Published online Mar 21, 2015. doi: 10.3748/wjg.v21.i11.3245
Revised: October 11, 2014
Accepted: December 14, 2014
Published online: March 21, 2015
Processing time: 223 Days and 2.5 Hours
AIM
To investigate the biological role of miR-1290 in esophageal squamous cell carcinoma (ESCC) progression and invasion and the underlying mechanism.
METHODS
Quantitative real-time polymerase chain reaction (qRT-PCR) was performed to evaluate miR-1290 expression in ESCC tissue samples. The roles of miR-1290 in cell proliferation, migration and invasion were identified using miR-1290 mimic-transfected cells. In addition, the regulatory effect of miR-1290 on suppressor of cancer cell invasion (SCAI) was evaluated using qRT-PCR, Western blot analysis and a dual luciferase reporter assay.
RESULTS
miR-1290 was significantly upregulated in ESCC tissue samples compared with normal adjacent tissues (9.213 ± 1.150
CONCLUSION
Our findings suggested that miR-1290 may play an oncogenic role in cellular processes of ESCC.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Core Tip: In this study, we reported the clinical significance and biological effects of miR-1290 in esophageal squamous cell carcinoma (ESCC). We found that miR-1290 was significantly up-regulated in ESCC tissues. Moreover, we showed that ectopic expression of miR-1290 significantly promoted ESCC cell growth, migration and invasion. Further investigation revealed that suppressor of cancer cell invasion was a downstream target of miR-1290.
- Citation: Li M, He XY, Zhang ZM, Li S, Ren LH, Cao RS, Feng YD, Ji YL, Zhao Y, Shi RH. MicroRNA-1290 promotes esophageal squamous cell carcinoma cell proliferation and metastasis. World J Gastroenterol 2015; 21(11): 3245-3255
- URL: https://www.wjgnet.com/1007-9327/full/v21/i11/3245.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i11.3245
Esophageal cancer accounts for 2% of all human malignant tumors and is the sixth leading cause of cancer death. Esophageal cancer morbidity evidently varies with geographic location; for example, this cancer type is common in China, Japan and Africa in both males and females[1]. In China, esophageal squamous cell carcinoma (ESCC) is currently the major histologic subtype of esophageal cancer. Despite advances in therapeutic methods, ESCC remains one of the most common malignancies in China with an overall five-year survival rate of 20% after surgery[2].
Tumor proliferation and metastasis are main factors responsible for ESCC mortality. However, the molecular mechanism of proliferation and metastasis remains unclear[3]. Studies have shown that microRNAs (miRNAs) play a considerable role in tumor dissemination[4,5]. miRNAs are a group of endogenous small non-coding RNAs of approximately 22 nucleotides that inversely regulate gene expression by imprecisely binding to a complementary sequence in the 3′-untranslated region (UTR) of their target mRNAs[6]. miRNAs play an important role in gene regulation, cell differentiation, proliferation, apoptosis and tumor genesis[7]. These miRNAs function as either oncogenes or tumor suppressors[8,9]. A study has shown that patients in the same pathological stage of esophageal cancer receiving identical surgical therapy by the same surgeon but with differences in miRNA expression may have distinct prognoses[10].
Previous study have shown that miR-1290 is upregulated in colon cancer cells and osteosarcoma cells[11,12]. These studies have also demonstrated that miR-1290 functions as an oncogenic miRNA. However, the association between miR-1290 and ESCC has not been evaluated yet, and the biological value of miR-1290 in ESCC remains poorly understood. Suppressor of cancer cell invasion (SCAI) is a tumor-suppressor gene that is downregulated in several human tumors. Decreased SCAI levels are tightly correlated with increased invasive cell migration[13]. Nevertheless, whether miR-1290 can regulate SCAI expression in human ESCC cells remains unknown. In the present study, we investigated the relative miR-1290 expression level between tumor and normal tissues; we further studied the possible mechanism of miR-1290 in ESCC metastasis. The results showed that the expression level of miR-1290 in ESCC tissues was higher than that in normal adjacent tissues. Moreover, miR-1290 overexpression promoted colony formation, proliferation, migration and invasion; miR-1290 overexpression also reduced SCAI mRNA and protein levels in Eca109 and TE13 human ESCC cells in vitro. Understanding the molecular mechanisms of miR-1290 in the initiation and progression of human ESCC could provide the basis for developing a treatment strategy for ESCC.
A total of 24 matched human ESCC tumor tissues and normal adjacent tissues (NAT) were collected directly after surgical resection was performed at the First Affiliated Hospital, Nanjing Medical University (China). ll of the tissue samples from patients with no prior neoadjuvant treatment were immediately frozen in liquid nitrogen and stored at -80 °C until miRNA was extracted. Clinicopathological information for all of the samples was available. ESCC tumors were graded according to the 2010 WHO classification of the tumors of digestive system[14]. Our research protocol was approved by the Ethics Review Committee of the Institutional Review Board of the hospital. Standard written consent was obtained from each patient. Total RNA was extracted from tissue samples and cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s protocol.
Human ESCC cell lines Eca109 and TE13 were purchased from the Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). All of the cell lines were maintained in Roswell Park Memorial Institute (RPMI)-1640 medium (Invitrogen) with 10% fetal bovine serum (FBS; Gibco, Gaithersburg, MD, United States) supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin (Invitrogen) at 37 °C in a humidified chamber with 5% CO2. Hsa-miR-1290 mimic (sense 5′-UGGAUUUUUGGAUCAGGGA-3′, antisense 5′-CCUGAUCCAAAAAUCCAUU-3′), negative control oligonucleotide (sense 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense 5′-ACGUGACACGUUCGGAGAATT-3′), has-miR-1290 inhibitor (sequence 5′-UCCCUGAUCCAAAAAUCCA-3′) and miRNA inhibitor negative control (sequence 5′-CAGUACUUUUGUGUAGUACAA-3′) were synthesized by Genepharma (Shanghai, China). Ectopic miR-1290 expression in the cells was achieved by performing transfection with has-miR-1290 mimic using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions.
For quantitative real-time PCR (qRT-PCR), 2 μg of total RNA was reverse transcribed with Bulge-LoopTM miRNA-specific reverse transcription primers (RiboBio, Guangzhou, China). Quantitative PCR was performed with SYBR Premix Ex Taq (Takara, Dalian, China) reagents and Bulge-LoopTM primers (RiboBio, Guangzhou, China) in an ABI PRISM 7900 system (Applied Biosystems, Carlsbad, CA, United States) with small nuclear RNA U6 as a normalization control. The mRNA levels of SCAI were determined by qRT-PCR using SYBR Premix Ex Taq (Takara, Dalian, China) in an ABI PRISM 7900 Sequence Detection System and normalized to GAPDH levels. The SCAI primers used were: forward, 5′-AAGCAGTGGCAGTCCTATTTTG-3′ and reverse, 5′-GCTTCAAGCCATACCGATTATCC-3′. GAPDH primers were: forward, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ and reverse, 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′ (HuaGene, Shanghai, China). All of the samples were normalized to internal controls, and fold changes were calculated using 2-∆∆Ct.
For cell proliferation assays, cells were seeded in 96-well plates with 3000 cells for Eca109 and 2000 cells for TE13 per well on 0 d (24 h after has-miR-1290 mimic or has-miR-1290 inhibitor was transfected and negative control was administered). On 1, 2, 3, 4 and 5 d, cell viability was determined using cell counting kit-8 (CCK-8, Beyotime, Haimen, China). In brief, 10 μL of CCK8 solution was added to each well of the 96-well plate, and the plate was incubated for 2 h in an incubator. After the incubation was performed, the plates were washed with phosphate-buffered saline (PBS). The absorbance of each well was read at a wavelength of 450 nm and a proliferation curve on the basis of absorbance and time was constructed. To perform flow cytometry analysis (FCM) of cell apoptosis, an Annexin V-FITC/PI apoptosis detection kit (KeyGEN, Nanjing, China) was used. At 48 h after transfection, cells were harvested and washed with PBS twice, incubated with Annexin V-FITC and propidium iodide for 15 min in a dark room and analyzed by flow cytometry according to the manufacturer’s instructions.
After transfection was performed, 300 cells were seeded in each well of a six-well culture plate and incubated for 14 d. Fresh culture medium was replaced at an interval of 3 d. The cells were fixed with 75% ethanol and stained with 0.5% crystal violet, and the number of colonies containing > 50 cells was counted.
To perform transwell migration or invasion assays, we placed Eca109 or TE13 cells (1 × 105 cells/well) transfected with has-miR-1290 mimic, has-miR-1290 inhibitor or negative control in 0.2 mL of RPMI-1640 without FBS in the upper chamber of each insert (8 μm pore size, BD Biosciences, United States) with or without 60 μL of 1 mg/mL Matrigel (BD, Biosciences, Bedford, MD, United States). The lower chamber was filled with 600 μL of RPMI1640 medium containing 10% FBS as a nutritional attractant. After 28 h, the cells on the top surface of the membrane were carefully removed using a cotton swab. Migrant cells attached to the lower surface were fixed with 75% methanol and stained with crystal violet for 30 min. The number of cells was counted in five different fields of view using an inverted microscope (magnification × 100).
The putative targets of miRNA were predicted using TargetScan (http://www.targetscan.org) and miRanda (http://www.microrna.org).
The 3′-UTR of SCAI containing the predicted miR-1290 binding seed sequence was synthesized and directly cloned downstream of the firefly luciferase gene at XbaI sites to create a pGL3-SCAI-3′UTR-wt plasmid (5′-ACCCUGAGAAGAGUAAAUCAUUUAUUUUUGUAUAAUGAGGUAAAUCCAACUCUUAUACUUGGACCUAAGUUAAAUGUCUGGAUUUGGA-3′; Invitrogen, Shanghai, China). The corresponding mutant reporter plasmid (pGL3-SCAI-3′UTR-mut; 5′-ACCCUGAGAAGAGUAAAUCAUUUAUUUUUGUAUAAUGAGGUUAGUCUAACUCUUAUACUUGGACCUAAGUUAAAUGUCUGGAUUUGGA-3′) was then synthesized. The pGL3-SCAI-3′UTR-wt/-mut reporter plasmid (800 ng) and phRL-TK vector (800 ng) expressing Renilla luciferase (Invitrogen) were co-transfected in Eca109 and TE13 cells with 80 ng has-miR-1290 mimic and negative control using Lipofectamine 2000 reagent (Invitrogen). After 48 h, cells were harvested and lysed with passive lysis buffer (Promega). Luciferase activity was determined using a dual-luciferase reporter assay system (Promega, Madison, WI, United States) according to the manufacturer’s protocol. Renilla luciferase activities were used for normalization.
Western blot analysis was performed to detect the protein expression of SCAI in ESCC tissues and cell lines. The cells were lysed 48 h post-transfection with RIPA lysis buffer (Beyotime, Jiangsu, China) containing protease inhibitor; the proteins were then harvested. Total protein content was quantified by BCA assay (Beyotime). Equal amounts of protein extracts (30 to 40 ng) were separated using 8% gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, United States). Afterwards, blots were blocked with 5% fat-free milk powder for 1 h. The membranes were incubated overnight at 4 °C in a 1:500 dilution of anti-human SCAI rabbit monoclonal antibody (Abcam, Cambridge, MA, United States). The blots were subsequently incubated with a horseradish peroxidase-conjugated secondary antibody (1 : 5000) and visualized using a super enhanced chemiluminescence detection reagent (Amersham Biosciences, Piscataway, NJ). Protein expression was assessed using Alpha Innotech imaging software (San Leandro, CA). GAPDH was used as an endogenous protein for normalization.
Data are presented as mean ± standard deviation (SD) from at least three independent experiments. Statistical analyses were performed with SPSS 18.0 software (SPSS Inc., Chicago, IL, United States). The difference between groups was analyzed using a two-tailed Student’s t-test to compare two independent groups only and ANOVA followed by Student-Newman-Keuls Q test to compare two groups among three groups. The relationship between miR-1290 and SCAI expression was explored by Spearman’s correlation analysis. Significant associations between miR-1290 changes and clinicopathological parameters were assessed using a χ2 test. Two-sided P values < 0.05 were considered statistically significant.
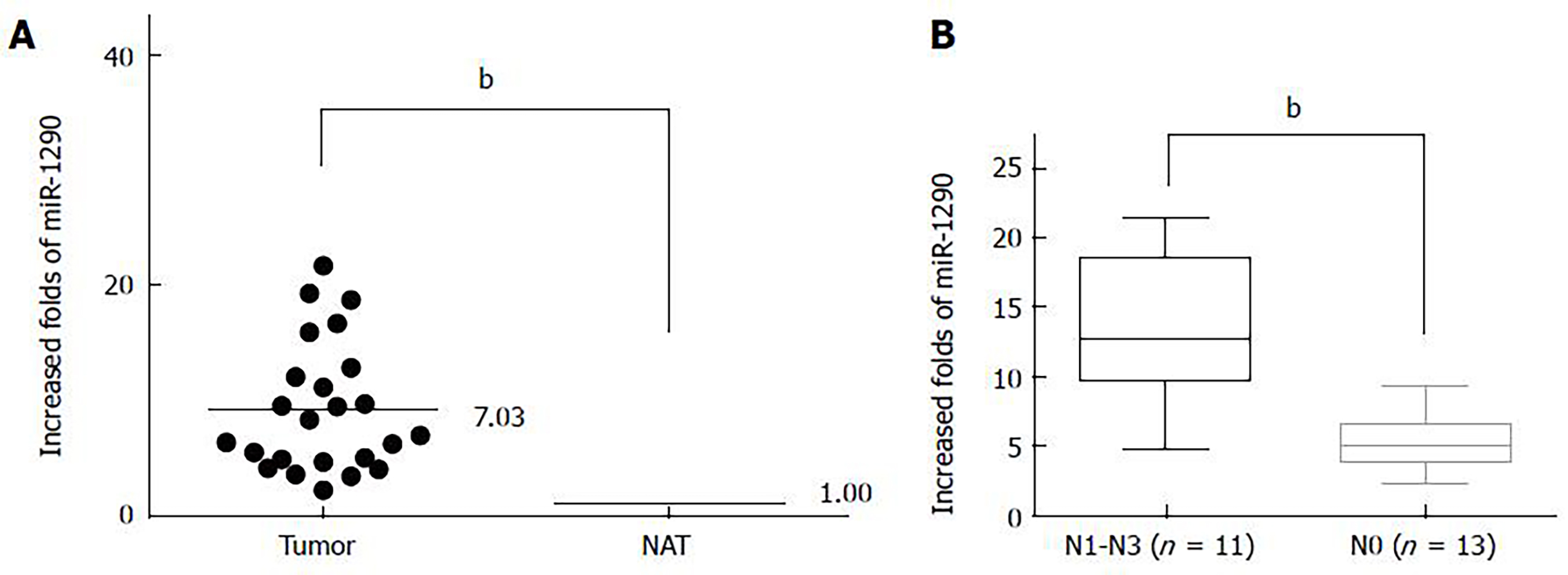
Relative miR-1290 expression level is specifically upregulated and correlated with lymph node metastasis and tumor-node-metastasis stage in patients with ESCC
Relative miR-1290 expression was detected by qRT-PCR between paired tumor tissues and normal adjacent tissues from 24 patients with ESCC. Our results demonstrated that the relative fold increases in miR-1290 expression were markedly upregulated in ESCC samples compared with the paired tumor-adjacent tissues (9.213 ± 1.150 vs 1.000 ± 0.0) (P < 0.01; Figure 1A). To evaluate the clinical value of miR-1290 in ESCC patients, we divided the patients into two groups according to the median value (6.6181) of miR-1290 level. The association between relative miR-1290 expression and clinicopathological information was then analyzed. A significant difference was observed between the two groups in terms of differentiation (P = 0.021), N classification (P = 0.006) and tumor-node-metastasis stage (P = 0.021) (Figure 1B, Table 1). No significant association was found between miR-1290 expression and other clinical characteristics, such as age, gender and T classification (Table 1). Hence, upregulated miR-1290 expression was closely related to ESCC metastasis.
| Characteristic | Total(n = 24) | miR-1290 Expression | P value | |
| Low | High | |||
| Age | ||||
| < 60 | 12 | 5 | 7 | 0.682 |
| ≥ 60 | 12 | 6 | 6 | |
| Gender | ||||
| Female | 10 | 5 | 5 | 0.729 |
| Male | 14 | 6 | 8 | |
| Differentiation | ||||
| High | 13 | 3 | 10 | 0.021 |
| Middle + Low | 11 | 8 | 3 | |
| T Classification | ||||
| T1 + T2 | 10 | 3 | 7 | 0.240 |
| T3 + T4 | 14 | 8 | 6 | |
| N Classification | ||||
| N0 | 13 | 3 | 10 | 0.006 |
| N1-N3 | 11 | 8 | 3 | |
| TNM Stage | ||||
| Ⅰ+Ⅱ | 15 | 4 | 11 | 0.021 |
| Ⅲ | 9 | 7 | 2 | |
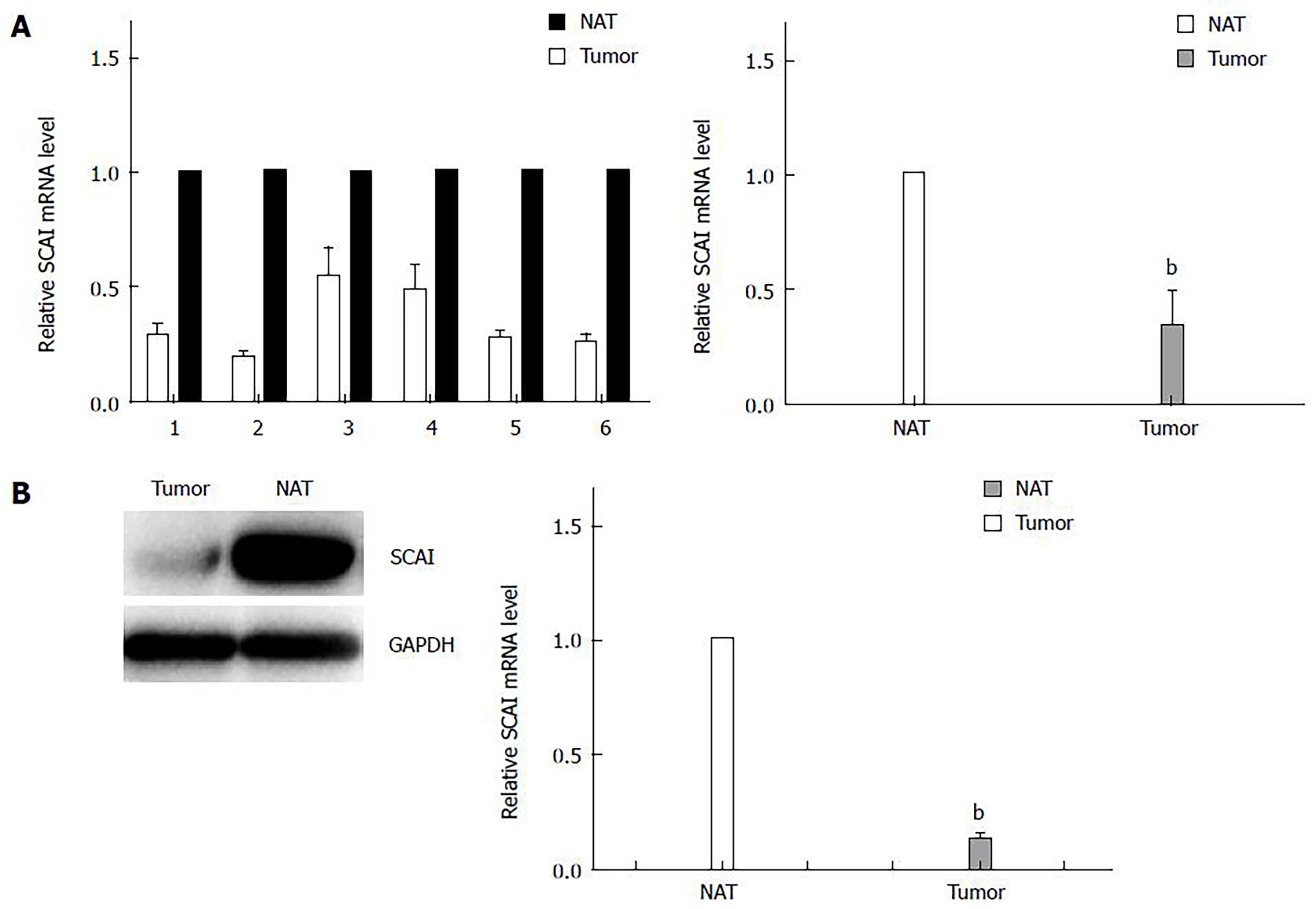
The mRNA and protein expression of SCAI in ESCC tissues was analyzed by qRT-PCR and Western blot analysis between paired tumor tissues and normal adjacent tissues from six patients with ESCC. These results showed that the relative mRNA and protein expression of SCAI was downregulated in ESCC tissues (P < 0.01; Figures 2A and 2B), which is in accordance with the results from a previous study[15].
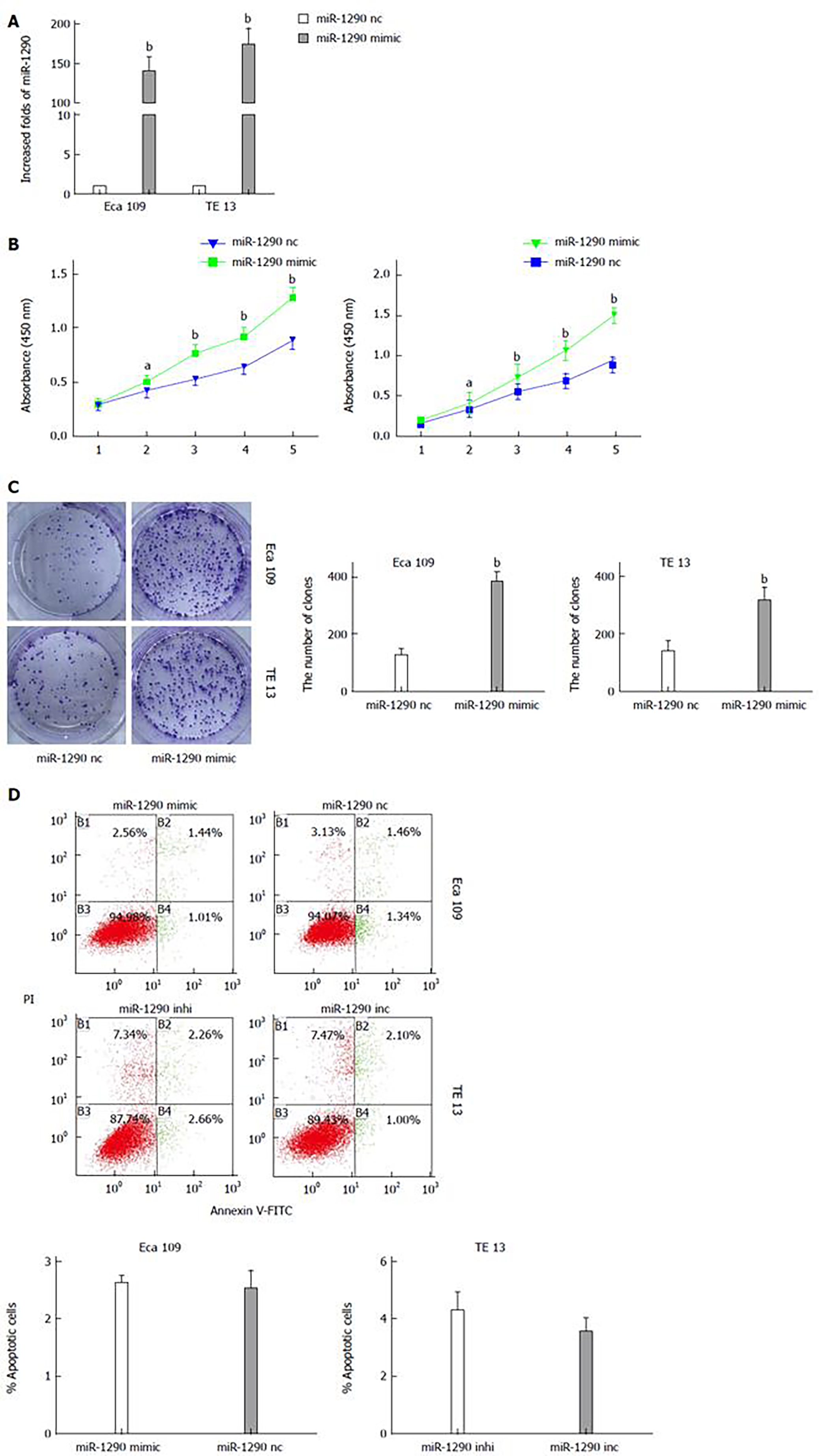
The significantly increased expression of miR-1290 in ESCC tissues prompted us to investigate the possible biological function of miR-1290 in tumorigenesis. qRT-PCR analysis results showed that miR-1290 expression increased by more than 100-fold in Eca109 and TE13 cells transfected with has-miR-1290 mimic compared with the control cells (P < 0.01; Figure 3A). A CCK8 staining assay revealed that miR-1290 promoted significant proliferation in Eca109 and TE13 cell lines transfected with has-miR-1290 mimic compared with the control cells (P < 0.01; Figure 3B). We also evaluated the ability of ECa109 and TE13 cell lines transfected with has-miR-1290 mimic to form colonies. Our data indicated that miR-1290 significantly stimulated ECa109 and TE13 cells to grow numerous and large colonies on soft agar (P < 0.01; Figure 3C). miR-1290 overexpression in Eca109 cells and miR-1290 under expression in TE13 cells did not significantly change the apoptotic ability of cells (Figure 3D). The results revealed that miR-1290 enhanced the proliferation ability of ESCC cells.
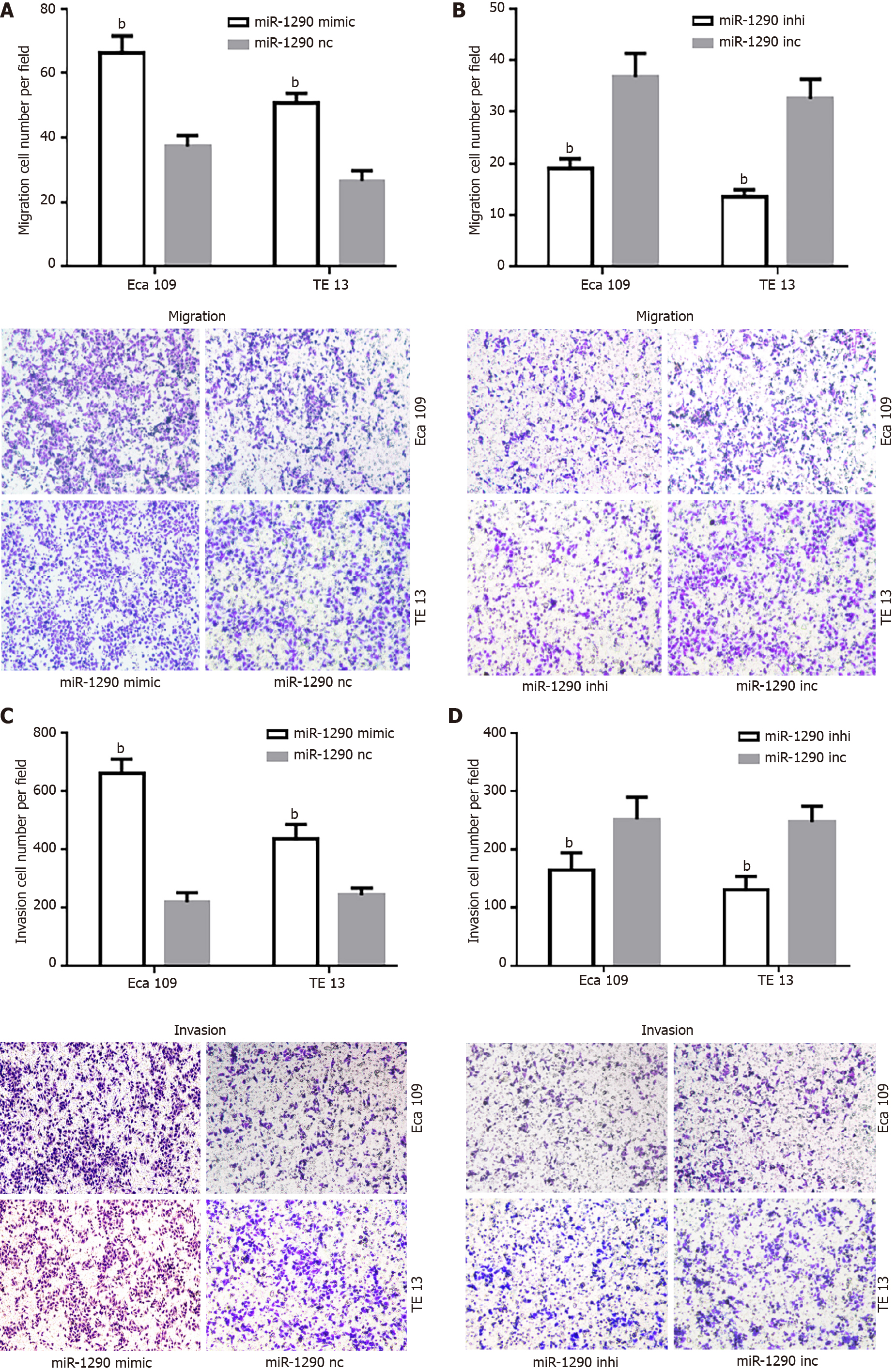
To understand the biological effects of miR-1290 overexpression on the migration and invasion of ESCC cell lines in vitro, we performed transwell assays by transfecting Eca109 and TE13 cell lines with has-miR-1290 mimic or inhibitor. Matrigel-coated (for invasion) or uncoated (for migration) Transwell assays revealed that miR-1290 overexpression markedly promoted the invasion and migration of Eca109 and TE13 cells (P < 0.01; Figures 4A and 4C). The effect of miR-1290 under expression was examined. As expected, miR-1290 under expression remarkably decreased the invasion capabilities of Eca109 and TE13 cells (P < 0.01; Figures 4B and 4D). These observations suggested that miR-1290 significantly promoted the in vitro migration and invasion of ESCC cell lines.
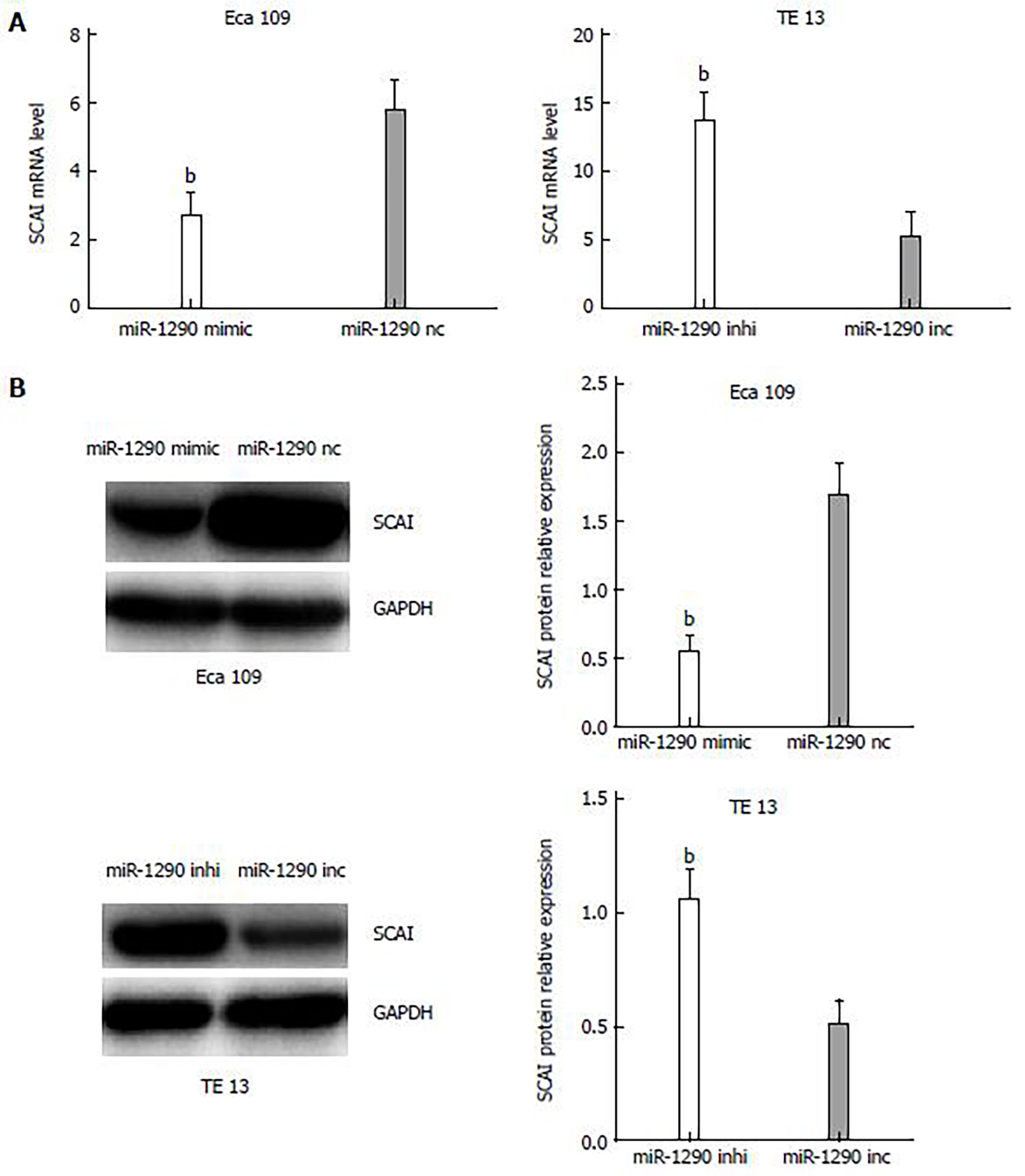
To further observe the correlation of miR-1290, SCAI mRNA and protein expression in Eca109 and TE13 cell lines, we performed qRT-PCR and Western blot analyses. qRT-PCR analysis results indicated a decrease in the mRNA expression of SCAI in miR-1290-overexpressing cells, whereas miR-1290- under-expressing cells had increased mRNA levels of SCAI (P < 0.01; Figure 5A). Similar changes were found in SCAI protein levels from Western blot assays (P < 0.01; Figure 5B). These findings confirmed that miR-1290 overexpression downregulated the mRNA and protein expression of SCAI in Eca109 and TE13 cell lines.
miR-1290 directly binds to the 3′-UTR of SCAI
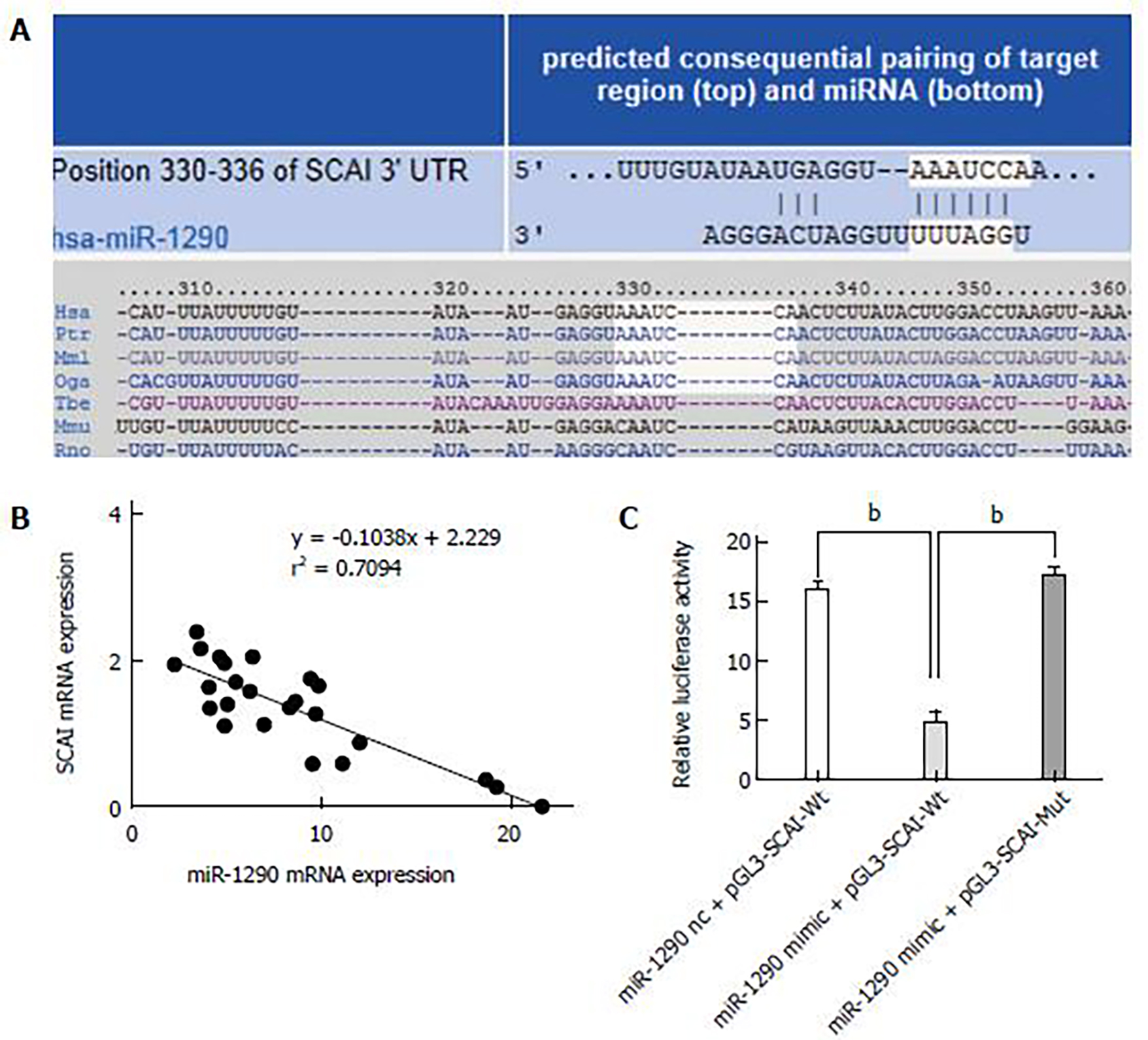
The putative miR-1290 target genes were predicted using the target prediction programs TargetScan and miRDB. SCAI was identified as a candidate miR-1290 target gene and sequence analysis results indicated that miR-1290 target sequence at 330 nt to 336 nt of the SCAI 3′-UTR was highly conserved across different species (Figure 6A). The relationship between the SCAI mRNA level and miR-1290 in 24 ESCC tissues was tested using qRT-PCR, and data showed an clearly negative relationship between expression SCAI and miR-1290 (r = –0.842, P = 0.000, Figure 6B). To validate whether SCAI is a valid target of miR-1290, we inserted wild-type or mutant SCAI 3′-UTR sequence in the downstream region of the luciferase reporter gene and co-expressed these sequences with either has-miR-1290 mimic or has-miR-1290 nc in Eca109 cells. miR-1290 overexpression caused an unambiguous decrease in relative luciferase activity (P < 0.01; Figure 5C); in contrast, activity did not decrease in the mutant 3′-UTR reporter, indicating that functionality depends on an intact seed sequence. Therefore, SCAI can be directly suppressed by miR-1290 via mRNA degradation and translation repression.
Evidence has shown anomalous miRNA expression in various types of human tumors[16,17]. Thus, studies have focused on tumor-associated miRNAs and specific target genes to elucidate biological mechanism. The identification of cancer-specific miRNAs and their target genes is necessary for understanding their role in tumor metastasis, which may be a requisite to define new therapeutic targets. miR-1290 is upregulated in several cancer forms [11,18]. Therefore, whether miR-1290 is upregulated in ESCC and whether miR-1290 can promote ESCC cell metastasis are postulated.
In the present study, miR-1290 expression in 24 pairs of human ESCC tumor tissues and normal adjacent tissues was determined by qRT-PCR. The miR-1290 expression was upregulated in ESCC. The upregulated miR-1290 in human ESCC may promote the tumor initiation and progression; this result indicates that miR-1290 may play an oncogenic role[11]. Before this study was conducted, however, the role of miR-1290 and its target genes was unclear in ESCC. Therefore, this study focused on the biological mechanisms of miR-1290 in human ESCC.
To characterize the functions of miR-1290 in human ESCC, we examined the changes in Eca109 and TE13 cell lines after miR-1290 was overexpressed and under-expressed. A CCK8 assay was performed to analyze the cell viability in Eca109 and TE13 cell lines; our results showed that miR-1290 enhanced cell viability. In the colony formation assay, however, miR-1290 promoted colony formation activities of Eca109 and TE13 cell lines. These findings showed that ectopic miR-1290 expression affected cell viability over a short time period and enhanced ESCC cell proliferation. Transwell assays with or without Matrigel were conducted to determine the functions of miR-1290 in the migration and invasion of Eca109 and TE13 cells. Our results demonstrated that miR-1290 overexpression significantly accelerated the migration and invasion of Eca109 and TE13 cells compared with the control group. Conversely, the migratory and invasive abilities were markedly decreased in Eca109 and TE13 cells transfected with has-miR-1290 inhibitor. In summary, miR-1290 promoted ESCC colony formation, migration and invasion in Eca109 and TE13 cell lines.
In this research, the role of miR-1290 in targeting SCAI in human Eca109 and TE13 cell lines was considered. Using bioinformatics technology, we confirmed SCAI as one of the target genes of miR-1290. SCAI functions in the RhoA-Dia1 signal transduction pathway and localizes in the nucleus, where it binds and inhibits myocardin-related transcription factor MAL by forming a ternary complex with the serum response factor (SRF)[13]. In a previous study[15], it was found that protein and mRNA expression of SCAI was significantly downregulated in glioma tissues and cell lines. SCAI silencing robustly promoted invasive and cancer stem cell-like phenotypes of glioma cells. Furthermore, SCAI downregulation activated Wnt/beta-catenin signaling and Wnt/beta-catenin pathway inhibition abrogated the effects of SCAI downregulation on glioma cell aggressiveness. SCAI acts as a transcriptional modulator to regulate cancer cell motility by suppressing MAL/SRF-dependent gene transcription[19]. These studies have suggested that SCAI may be involved in cancer development. Thus, a lower SCAI expression level was observed in ESCC than in adjacent normal tissues; miR-1290 expression was inversely correlated with SCAI expression in tumor tissues. Furthermore, the relative fluorescence intensity of pGL3-SCAI-3′UTR-wt was specifically responsive to miR-1290 overexpression. A mutation in the miR-1290 binding site abolished the effect of miR-1290 on the regulation of fluorescence intensity. An increase in mRNA and protein levels of SCAI was found in Eca109 and TE13 cells transfected with a miR-1290 inhibitor. These results suggested that SCAI is a target of miR-1290 and is negatively regulated. SCAI may also exhibit anti-proliferative and anti-malignant transformation effects in Eca109 and TE13 cell lines.
In summary, miR-1290 expression was upregulated in ESCC tissues; miR-1290 elicited oncogenic effects, including the promotion of ESCC cell proliferation, migration and invasion, by targeting the anti-oncogene SCAI, highlighting the function of miR-1290 in tumor progression.
Esophageal cancer is the sixth leading cause of cancer-related deaths in China. Although recent developments in therapeutic strategies have helped cure many patients with early stage disease, the prognosis of patients with advanced disease and metastasis remains poor.
MiRNAs have been found to be involved in the regulation of multiple pathological processes that contribute to tumorigenesis and metastasis, such as tumor cell proliferation, differentiation, apoptosis, and invasion. In esophageal squamous cell carcinoma (ESCC), studies have indicated that miRNAs play important roles in regulating tumor invasion and metastasis. Previously, Zhang et al reported that miR-100 promoted migration and invasion through mammalian target of rapamycin in ESCC. However, the role of miR-1290 in ESCC progression and metastasis remains unclear and needs further exploration.
The authors found that the level of miR-1290 was significantly up-regulated in ESCC tissues compared with normal adjacent tissues. Ectopic expression of miR-1290 markedly promoted the proliferation, invasion and metastasis in ESCC cell lines. Further analysis indicated that the suppressor of cancer cell invasion (SCAI) was a direct downstream target of miR-1290. Collectively, these results demonstrated that miR-1290 promoted cell invasion and metastasis by targeting SCAI, thus providing a valuable target for cancer therapy.
The findings in this study indicated that miR-1290 was significantly up-regulated in ESCC with distant metastases. Further investigation identified that the SCAI was a direct target of miR-1290. Taken together, these data implicate that miR-1290 might be used a prognostic indicator and therapeutic target in ESCC patients.
MicroRNAs: A group of small non-coding RNA molecules (approximately 22 nucleotides in length) found in plants, animals, and some viruses that function in transcriptional and post-transcriptional regulation of gene expression.
The authors used clinical samples for expression analyses, and a number of experimental state-of-the-art techniques to investigate the respective questions in an in vitro model. The different steps of the manuscript are logical, and the results present some very interesting findings about the role of miR-1290 in esophageal squamous cell carcinoma.
P-Reviewer: Franzen T, Hummel R S-Editor: Ma YJ L-Editor: A P-Editor: Liu XM
| 1. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23762] [Cited by in RCA: 25541] [Article Influence: 1824.4] [Reference Citation Analysis (7)] |
| 2. | McCann J. Esophageal cancers: changing character, increasing incidence. J Natl Cancer Inst. 1999;91:497-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 3. | Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell. 2006;127:679-695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2932] [Cited by in RCA: 3209] [Article Influence: 168.9] [Reference Citation Analysis (0)] |
| 4. | Zhu M, Zhang N, He S, Lui Y, Lu G, Zhao L. MicroRNA-106a targets TIMP2 to regulate invasion and metastasis of gastric cancer. FEBS Lett. 2014;588:600-607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 5. | Yang MH, Yu J, Jiang DM, Li WL, Wang S, Ding YQ. microRNA-182 targets special AT-rich sequence-binding protein 2 to promote colorectal cancer proliferation and metastasis. J Transl Med. 2014;12:109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 65] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 6. | Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1638] [Cited by in RCA: 1781] [Article Influence: 127.2] [Reference Citation Analysis (0)] |
| 7. | Sotillo E, Thomas-Tikhonenko A. Shielding the messenger (RNA): microRNA-based anticancer therapies. Pharmacol Ther. 2011;131:18-32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 8. | Peng Y, Liu YM, Li LC, Wang LL, Wu XL. MicroRNA-338 inhibits growth, invasion and metastasis of gastric cancer by targeting NRP1 expression. PLoS One. 2014;9:e94422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 9. | Fenger JM, Bear MD, Volinia S, Lin TY, Harrington BK, London CA, Kisseberth WC. Overexpression of miR-9 in mast cells is associated with invasive behavior and spontaneous metastasis. BMC Cancer. 2014;14:84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 10. | Zhao BS, Liu SG, Wang TY, Ji YH, Qi B, Tao YP, Li HC, Wu XN. Screening of microRNA in patients with esophageal cancer at same tumor node metastasis stage with different prognoses. Asian Pac J Cancer Prev. 2013;14:139-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 57] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 11. | Wu J, Ji X, Zhu L, Jiang Q, Wen Z, Xu S, Shao W, Cai J, Du Q, Zhu Y, Mao J. Up-regulation of microRNA-1290 impairs cytokinesis and affects the reprogramming of colon cancer cells. Cancer Lett. 2013;329:155-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 12. | Dai N, Zhong ZY, Cun YP, Qing Y, Chen Ch, Jiang P, Li MX, Wang D. Alteration of the microRNA expression profile in human osteosarcoma cells transfected with APE1 siRNA. Neoplasma. 2013;60:384-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 13. | Brandt DT, Baarlink C, Kitzing TM, Kremmer E, Ivaska J, Nollau P, Grosse R. SCAI acts as a suppressor of cancer cell invasion through the transcriptional control of beta1-integrin. Nat Cell Biol. 2009;11:557-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 116] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 14. | Montgomery E, Field JK, Boffetta P, Daigo Y, Shimizu M, Shimoda T. Squamous cell carcinoma of the oesophagus. In: Bosman FT, Carneiro F, Hruban RH, Theise ND. WHO classification of tumors of the digestive system. Lyon: IARC Press, 2010: 18-24. |
| 15. | Chen X, Hu W, Xie B, Gao H, Xu C, Chen J. Downregulation of SCAI enhances glioma cell invasion and stem cell like phenotype by activating Wnt/β-catenin signaling. Biochem Biophys Res Commun. 2014;448:206-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 16. | Zhang N, Wang X, Huo Q, Sun M, Cai C, Liu Z, Hu G, Yang Q. MicroRNA-30a suppresses breast tumor growth and metastasis by targeting metadherin. Oncogene. 2014;33:3119-3128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 127] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 17. | Yu SJ, Hu JY, Kuang XY, Luo JM, Hou YF, Di GH, Wu J, Shen ZZ, Song HY, Shao ZM. MicroRNA-200a promotes anoikis resistance and metastasis by targeting YAP1 in human breast cancer. Clin Cancer Res. 2013;19:1389-1399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 128] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 18. | Li A, Yu J, Kim H, Wolfgang CL, Canto MI, Hruban RH, Goggins M. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin Cancer Res. 2013;19:3600-3610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 241] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 19. | Kreßner C, Nollau P, Grosse R, Brandt DT. Functional interaction of SCAI with the SWI/SNF complex for transcription and tumor cell invasion. PLoS One. 2013;8:e69947. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |