] |
| 19. | Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, Shimizu M, Tili E, Rossi S, Taccioli C, Pichiorri F, Liu X, Zupo S, Herlea V, Gramantieri L, Lanza G, Alder H, Rassenti L, Volinia S, Schmittgen TD, Kipps TJ, Negrini M, Croce CM. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 564] [Cited by in RCA: 557] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 20. | Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 372] [Cited by in RCA: 373] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 21. | Qian XX, Peng JC, Xu AT, Zhao D, Qiao YQ, Wang TR, Shen J, Ran ZH. Noncoding Transcribed Ultraconserved Region (T-UCR) uc.261 Participates in Intestinal Mucosa Barrier Damage in Crohn's Disease. Inflamm Bowel Dis. 2016;22:2840-2852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 22. | Zhang Y, Wang S, Qian W, Ji D, Wang Q, Zhang Z, Ji B, Fu Z, Sun Y. uc.338 targets p21 and cyclin D1 via PI3K/AKT pathway activation to promote cell proliferation in colorectal cancer. Oncol Rep. 2018;40:1119-1128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (1)] |
| 23. | Pereira Zambalde E, Mathias C, Rodrigues AC, de Souza Fonseca Ribeiro EM, Fiori Gradia D, Calin GA, Carvalho de Oliveira J. Highlighting transcribed ultraconserved regions in human diseases. Wiley Interdiscip Rev RNA. 2020;11:e1567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 24. | Mestdagh P, Fredlund E, Pattyn F, Rihani A, Van Maerken T, Vermeulen J, Kumps C, Menten B, De Preter K, Schramm A, Schulte J, Noguera R, Schleiermacher G, Janoueix-Lerosey I, Laureys G, Powel R, Nittner D, Marine JC, Ringnér M, Speleman F, Vandesompele J. An integrative genomics screen uncovers ncRNA T-UCR functions in neuroblastoma tumours. Oncogene. 2010;29:3583-3592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 114] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 25. | Hudson RS, Yi M, Volfovsky N, Prueitt RL, Esposito D, Volinia S, Liu CG, Schetter AJ, Van Roosbroeck K, Stephens RM, Calin GA, Croce CM, Ambs S. Transcription signatures encoded by ultraconserved genomic regions in human prostate cancer. Mol Cancer. 2013;12:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 26. | Sana J, Hankeova S, Svoboda M, Kiss I, Vyzula R, Slaby O. Expression levels of transcribed ultraconserved regions uc.73 and uc.388 are altered in colorectal cancer. Oncology. 2012;82:114-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 27. | Liz J, Portela A, Soler M, Gómez A, Ling H, Michlewski G, Calin GA, Guil S, Esteller M. Regulation of pri-miRNA processing by a long noncoding RNA transcribed from an ultraconserved region. Mol Cell. 2014;55:138-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 123] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 28. | Ding L, Gong C, Zhao J, Liu X, Li T, Rao S, Wang S, Liu Y, Peng S, Xiao W, Xiong C, Wang R, Liang S, Xu H. Noncoding transcribed ultraconserved region (T-UCR) UC.48+ is a novel regulator of high-fat diet induced myocardial ischemia/reperfusion injury. J Cell Physiol. 2019;234:9849-9861. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 29. | Terracciano D, Terreri S, de Nigris F, Costa V, Calin GA, Cimmino A. The role of a new class of long noncoding RNAs transcribed from ultraconserved regions in cancer. Biochim Biophys Acta Rev Cancer. 2017;1868:449-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 30. | Fabris L, Calin GA. Understanding the Genomic Ultraconservations: T-UCRs and Cancer. Int Rev Cell Mol Biol. 2017;333:159-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 31. | Peng JC, Shen J, Ran ZH. Transcribed ultraconserved region in human cancers. RNA Biol. 2013;10:1771-1777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 57] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 32. | Johnsson P, Lipovich L, Grandér D, Morris KV. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta. 2014;1840:1063-1071. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 501] [Cited by in RCA: 532] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 33. | Katzman S, Kern AD, Bejerano G, Fewell G, Fulton L, Wilson RK, Salama SR, Haussler D. Human genome ultraconserved elements are ultraselected. Science. 2007;317:915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 194] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 34. | Scaruffi P. The transcribed-ultraconserved regions: a novel class of long noncoding RNAs involved in cancer susceptibility. ScientificWorldJournal. 2011;11:340-352. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 35. | Luo HL, Chen J, Luo T, Wu FX, Liu JJ, Wang HF, Chen M, Li LQ, Li H. Downregulation of Macrophage-Derived T-UCR uc.306 Associates with Poor Prognosis in Hepatocellular Carcinoma. Cell Physiol Biochem. 2017;42:1526-1539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 36. | Carotenuto P, Fassan M, Pandolfo R, Lampis A, Vicentini C, Cascione L, Paulus-Hock V, Boulter L, Guest R, Quagliata L, Hahne JC, Ridgway R, Jamieson T, Athineos D, Veronese A, Visone R, Murgia C, Ferrari G, Guzzardo V, Evans TRJ, MacLeod M, Feng GJ, Dale T, Negrini M, Forbes SJ, Terracciano L, Scarpa A, Patel T, Valeri N, Workman P, Sansom O, Braconi C. Wnt signalling modulates transcribed-ultraconserved regions in hepatobiliary cancers. Gut. 2017;66:1268-1277. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 71] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 37. | Kogure T, Yan IK, Lin WL, Patel T. Extracellular Vesicle-Mediated Transfer of a Novel Long Noncoding RNA TUC339: A Mechanism of Intercellular Signaling in Human Hepatocellular Cancer. Genes Cancer. 2013;4:261-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 262] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 38. | Jiang J, Azevedo-Pouly AC, Redis RS, Lee EJ, Gusev Y, Allard D, Sutaria DS, Badawi M, Elgamal OA, Lerner MR, Brackett DJ, Calin GA, Schmittgen TD. Globally increased ultraconserved noncoding RNA expression in pancreatic adenocarcinoma. Oncotarget. 2016;7:53165-53177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 39. | Liu C, Wang J, Yuan X, Qian W, Zhang B, Shi M, Xie J, Shen B, Xu H, Hou Z, Chen H. Long noncoding RNA uc.345 promotes tumorigenesis of pancreatic cancer by upregulation of hnRNPL expression. Oncotarget. 2016;7:71556-71566. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 40. | Terreri S, Mancinelli S, Ferro M, Vitale MC, Perdonà S, Castaldo L, Gigantino V, Mercadante V, De Cecio R, Aquino G, Montella M, Angelini C, Del Prete E, Aprile M, Ciaramella A, Liguori GL, Costa V, Calin GA, La Civita E, Terracciano D, Febbraio F, Cimmino A. Subcellular Localization of uc.8+ as a Prognostic Biomarker in Bladder Cancer Tissue. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 41. | Sekino Y, Sakamoto N, Ishikawa A, Honma R, Shigematsu Y, Hayashi T, Sentani K, Oue N, Teishima J, Matsubara A, Yasui W. Transcribed ultraconserved region Uc.63+ promotes resistance to cisplatin through regulation of androgen receptor signaling in bladder cancer. Oncol Rep. 2019;41:3111-3118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 42. | Zheng Z, Hong D, Zhang X, Chang Y, Sun N, Lin Z, Li H, Huang S, Zhang R, Xie Q, Huang H, Jin H. uc.77- Downregulation Promotes Colorectal Cancer Cell Proliferation by Inhibiting FBXW8-Mediated CDK4 Protein Degradation. Front Oncol. 2021;11:673223. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 43. | Wang C, Wang Z, Zhou J, Liu S, Wu C, Huang C, Ding Y. TUC.338 promotes invasion and metastasis in colorectal cancer. Int J Cancer. 2017;140:1457-1464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 44. | Sekino Y, Sakamoto N, Goto K, Honma R, Shigematsu Y, Sentani K, Oue N, Teishima J, Matsubara A, Yasui W. Transcribed ultraconserved region Uc.63+ promotes resistance to docetaxel through regulation of androgen receptor signaling in prostate cancer. Oncotarget. 2017;8:94259-94270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 45. | Bao BY, Lin VC, Yu CC, Yin HL, Chang TY, Lu TL, Lee HZ, Pao JB, Huang CY, Huang SP. Genetic variants in ultraconserved regions associate with prostate cancer recurrence and survival. Sci Rep. 2016;6:22124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 46. | Li Q, Li X, Wang C. Uc.206 regulates cell proliferation and apoptosis by targeting P53 in cervical cancer cells. Neoplasma. 2016;63:411-418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 47. | Pereira Zambalde E, Bayraktar R, Schultz Jucoski T, Ivan C, Rodrigues AC, Mathias C, Knutsen E, Silveira de Lima R, Fiori Gradia D, de Souza Fonseca Ribeiro EM, Hannash S, Adrian Calin G, Carvalhode Oliveira J. A novel lncRNA derived from an ultraconserved region: lnc-uc.147, a potential biomarker in luminal A breast cancer. RNA Biol. 2021;18:416-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 48. | Liu X, Zhou X, Deng CJ, Zhao Y, Shen J, Wang Y, Zhang YL. Comprehensive analyses of T-UCR expression profiles and exploration of the efficacy of uc.63- and uc.280+ as biomarkers for lung cancer in Xuanwei, China. Pathol Res Pract. 2020;216:152978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 49. | Gao X, Gao X, Li C, Zhang Y, Gao L. Knockdown of Long Noncoding RNA uc.338 by siRNA Inhibits Cellular Migration and Invasion in Human Lung Cancer Cells. Oncol Res. 2016;24:337-343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 50. | Tian Y, Feng Y. Up-regulation of long noncoding RNA uc.338 predicts poor survival in non-small cell lung cancer. Cancer Biomark. 2018;22:781-785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 51. | das Chagas PF, de Sousa GR, Kodama MH, de Biagi Junior CAO, Yunes JA, Brandalise SR, Calin GA, Tone LG, Scrideli CA, de Oliveira JC. Ultraconserved long non-coding RNA uc.112 is highly expressed in childhood T versus B-cell acute lymphoblastic leukemia. Hematol Transfus Cell Ther. 2021;43:28-34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 52. | Lujambio A, Portela A, Liz J, Melo SA, Rossi S, Spizzo R, Croce CM, Calin GA, Esteller M. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 2010;29:6390-6401. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 147] [Cited by in RCA: 147] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 53. | Xiao L, Wu J, Wang JY, Chung HK, Kalakonda S, Rao JN, Gorospe M. Long Noncoding RNA uc.173 Promotes Renewal of the Intestinal Mucosa by Inducing Degradation of MicroRNA 195. Gastroenterology. 2018;154:599-611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 94] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 54. | Wang JY, Cui YH, Xiao L, Chung HK, Zhang Y, Rao JN, Gorospe M, Wang JY. Regulation of Intestinal Epithelial Barrier Function by Long Noncoding RNA uc.173 through Interaction with MicroRNA 29b. Mol Cell Biol. 2018;38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 55. | Terreri S, Durso M, Colonna V, Romanelli A, Terracciano D, Ferro M, Perdonà S, Castaldo L, Febbraio F, de Nigris F, Cimmino A. New Cross-Talk Layer between Ultraconserved Non-Coding RNAs, MicroRNAs and Polycomb Protein YY1 in Bladder Cancer. Genes (Basel). 2016;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 56. | Bhol CS, Panigrahi DP, Praharaj PP, Mahapatra KK, Patra S, Mishra SR, Behera BP, Bhutia SK. Epigenetic modifications of autophagy in cancer and cancer therapeutics. Semin Cancer Biol. 2020;66:22-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 57. | Patel TN, Roy S, Ravi R. Gastric cancer and related epigenetic alterations. Ecancermedicalscience. 2017;11:714. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 58. | Choi SJ, Jung SW, Huh S, Chung YS, Cho H, Kang H. Alteration of DNA Methylation in Gastric Cancer with Chemotherapy. J Microbiol Biotechnol. 2017;27:1367-1378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 59. | Song Y, Tu J, Cheng Y, Zhou F, Liu P, Zhou S, Gu Y, Sun Y. HHIP Overexpression Suppresses Human Gastric Cancer Progression and Metastasis by Reducing Its CpG Island Methylation. Front Oncol. 2020;10:1667. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 60. | Issa JP. CpG-island methylation in aging and cancer. Curr Top Microbiol Immunol. 2000;249:101-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 119] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 61. | Shigeyasu K, Nagasaka T, Mori Y, Yokomichi N, Kawai T, Fuji T, Kimura K, Umeda Y, Kagawa S, Goel A, Fujiwara T. Clinical Significance of MLH1 Methylation and CpG Island Methylator Phenotype as Prognostic Markers in Patients with Gastric Cancer. PLoS One. 2015;10:e0130409. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 62. | Tedaldi G, Molinari C, São José C, Barbosa-Matos R, André A, Danesi R, Arcangeli V, Ravegnani M, Saragoni L, Morgagni P, Rebuzzi F, Canale M, Pignatta S, Ferracci E, Martinelli G, Ranzani GN, Oliveira C, Calistri D, Ulivi P. Genetic and Epigenetic Alterations of CDH1 Regulatory Regions in Hereditary and Sporadic Gastric Cancer. Pharmaceuticals (Basel). 2021;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 63. | Loh M, Liem N, Vaithilingam A, Lim PL, Sapari NS, Elahi E, Mok ZY, Cheng CL, Yan B, Pang B, Salto-Tellez M, Yong WP, Iacopetta B, Soong R. DNA methylation subgroups and the CpG island methylator phenotype in gastric cancer: a comprehensive profiling approach. BMC Gastroenterol. 2014;14:55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 64. | Curtin K, Slattery ML, Samowitz WS. CpG island methylation in colorectal cancer: past, present and future. Patholog Res Int. 2011;2011:902674. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 97] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 65. | Draht MX, Riedl RR, Niessen H, Carvalho B, Meijer GA, Herman JG, van Engeland M, Melotte V, Smits KM. Promoter CpG island methylation markers in colorectal cancer: the road ahead. Epigenomics. 2012;4:179-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 66. | Toyota M, Ahuja N, Suzuki H, Itoh F, Ohe-Toyota M, Imai K, Baylin SB, Issa JP. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res. 1999;59:5438-5442. [PubMed] |
| 67. | Nazemalhosseini Mojarad E, Kuppen PJ, Aghdaei HA, Zali MR. The CpG island methylator phenotype (CIMP) in colorectal cancer. Gastroenterol Hepatol Bed Bench. 2013;6:120-128. [PubMed] |
| 68. | De Carvalho DD, You JS, Jones PA. DNA methylation and cellular reprogramming. Trends Cell Biol. 2010;20:609-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 159] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 69. | Ehrlich M. DNA hypermethylation in disease: mechanisms and clinical relevance. Epigenetics. 2019;14:1141-1163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 233] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 70. | Malta TM, de Souza CF, Sabedot TS, Silva TC, Mosella MS, Kalkanis SN, Snyder J, Castro AVB, Noushmehr H. Glioma CpG island methylator phenotype (G-CIMP): biological and clinical implications. Neuro Oncol. 2018;20:608-620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 196] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 71. | Kang BW, Chau I. Current status and future potential of predictive biomarkers for immune checkpoint inhibitors in gastric cancer. ESMO Open. 2020;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 72. | Gu H, Zhong Y, Liu J, Shen Q, Wei R, Zhu H, Zhang X, Xia X, Yao M, Ni M. The Role of miR-4256/HOXC8 Signaling Axis in the Gastric Cancer Progression: Evidence From lncRNA-miRNA-mRNA Network Analysis. Front Oncol. 2021;11:793678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 73. | Liu J, Wang H, Liao X. MicroRNA-223-5p targets long non-coding RNA TP73 antisense RNA1 to promote the invasion of gastric cancer. Hum Cell. 2020;33:676-682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 74. | Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis. 2015;35:3-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 640] [Cited by in RCA: 846] [Article Influence: 84.6] [Reference Citation Analysis (0)] |
| 75. | Andrés-León E, Cases I, Alonso S, Rojas AM. Novel miRNA-mRNA interactions conserved in essential cancer pathways. Sci Rep. 2017;7:46101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |