Published online Mar 6, 2021. doi: 10.12998/wjcc.v9.i7.1600
Peer-review started: November 23, 2020
First decision: December 8, 2020
Revised: December 10, 2020
Accepted: December 24, 2020
Article in press: December 24, 2020
Published online: March 6, 2021
Processing time: 97 Days and 16.4 Hours
There are many factors that lead to dwarfism, and the mechanism has not yet been elucidated. Next-generation sequencing may identify candidate-related gene mutations, which may clarify the molecular cause.
To analyze genetic variation by using a constructed panel related to dwarfism by utilizing next-generation sequencing platform sequencing analysis to screen candidate-related gene mutations.
Physical and laboratory characteristics, including clinical examination, growth hormone drug challenge test, serum insulin-like growth factor-1 (IGF-1), IGF binding protein 3, other related tests, imaging examination, and chromosome karyotyping, were analyzed. Next-generation sequencing was performed to analyze pathogenicity variability.
In the 39 dwarfism patients, 10 had pathogenicity variability. Gene variation was found in the OBSL1, SLC26A2, PTPN11, COL27AI, HDAC6, CUL7, FGFR3, DYNC2H1, GH1, and ATP7B genes. Of the 10 patients with pathogenicity variability, the related physical characteristics included double breast development and growth hormone deficiency, enuresis and indirect inguinal hernia on the left, two finger distance of 70.2 cm, head circumference of 49.2 cm, ischium/lower body length of 1.8 cm, weak limb muscles, and partial growth hormone deficiency. After 6 mo of growth hormone therapy, the concentrations of IGF-1 and IGF binding protein 3 increased from 215.2 ± 170.3 to 285.0 ± 166.0 and 3.9 ± 1.4 to 4.2 ± 1.1, respectively.
OBSL1, SLC26A2, PTPN11, COL27AI, HDAC6, CUL7, FGFR3, DYNC2H1, GH1, and ATP7B genes may be related to the incidence of dwarfism, and more research needs to be performed to elucidate the mechanism.
Core Tip: Genetic variation may relate to the incidence of dwarfism. After Solexa sequencing, the OBSL1, SLC26A2, PTPN11, COL27AI, HDAC6, CUL7, FGFR3, DYNC2H1, GH1, and ATP7B genes may be related to dwarfism and clarify the molecular cause.
- Citation: Yang LL, Liang SS. Study on pathogenic genes of dwarfism disease by next-generation sequencing. World J Clin Cases 2021; 9(7): 1600-1609
- URL: https://www.wjgnet.com/2307-8960/full/v9/i7/1600.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i7.1600
Dwarfism is one of the most common diseases in the endocrine system of children. Dwarfism is defined as a height lower than the average height of the average population by two standard deviations (-2 SD) or lower than the third percentile (-1.88 SD) in children of the same race, sex, and age[1]. Human height is related to a combination of genetics, hormones, nutrition, environment, and other factors. It is a complex process involving multiple genes and multiple factors. Genetic factors are the main factors affecting individual height differences, and the heritability of human height accounts for approximately 80%[2]. There are many factors that lead to dwarfism. Genetic, nutritional, environmental, mental and mental diseases, intrauterine growth retardation, hypothalamic-pituitary-insulin-like growth factor growth axis dysfunction, chromosomal aberrations, systemic chronic diseases, genetic metabolic diseases and endocrine hormones, and other factors are related to the incidence of dwarfism[3]. The mechanism has not yet been clarified.
With the rapid development of genetic technology, the causes of dwarfism include endocrine factors and nonendocrine factors. Endocrine factors include growth hormone deficiency, primary hypothyroidism, adrenal hyperplasia congenital adrenal hyperplasia, pseudoparathyroidism, Williams syndrome, and other diseases. Nonendocrine factors include idiopathic short stature, familial shortness, constitutional puberty, delay of puberty, intrauterine growth retardation, congenital ovarian hypoplasia syndrome, and other diseases[4].
Due to the diverse clinical manifestations of dwarfism, the existence of phenotypic overlap between diseases and high genetic heterogeneity, the etiology cannot be clarified simply by asking about medical history, clinical manifestations, and laboratory tests[5]. The rapid development of emerging technologies has resulted in increased knowledge on the related molecular etiological mechanisms. Some of the unexplained cases of short stature can be clarified by studying their genetic background by using related genetic testing methods[6]. Molecular diagnosis methods have the characteristics of high sensitivity and good specificity. They can not only provide clear diagnostic evidence for the clinic to reduce the misdiagnosis and missed diagnosis of the disease but also guide genetic counseling to provide a reliable basis for prenatal diagnosis[7]. Efficient and highly sensitive diagnosis methods have become the focus of dwarfism research in recent years.
In our study, we aimed to analyze the genetic variation by using a constructed panel related to dwarfism through next-generation sequencing platform sequencing analysis and screened candidate-related gene mutations that may clarify the molecular cause and provide a scientific basis for clinical treatment.
The samples of patients with dwarfism disease in this study came from the Quanzhou First Hospital, and a total of 39 dwarf patients were enrolled. This study was reviewed and approved by the Quanzhou First Hospital Review Board. Informed consent was obtained from the parents or guardians of the children prior to examinations. The inclusion criteria of primary growth hormone deficiency included the following: (1) The height lags behind the average height of normal healthy children of the same area, age, and sex -2.5 SD; (2) Short shape; (3) No history of birth injury/birth suffocation, head trauma/tumor, etc.; (4) The peak growth hormone (GH) of two GH drug challenge tests was < 10 μg/L; and (5) Simple growth hormone deficiency and multiple pituitary hormones or total pituitary hormone deficiency. The inclusion criteria of idiopathic short stature included the following: (1) The height lags behind the average height of normal healthy children of the same area, age, and sex -2.5 SD; (2) GH drug challenge test GH peak value ≥ 10 μg/L; and (3) No chronic systemic diseases (digestion, kidney, heart, blood system diseases, etc.), nutritional diseases, genetic diseases, other endocrine diseases, chromosome abnormalities, bone diseases with definite genetic diagnosis (such as cartilage dysplasia, hypophosphatemia rickets, etc.), and other causes of short stature; and (4) No familial short stature. The exclusion criteria were as follows: (1) Chronic systemic diseases, such as congenital heart disease, chronic kidney disease, blood system diseases, disease, malnutrition, and long-term treatment of children with glucocorticoids; (2) Hereditary-familial shortness; (3) Constitutive growth delay; (4) Acquired hypothyroidism; (5) Cushing's syndrome; and (6) Dwarfism caused by tumors, trauma, surgery, radiotherapy, and chemotherapy.
Clinical examination included posture, body shape, facial features, gait, skin color, nutritional status (abdominal subcutaneous fat), bone development (limbs, spine, etc.), thoracic frame, head and neck, cardiopulmonary and abdominal examinations, and evaluation of gonads and secondary sexual characteristics. Growth hormone drug challenge tests included clonin challenge tests and arginine challenge tests. The concentrations of serum insulin-like growth factor-1 (IGF-1) and IGF binding protein 3 (IGFBP3) were detected by an enzyme-linked immunoassay test kit. Other related tests included routine hematuria and biochemical tests, such as liver function, kidney function, electrolytes, blood glucose, and insulin determination. Imaging examinations included detecting left-hand orthotopic X-ray bone age and using the Greulich-Pyle atlas method (referred to as the G-P method for short) to judge the maturity of bone age; magnetic resonance imaging of the cranial saddle area; X-rays of the spine, pelvis, and extremities if necessary; and B-mode ultrasound. Chromosome karyotypes were analyzed by the G-banding method.
After informed consent was obtained from the family members of the children, 2 mL of venous blood from the children and their parents was collected in ethylenediami-netetraacetic acid tubes, and the basic information of the child (name, case number, sex, age, diagnosis, etc.) was marked on the ethylenediaminetetraacetic acid tubes. Deoxyribonucleic acid (DNA) was extracted according to the instructions of the blood/cell/tissue gene extraction DNA kit. For ribonucleic acid probe preparation, refer to UC Santa Cruz. Polymerase chain reaction was performed to prepare a whole-genome sample library. A second-generation sequencing library construction kit was used to construct the sample DNA library, which was then captured by xGen Exome research panel v1.0 (IDT) and sequenced. The final product was analyzed on the HiSeq 4000 platform (Illumina, San Diego, CA, United States). The steps to filter the target area screening items were the 1000 Genomes Project (≤ 0.01), ExAC (≤ 0.01), Exome Variant Server (≤ 0.01), and an in-house database (≤ 0.05).
Statistical analyses were performed with Statistical Analysis System software SPSS 16.0 (Chicago, IL, United States), and data are presented as the mean ± SD. P < 0.05 was considered as significantly difference.
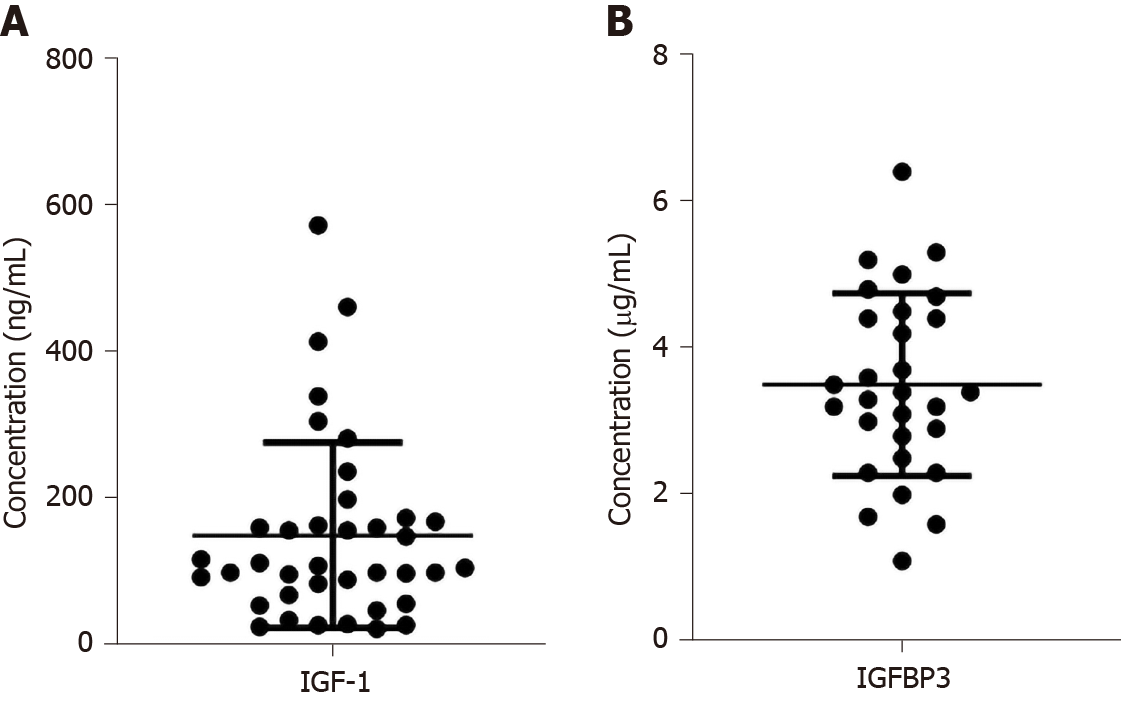
The clinical characteristics are shown in Table 1. A total of 39 dwarf patients were enrolled in our study. The 39 dwarfism patients included 27 male and 12 female patients. The age ranged from 2 to 14 years. The mean height was 107.3 ± 16.4 cm. The mean weight was 17.7 ± 5.8 kg. The mean height stand error was 30.8. The mean arginine peak was 3.9 ± 4.0, and the mean L-dopa peak was 5.6 ± 6.1. As shown in Figure 1, the concentrations of IGF-1 and IGFBP3 were 151.4 ± 125.5 and 3.5 ± 1.2, respectively.
| Number | Gender | Age | Height | Weight | Height SD | AP | LDP |
| 1 | Male | 11 | 132 | 27.7 | -2.55 | 6.81 | 4.96 |
| 2 | Male | 11 | 124.7 | 28 | -3.12 | 5.32 | 2.99 |
| 3 | Female | 4 | 94.3 | 14 | -2.26 | 1.65 | 3.74 |
| 4 | Male | 4 | 91 | 12 | -3.96 | 0.83 | 4.26 |
| 5 | Male | 8 | 118 | 19.5 | -2.94 | 3.7 | 4.9 |
| 6 | Male | 5 | 101.5 | 14 | -3.13 | 5.55 | 6.04 |
| 7 | Male | 14 | 124.8 | 12 | -3.39 | 0.89 | 0.56 |
| 8 | Male | 4 | 90.5 | 11 | -3.9 | 1.77 | 1.12 |
| 9 | Male | 10 | 125.8 | 21.6 | -2.55 | 2.05 | 5.52 |
| 10 | Male | 10 | 122.2 | 24 | -2.9 | 0.96 | 9.95 |
| 11 | Female | 5 | 95 | 13.5 | -3.62 | 9.54 | 0.69 |
| 12 | Male | 7 | 114 | 19.5 | -2.75 | 3.62 | 4.18 |
| 13 | Male | 14 | 145 | 34.6 | -2.9 | 15.18 | 27.24 |
| 14 | Female | 6 | 103 | 17.1 | -3.11 | 2.08 | 9.71 |
| 15 | Male | 4 | 93 | 11.5 | -3.74 | 0.63 | 3.35 |
| 16 | Female | 10 | 121.4 | 23.9 | -3.1 | 9.97 | 12.07 |
| 17 | Male | 14 | 138 | 29.5 | -3.12 | 1.63 | 1.99 |
| 18 | Male | 6 | 107 | 19 | -2.45 | 0.45 | 1.18 |
| 19 | Female | 5 | 101 | 17 | -2.51 | 6.62 | 6.17 |
| 20 | Male | 4 | 90.1 | 12 | -3.59 | 6.47 | 1.99 |
| 21 | Male | 13 | 135.5 | 25 | -3.2 | 2.42 | 0.65 |
| 22 | Male | 5 | 97.4 | 15.8 | -4 | 5.31 | 3.08 |
| 23 | Male | 5 | 101 | 17 | -3.04 | 1.84 | 3.57 |
| 24 | Male | 6 | 103 | 18 | -3.48 | 5.16 | 4.75 |
| 25 | Male | 7 | 109 | 18 | -2.49 | 2.4 | 12.6 |
| 26 | Male | 3 | 90.7 | 13.5 | -2.84 | 3.98 | 7.52 |
| 27 | Male | 5 | 100 | 15.5 | -2.63 | 0.83 | 8.49 |
| 28 | Female | 8 | 114.3 | 17 | -2.56 | 3.59 | 3.75 |
| 29 | Female | 5 | 98 | 15 | -3.43 | 1.37 | 1.42 |
| 30 | Male | 7 | 95 | 15 | -6 | 0.44 | 0.48 |
| 31 | Female | 4 | 85 | 11.5 | -5.8 | 1.18 | 3.55 |
| 32 | Female | 2 | 81.6 | 11 | -2.98 | 3.42 | 12.95 |
| 33 | Male | 13 | 135.5 | 25 | 3.2 | 2.42 | 0.69 |
| 34 | Female | 9 | 120 | 22 | -3 | 5 | 6 |
| 35 | Male | 4 | 95 | 14 | -2.98 | 19.27 | 26.3 |
| 36 | Male | 5 | 91 | 10 | -3.5 | 6.55 | 6.55 |
| 37 | Female | 5 | 99 | 15 | -2.67 | 1.14 | 1.57 |
| 38 | Female | 7 | 101 | 15 | -4.08 | 1.14 | 0.19 |
| 39 | Male | 4 | 101 | 16 | -3.13 | 0.38 | 1.52 |
All 39 patients were analyzed by Solexa sequencing. Of the 39 patients, 10 had pathogenicity variability, and the other 29 patients did not have pathogenicity variability. As shown in Table 2, patient 20 had the c.252delA genetic variation of NM-015093.5 in the TAB2 gene, which caused the Ser84fs amino acid variation. In addition, patient 20 also had the c.254insCCATGGAAGAGAAG genetic variation of NM-015093.5 in the TAB2 gene, which caused the Gln85fs amino acid variation. Patient 29 had the c.3337C > T genetic variation of NM-015311.2 in the OBSL1 gene, which caused the Arg1113Cys amino acid variation. In addition, patient 29 also had the c.82G > A genetic variation of NM-001173408.1 in the OBSL1 gene, which caused the Glu28Lys amino acid variation. Patient 31 had the c.1365-1387dup genetic variation of NM-015311.2 in the OBSL1 gene, which caused the Arg463fs amino acid variation. In addition, patient 31 also had the c.458dupG genetic variation of NM-015311.2 in the OBSL1 gene, which caused the Leu154fs amino acid variation. Patient 32 had the c.485-486delTG genetic variation of NM-000112.3 in the SLC26A2 gene, which caused the amino acid variation Val162fs. In addition, patient 32 also had the c.484G > T genetic variation of NM-000112.3 in the SLC26A2 gene, which caused the amino acid variation Val162Leu. Patient 34 had the C.844A > G genetic variation of NM-002834.3 in the PTPN11 gene, which caused the amino acid variation Ile282Val. The other patient numbers, genes, gene locations, gene variations, and amino acid variations were 35, COL27AI, NM-032888.3, c.2113C > T, and Pro705Ser, respectively, and COL27AI, NM-032888.3, c.4066C > C, Asp1356His; 36, COL27AI, NM-032888.3, c.1163C > T, Thr388Ile. COL27AI, NM-032888.3, c.2113C > T, Pro705Ser. HDAC6, NM-006044.2, c.3049G>A, Glu1017Lys; 37, CUL7, NM-014780.4, c.4898C > T, Thr1633Me. CUL7, NM-014780.4, c.4261A > G, Thr1421Ala. FGFR3, NM-000142.4, c.1738G > A, Asp580Asn. DYNC2H1, NM-001377.2, c.12316T > G, Leu4106Va; 38, GH1, NM-000515.4, c.291+28G>A; 39. ATP7B, NM-000053.3, c.3889G>A, V1297I. ATP7B, NM-000053.3, c.2785A > G, I929V.
| Number | Gene | Location | Base change | Amino acid change | Type |
| 20 | TAB2 | NM-015093.5 | c.252delA | Ser84fs | Heterozygote |
| NM-015093.5 | c.254insCCATGGAAGAGAAG | Gln85fs | Heterozygote | ||
| 29 | OBSL1 | NM-015311.2 | c.3337C > T | Arg1113Cys | Heterozygote |
| OBSL1 | NM-001173408.1 | c.82G > A | Glu28Lys | Heterozygote | |
| 31 | OBSL1 | NM-015311.2 | c.1365-1387dup | Arg463fs | Heterozygote |
| OBSL1 | NM-015311.2 | c.458dupG | Leu154fs | Heterozygote | |
| 32 | SLC26A2 | NM-000112.3 | c.485-486delTG | Val162fs | Heterozygote |
| SLC26A2 | NM-000112.3 | c.484G > T | Val162Leu | Heterozygote | |
| 34 | PTPN11 | NM-002834.3 | C.844A > G | Ile282Val | Heterozygote |
| 35 | COL27A1 | NM-032888.3 | c.2113C > T | Pro705Ser | Heterozygote |
| COL27A1 | NM-032888.3 | c.4066C > C | Asp1356His | Heterozygote | |
| 36 | COL27A1 | NM-032888.3 | c.1163C > T | Thr388Ile | Heterozygote |
| COL27A1 | NM-032888.3 | c.2113C > T | Pro705Ser | Heterozygote | |
| HDAC6 | NM-006044.2 | c.3049G > A | Glu1017Lys | Heterozygote | |
| 37 | CUL7 | NM-014780.4 | c.4898C > T | Thr1633Me | Heterozygote |
| CUL7 | NM-014780.4 | c.4261A > G | Thr1421Ala | Heterozygote | |
| FGFR3 | NM-000142.4 | c.1738G > A | Asp580Asn | Heterozygote | |
| DYNC2H1 | NM-001377.2 | c.12316T > G | Leu4106Va | Heterozygote | |
| 38 | GH1 | NM-000515.4 | c.291 + 28G > A | z heterozygote | |
| 39 | ATP7B | NM-000053.3 | c.3889G > A | V1297I | Heterozygote |
| ATP7B | NM-000053.3 | c.2785A > G | I929V | Heterozygote |
Of the 10 dwarfism disease patients with pathogenicity variability, patients 20, 33, and 39 had no related physical characteristic variation. Patient 29 had double breast development and growth hormone deficiency. Patient 30 had enuresis and indirect inguinal hernia on the left. Patient 31 had serrated teeth. Patient 32 showed symptoms in which the distance between the two fingers was 70.2 cm, the head circumference was 49.2 cm, the ischium/lower body length was 1.8 cm, and the limb muscles were weak. Patients 34, 35, and 36 had partial growth hormone deficiency; in addition, patient 36 had pituitary dysplasia symptoms. Patients 37 and 38 had symptoms of growth hormone deficiency.
Of the 39 patients enrolled in our study, 14 patients had IGF-1 and IGFBP3 data before therapy and 6 mo after completing growth hormone therapy, and the other 25 patients lacked partial data, so the 14 patients with complete data were chosen for further analysis. As shown in Table 3, of the 14 patients, 11 did not have pathogenicity variation, and the other three patients had pathogenicity variation. The concentrations of IGF-1 before therapy and 6 mo after completing growth hormone therapy in the 14 patients were 215.2 ± 170.3 and 285.0 ± 166.0, respectively. After therapy, IGF-1 increased 1.9 ± 1.5-fold. The concentrations of IGFBP3 before therapy and 6 mo after completing growth hormone therapy were 3.9 ± 1.4 and 4.2 ± 1.1, respectively. After therapy, IGFBP3 increased 1.2 ± 0.5-fold. Among the 14 patients, the concentrations of IGF-1 before therapy and 6 mo after completing growth hormone therapy in the 11 patients who did not have pathogenicity variation were 217.9 ± 173.3 and 281.4 ± 177.3, respectively. After therapy, IGF-1 increased 1.5 ± 0.5-fold. The concentrations of IGFBP3 before therapy and 6 mo after completing growth hormone therapy were 4.0 ± 1.4 and 4.3 ± 1.2, respectively. After therapy, IGFBP3 increased 1.2 ± 0.4-fold. The concentrations of IGF-1 before therapy and 6 mo after completing growth hormone therapy in the three patients who were diagnosed with pathogenicity variation were 205.5 ± 195.1 and 298.0 ± 147.1, respectively. After therapy, IGF-1 increased 3.1 ± 3.2-fold. The concentrations of IGFBP3 before therapy and 6 mo after completing growth hormone therapy were 3.7 ± 1.5 and 4.0 ± 0.8, respectively. After therapy, IGFBP3 increased 1.4 ± 0.9-fold.
| Number | IGF-1 | After therapy | Fold change | IGFBP3 | After therapy | Fold change |
| 3 | 283 | 453 | 1.60 | 4.4 | 6.3 | 1.43 |
| 5 | 101 | 266 | 2.63 | 3.4 | 4.8 | 1.41 |
| 6 | 98.7 | 149 | 1.51 | 2.9 | 3 | 1.03 |
| 7 | 461 | 527 | 1.14 | 5.2 | 5.3 | 1.02 |
| 8 | 31.2 | 63.5 | 2.04 | 1.1 | 2.3 | 2.09 |
| 11 | 238 | 220 | 0.92 | 4.5 | 4.1 | 0.91 |
| 17 | 572 | 603 | 1.05 | 6.4 | 5.6 | 0.88 |
| 18 | 306 | 340 | 1.11 | 5.3 | 4.8 | 0.91 |
| 22 | 101 | 118 | 1.17 | 3.6 | 3.2 | 0.89 |
| 24 | 86 | 160 | 1.86 | 3.5 | 3.7 | 1.06 |
| 27 | 119 | 196 | 1.65 | 3.2 | 4.7 | 1.47 |
| 29 | 414 | 464 | 1.12 | 4.8 | 3.2 | 0.67 |
| 31 | 175 | 246 | 1.41 | 4.2 | 4.1 | 0.98 |
| 38 | 27.4 | 184 | 6.72 | 2 | 4.8 | 2.40 |
With the rapid development of gene detection, next-generation sequencing can provide more information on gene variation. In our study, we aimed to explore the pathogenicity variability related to the incidence of dwarfism. Obscurin-like 1 (OBSL1) is a new member of the UNC-89/obscurin gene family. It contains 22 exons and is located on the nuclear membrane. OBSL1 is located in the intercalated disc of the myocardium, the nucleus of myocardial cells, and the Z line, but it is expressed at low levels in the M zone. OBSL1 functions as a cytoplasmic support network as a cytoskeletal adaptor protein connecting membrane-bound nucleoproteins[8]. OBSL1 is expressed in many different cell types, and OBSL1 acts as a scaffolding protein. The fibronectin domain of OBSL1 is closely related to the amino-terminal fibronectin domain of obscurin[9]. Studies have found that a mutation in the OBSL1 gene is the cause of 3M syndrome, which plays a greater role in the CUL7 ubiquitination pathway. Three M syndrome is a rare autosomal stealth genetic disease characterized by low birth weight and postpartum developmental delays associated with a series of minor developmental abnormalities[10].
A study found that solute carrier 26 (SLC26) members had a total of 11 subtypes. The four most common SLC26A2 pathogenic variants account for approximately 70% of all SLC26A2-related dysplasia alleles[11]. P.Arg279Trp is the most common pathogenic variant of SLC26A2 (accounting for 45% of alleles), except in Finland. It is a mild pathogenic variant that results in the mild recessive multiple epiphyseal dysplasia phenotype when homozygous and is mainly malformed in the complex dysplasia phenotype. P.Arg178Ter is the second most common pathogenic variant (accounting for 9% of alleles) and is associated with a more severe diastrophic dysplasia phenotype or even the perinatal lethal dysplasia type 2 phenotype. P.Cys653Ser and c.-26+2T > C are the third most common pathogenic variants (each accounts for 8% of alleles)[12,13].
The PTPN11 gene encodes the protein tyrosine phosphatase src homology phosphatase 2 and is located on chromosome 12 q24.13 and contains 16 exosomes[14]. Approximately 50% of Noonan syndrome cases are caused by missense mutations in the PTPN11 gene. Noonan syndrome is a relatively common autosomal dominant inheritance disease, and the main clinical manifestations include congenital heart disease, special face, short stature, chest deformity, and mental retardation[15].
Collagen type XXVII alpha 1 chain plays a role during the calcification of cartilage and the transition of cartilage to bone. It is closely related to Steel Syndrome, which is characterized by dislocated hips and radial heads, fusion of carpal bones, short stature, scoliosis, and cervical spine anomalies[16]. Facial features include prominent forehead, long oval-shaped face, hypertelorism, and a broad nasal bridge.
Histone deacetylase 6 is a cytoplasmic enzyme containing two N-terminal deacetylase domains and a C-terminal ubiquitin-binding domain. It has a relative molecular weight of 131 kDa and contains 1215 amino acid residues. It is involved in the occurrence and development of diseases such as tumors, neurodegenerative diseases, and cardiovascular diseases[17]. Histone deacetylase 6 is also considered a potential therapeutic target for related diseases. However, to date, there has been no evidence demonstrating that it is related to the incidence of dwarfism.
The CUL7 gene is located in the 6p21.1 region and contains 26 exons. It is a member of the Cullin family. It forms ubiquitin ligase E3 with SKP1-FBX29 and RoCl. Nonsense or missense mutations in the CUL7 gene prevent the cullin-7 protein from recruiting RoCl, thereby preventing the substrate from ubiquitination and degradation, resulting in accumulation in the body[18]. CUL7 gene knockout mouse embryos showed accumulation of insulin receptor substrate 1, which caused the activation of the downstream substrates Akt and mitogen-activated protein kinase kinase/extracellular signal-regulated kinase to increase, resulting in growth retardation[19].
FGFR3 is a physiologically negative regulator of bone growth. As a member of the tyrosine kinase receptor family, mutations cause fibroblast growth factor receptor 3 to be rapidly ubiquitinated and the subsequent degradation of the protease body. Signal transmission is terminated immediately, which can be inhibited by varying degrees to chondrocyte proliferation[20]. Studies found that the gene mutation was closely related to achondroplasia[21].
The DYNC2H1 gene encodes dynein, cytoplasmic 2, and heavy chain 1 protein. It functions as a motor for retrograde intraflagellar transport, a process required for the assembly and maintenance of cilia[22]. Mutations in this gene cause a heterogeneous spectrum of conditions related to altered primary cilium function and often involve polydactyly, abnormal skeletogenesis, and polycystic kidneys[23]. Alternative splicing results in multiple transcript variants encoding distinct proteins.
The GH1 gene provides instructions for making the growth hormone protein. Growth hormone is produced in the growth-stimulating somatotropic cells of the pituitary gland[24]. Growth hormone is necessary for the normal growth of the body's bones and tissues. The production of growth hormone is triggered when two other hormones are turned on. The release of growth hormone into the body peaks during puberty and reaches a low point at approximately age 55.
The ATP7B gene provides instructions for making a protein called copper-transporting ATPase 2, which is part of the P-type ATPase family. Copper plays important roles in certain enzymes that maintain normal cell functions. Copper-transporting ATPase 2 is also important for the removal of excess copper from the body[25]. With a shortage of ATPase 2, the removal of copper is impaired. When it accumulates to toxic levels, it can damage tissues and organs, particularly the liver and brain[26].
However, there are still some limitations in our study. First, the sample size of our study was relatively small and cannot effectively reflect the pathogenicity variability related to dwarfism. Second, after next-generation sequencing, Sanger sequencing was not performed to validate the results, which may cause some bias in the results. Third, although some pathogenicity variability sites were screened, the mechanism of the relationship between genes and dwarfism was not explored.
Pathogenicity variability in the OBSL1, SLC26A2, PTPN11, COL27AI, HDAC6, CUL7, FGFR3, DYNC2H1, GH1, and ATP7B genes was screened, and it may be related to dwarfism incidence, which may clarify the molecular cause and provide the basis for personal therapy in the future.
Dwarfism is one of the most common diseases in the endocrine system of children. It is a complex process involving multiple genes and multiple factors. Genetic factors are the main factors affecting individual height differences, and the heritability of human height accounts for approximately 80%. The mechanism has not yet been clarified.
The rapid development of emerging technologies has resulted in increased understanding of on the related molecular etiological mechanisms. Some unexplained cases of short stature can be clarified by studying their genetic background by using related genetic testing methods. Efficient and highly sensitive diagnosis methods have become the focus of dwarfism research.
To analyze retrospectively the genetic variation by using a constructed panel related to dwarfism through next-generation sequencing platform sequencing analysis in order to screen candidate-related gene mutations that may clarify the molecular cause and provide a scientific basis for clinical treatment.
Data from 39 dwarf patients in Quanzhou First Hospital were collected according to the inclusion and exclusion criteria, then the clinical examination, growth hormone drug challenge test, serum insulin-like growth factor-1 (IGF-1) and IGF binding protein 3 (IGFBP3) levels, other related tests, imaging examination, and chromosome karyotyping were analyzed. Next-generation sequencing was also performed to analyze the pathogenicity variability.
Of the 39 dwarfism patients, 10 had pathogenicity variability. Gene variation was found in the OBSL1, SLC26A2, PTPN11, COL27AI, HDAC6, CUL7, FGFR3, DYNC2H1, GH1, and ATP7B genes. Of the 10 patients with pathogenicity variability, the related physical characteristics included double breast development and growth hormone deficiency, enuresis and indirect inguinal hernia on the left, two finger distance of 70.2 cm, head circumference of 49.2 cm, ischium/lower body length of 1.8 cm, weak limb muscles, and partial growth hormone deficiency. After 6 mo of growth hormone therapy, the concentrations of IGF-1 and IGF binding protein 3 increased from 215.2 ± 170.3 to 285.0 ± 166.0 and 3.9 ± 1.4 to 4.2 ± 1.1, respectively.
Pathogenicity variability in the OBSL1, SLC26A2, PTPN11, COL27AI, HDAC6, CUL7, FGFR3, DYNC2H1, GH1, and ATP7B genes was screened, and it may be related to dwarfism incidence.
This study adds to the evidence-base and will clarify the molecular cause of dwarf diseases; however, larger sample size and multi-center studies are still needed in the future.
Manuscript source: Unsolicited manuscript
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Tinsley A S-Editor: Gao CC L-Editor: Filipodia P-Editor: Ma YJ
| 1. | Alkuraya FS. Primordial dwarfism: an update. Curr Opin Endocrinol Diabetes Obes. 2015;22:55-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 2. | Ornitz DM, Legeai-Mallet L. Achondroplasia: Development, pathogenesis, and therapy. Dev Dyn. 2017;246:291-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 155] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 3. | Pauli RM. Achondroplasia: a comprehensive clinical review. Orphanet J Rare Dis. 2019;14:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 201] [Cited by in RCA: 269] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 4. | Léger J. How should we investigate children with growth failure? Ann Endocrinol (Paris). 2017;78:106-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 5. | Wassner AJ. Pediatric Hypothyroidism: Diagnosis and Treatment. Paediatr Drugs. 2017;19:291-301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 6. | Boegheim IJM, Leegwater PAJ, van Lith HA, Back W. Current insights into the molecular genetic basis of dwarfism in livestock. Vet J. 2017;224:64-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 7. | Khetarpal P, Das S, Panigrahi I, Munshi A. Primordial dwarfism: overview of clinical and genetic aspects. Mol Genet Genomics. 2016;291:1-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 8. | Hanson D, Murray PG, Black GC, Clayton PE. The genetics of 3-M syndrome: unravelling a potential new regulatory growth pathway. Horm Res Paediatr. 2011;76:369-378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 9. | Clayton PE, Hanson D, Magee L, Murray PG, Saunders E, Abu-Amero SN, Moore GE, Black GC. Exploring the spectrum of 3-M syndrome, a primordial short stature disorder of disrupted ubiquitination. Clin Endocrinol (Oxf). 2012;77:335-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 10. | Irving M, Holder-Espinasse M. Three M Syndrome. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A. GeneReviews((R)). Seattle (WA); 1993. |
| 11. | Bonafe L, Mittaz-Crettol L, Ballhausen D, Superti-Furga A. Atelosteogenesis Type 2. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A. GeneReviews((R)). Seattle (WA); 1993. |
| 12. | Bonafe L, Mittaz-Crettol L, Ballhausen D, Superti-Furga A. Multiple Epiphyseal Dysplasia, Recessive. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A. GeneReviews((R)). Seattle (WA); 1993. |
| 13. | Anthony S, Munk R, Skakun W, Masini M. Multiple epiphyseal dysplasia. J Am Acad Orthop Surg. 2015;23:164-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 14. | El Bouchikhi I, Belhassan K, Moufid FZ, Iraqui Houssaini M, Bouguenouch L, Samri I, Atmani S, Ouldim K. Noonan syndrome-causing genes: Molecular update and an assessment of the mutation rate. Int J Pediatr Adolesc Med. 2016;3:133-142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 54] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 15. | Allanson JE, Roberts AE. Noonan Syndrome. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A. GeneReviews((R)). Seattle (WA); 1993. |
| 16. | Roet KC, Bossers K, Franssen EH, Ruitenberg MJ, Verhaagen J. A meta-analysis of microarray-based gene expression studies of olfactory bulb-derived olfactory ensheathing cells. Exp Neurol. 2011;229:10-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 17. | Liang T, Fang H. Structure, Functions and Selective Inhibitors of HDAC6. Curr Top Med Chem. 2018;18:2429-2447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 18. | Deeb A, Afandi O, Attia S, Fatih AE. 3-M syndrome: a novel CUL7 mutation associated with respiratory distress and a good response to GH therapy. Endocrinol Diabetes Metab Case Rep. 2015;2015:150012. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 19. | Hasegawa K, Tanaka H, Higuchi Y, Yamashita M, Tsukahara H. Changes in facial appearance from neonate to adult in 3-M syndrome patient with novel CUL7 gene mutations. J Pediatr Endocrinol Metab. 2016;29:241-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 20. | Saint-Laurent C, Garcia S, Sarrazy V, Dumas K, Authier F, Sore S, Tran A, Gual P, Gennero I, Salles JP, Gouze E. Early postnatal soluble FGFR3 therapy prevents the atypical development of obesity in achondroplasia. PLoS One. 2018;13:e0195876. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 21. | Klag KA, Horton WA. Advances in treatment of achondroplasia and osteoarthritis. Hum Mol Genet. 2016;25:R2-R8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 22. | Qiao Y, Wen J, Tang F, Martell S, Shomer N, Leung PC, Stephenson MD, Rajcan-Separovic E. Whole exome sequencing in recurrent early pregnancy loss. Mol Hum Reprod. 2016;22:364-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 65] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 23. | Fujita A, Higashijima T, Shirozu H, Masuda H, Sonoda M, Tohyama J, Kato M, Nakashima M, Tsurusaki Y, Mitsuhashi S, Mizuguchi T, Takata A, Miyatake S, Miyake N, Fukuda M, Kameyama S, Saitsu H, Matsumoto N. Pathogenic variants of DYNC2H1, KIAA0556, and PTPN11 associated with hypothalamic hamartoma. Neurology. 2019;93:e237-e251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 24. | Majewska KA, Kedzia A, Kontowicz P, Prauzinska M, Szydlowski J, Switonski M, Nowacka-Woszuk J. Polymorphism of the growth hormone gene GH1 in Polish children and adolescents with short stature. Endocrine. 2020;69:157-164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 25. | Asada H, Chambers JK, Kojima M, Goto-Koshino Y, Nakagawa T, Yokoyama N, Tsuboi M, Uchida K, Tsujimoto H, Ohno K. Variations in ATP7B in cats with primary copper-associated hepatopathy. J Feline Med Surg. 2020;22:753-759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 26. | Shanmugavel KP, Wittung-Stafshede P. Copper relay path through the N-terminus of Wilson disease protein, ATP7B. Metallomics. 2019;11:1472-1480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |