Published online Oct 26, 2021. doi: 10.12998/wjcc.v9.i30.9011
Peer-review started: March 4, 2021
First decision: June 3, 2021
Revised: June 19, 2021
Accepted: August 18, 2021
Article in press: August 18, 2021
Published online: October 26, 2021
Processing time: 231 Days and 0.4 Hours
Primary small cell carcinoma of the esophagus (PSCE) is a highly invasive malignant tumor with a poor prognosis compared with esophageal squamous cell carcinoma. Due to the limited samples size and the short follow-up time, there are few reports on elucidating the prognosis of PSCE, especially on the establishment and validation of a survival prediction nomogram model covering general information, pathological factors and specific biological proteins of PSCE patients.
To establish an effective nomogram to predict the overall survival (OS) probability for PSCE patients in China.
The nomogram was based on a retrospective study of 256 PSCE patients. Univariate analysis and multivariate Cox proportional hazards regression analysis were used to examine the prognostic factors associated with PSCE, and establish the model for predicting 1-, 3-, and 5-year OS based on the Akaike information criterion. Discrimination and validation were assessed by the concordance index (C-index) and calibration curve and decision curve analysis (DCA). Histology type, age, tumor invasion depth, lymph node invasion, detectable metastasis, chromogranin A, and neuronal cell adhesion molecule 56 were integrated into the model.
The C-index was prognostically superior to the 7th tumor node metastasis (TNM) staging in the primary cohort [0.659 (95%CI: 0.607-0.712) vs 0.591 (95%CI: 0.517-0.666), P = 0.033] and in the validation cohort [0.700 (95%CI: 0.622-0.778) vs 0.605 (95%CI: 0.490-0.721), P = 0.041]. Good calibration curves were observed for the prediction probabilities of 1-, 3-, and 5-year OS in both cohorts. DCA analysis showed that our nomogram model had a higher overall net benefit compared to the 7th TNM staging .
Our nomogram can be used to predict the survival probability of PSCE patients, which can help clinicians to make individualized survival predictions.
Core Tip: Up to now, the main prognostic index of primary small cell carcinoma of the esophagus (PSCE) is still the disease stage, which is mainly based on the American Joint Committee on Cancer tumor node metastasis classification of esophageal squamous cell carcinoma. A large amount of evidence shows that the staging system cannot well evaluate the clinical outcome of PSCE. Compared with the traditional staging system, our nomogram model can accurately evaluate the overall survival probability at 1, 3, and 5 years for PSCE patients in China.
- Citation: Zhang DY, Huang GR, Ku JW, Zhao XK, Song X, Xu RH, Han WL, Zhou FY, Wang R, Wei MX, Wang LD. Development and validation of a prognostic nomogram model for Chinese patients with primary small cell carcinoma of the esophagus. World J Clin Cases 2021; 9(30): 9011-9022
- URL: https://www.wjgnet.com/2307-8960/full/v9/i30/9011.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i30.9011
Primary small cell carcinoma of the esophagus (PSCE) is a rare, highly invasive malignant tumor associated with early metastasis, and its incidence accounts for only 0.05%-4% of all esophageal cancers[1]. Similar to small cell lung cancer, the diagnosis of PSCE mainly depends on immunohistochemical staining for several neuroendocrine markers, including synaptophysin (Syn), neuronal cell adhesion molecule 56 (CD56), and chromogranin A (CgA)[2]. However, their impact on the overall survival (OS) of PSCE patients has not been thoroughly studied. Related studies have reported some prognostic factors for PSCE, but the results are controversial. The main prognostic indicator is still the stage of the disease[3].
The tumor node metastasis (TNM) staging system by the American Joint Committee on Cancer (AJCC) is currently widely used in the clinical treatment and prognosis of cancer patients[4]. PSCE is generally staged based on the AJCC TNM classification of esophageal squamous cell carcinoma (ESCC)[5]. The 7th AJCC TNM system includes tumor invasion depth (T), lymph node invasion (N), detectable metastasis (M), and tumor location and grade. A previous study which focused on the OS effect of CgA on 125 PSCE patients, demonstrated that the TNM staging system for ESCC may not be a good predictor of prognosis for PSCE[6]. Other clinicopathological factors that are not included in the 7th TNM staging may also affect OS[7]. Furthermore, a growing body of evidence has suggested that PSCE has a poorer prognosis compared to ESCC. A more accurate prognostic prediction model is needed for PSCE patients[8,9].
Nomograms are currently widely used in graphical representations of complex mathematical formulas[10]. A nomogram model has obvious advantages in quantifying risk compared with TNM staging, which generally takes all known clinicopathological variables into account and allows personalized prognosis prediction[11,12]. The first nomogram was developed in 2021 by Qie et al[12], based on the patients in the United States in the public surveillance, epidemiology, and end results (SEER) database to predict patient OS probability. Of note, this study did not involve the Chinese population and relevant neuroendocrine markers were not included in the model[13]. The present study, thus, aimed to build a prognostic predictive nomogram model including clinicopathological factors and neuroendocrine biomarkers for Chinese PSCE patients. It was also determined whether the nomogram model can predict OS more accurately than the 7th TNM staging system.
A retrospective observational study was conducted on 343 eligible patients, who were enrolled from our esophageal and gastric cardiac carcinoma database containing 500000 cases[14]. A total of 256 eligible patients were finally enrolled using the following inclusion criteria: Pathologically diagnosed with primary PSCE, no preoperative radiotherapy and/or chemotherapy, survival time more than 1 mo, and detailed clinical baseline records. Patients with other malignant disease or a history of anticancer treatment were excluded.
The demographic and clinicopathological parameters of the patients included gender, age, smoking history, alcohol history, histology type, tumor location, T stage, N stage, M, and treatment history. Immunostained biomarkers including CgA, Syn, and CD56 were also identified from clinical medical records. All eligible patients were followed every year by telephone or home interview. The latest follow-up was conducted at the end of June 2019. The OS was measured from the time of surgery to the most recent follow-up time or death. Survival status was determined on the last day of follow-up. The median follow-up time was 24 mo (range, 2 to 154.8 mo). The study protocol was approved by the Medical Ethics Committee of the First Affiliated Hospital of Zhengzhou University.
Statistical analyses were conducted using R version 4.0.3 (http://www.r-project.org/) and Stata version 15.0 (http://www.stata.com). The Kaplan-Meier (K-M) method was used to draw survival curves, which were compared by the log-rank test. Clinical related parameters (based on clinical experience and literature reports) and baseline variables with statistical correlation in univariate analysis (P < 0.2) were included in the multivariate Cox proportional hazard regression analysis[15,16]. A predictive nomogram of 1-year, 3-year, and 5-year OS based on the Akaike information criterion (AIC) was developed using the Cox regression model and the "RMS" package in R[17]. The concordance index (C-index)[18], which was calculated by a bootstrap approach with 1000 resamples[19], was used to determine discrimination. By mapping the nomogram predicted probabilities of OS with the actual OS observed at 1, 3, and 5 years, the calibration curve was graphically evaluated, and the 45° line indicated the best predicted value[20]. Decision curve analysis (DCA) was used to evaluate the clinical application of the prediction model by quantifying net benefits[21]. In addition, we divided the model into three groups according to the cutoff points of the total prognostic scores (TPS), which were determined by the minimum P value obtained using the X-tile program[22].
According to the random numbers generated by the computer, 70% of the eligible patients were randomly assigned to a training cohort (n = 179) and the remaining 30% to a validation cohort (n = 77). The clinical characteristics of these patients are summarized in Table 1. The male to female ratio in the training and validation cohorts was 1.84:1 (116/63) and 1.96:1 (51/26), respectively. The median age in each cohort was 62.19 years (range, 37-88 years) and 61.83 years (range, 46-77 years), respectively. The median survival time in the primary cohort was 23.59 mo (range, 1-105 mo) and the 1-, 3-, and 5-year OS rates were 67.6%, 35.1%, and 24.3%, respectively. In the validation cohort, the median survival time was 24.63 mo (range, 1-108 mo), and the
| Variable | Primary cohort | Validation cohort | ||
| n | % | n | % | |
| Gender | ||||
| Female | 63 | 35.2 | 26 | 33.8 |
| Male | 116 | 64.8 | 51 | 66.2 |
| Age, yr | ||||
| ≤ 60 | 74 | 41.3 | 32 | 41.6 |
| > 60 | 105 | 58.7 | 45 | 58.4 |
| Smoking history | ||||
| No | 122 | 68.2 | 59 | 76.6 |
| Yes | 57 | 31.8 | 18 | 23.4 |
| Drinking history | ||||
| No | 134 | 74.9 | 64 | 83.1 |
| Yes | 45 | 25.1 | 13 | 16.9 |
| Tumor location | ||||
| Upper | 23 | 12.9 | 8 | 10.4 |
| Middle | 111 | 62.0 | 46 | 59.7 |
| Lower | 45 | 25.1 | 23 | 29.9 |
| Histology | ||||
| Pure PSCE | 106 | 59.2 | 57 | 74.1 |
| Mixed PSCE | 73 | 40.8 | 20 | 25.9 |
| T stage | ||||
| T1a/T1b | 34 | 19.0 | 19 | 24.7 |
| T2 | 63 | 35.2 | 31 | 40.2 |
| T3/T4 | 82 | 45.8 | 27 | 35.1 |
| N stage | ||||
| N0 | 87 | 48.6 | 35 | 45.4 |
| N1 | 64 | 35.8 | 30 | 39.0 |
| N2/N3 | 28 | 15.6 | 12 | 15.6 |
| Metastasis | ||||
| No | 136 | 75.9 | 60 | 77.9 |
| Yes | 43 | 24.1 | 17 | 22.1 |
| Treatment methods | ||||
| Surgery | 155 | 86.6 | 68 | 88.3 |
| Others | 24 | 13.4 | 9 | 11.7 |
| CD56 | ||||
| Negative | 45 | 25.1 | 15 | 19.5 |
| Positive | 124 | 74.9 | 62 | 80.5 |
| Syn | ||||
| Negative | 40 | 22.4 | 17 | 22.1 |
| Positive | 139 | 77.6 | 60 | 77.9 |
| CgA | ||||
| Negative | 112 | 62.6 | 47 | 61.1 |
| Positive | 67 | 37.4 | 30 | 38.9 |
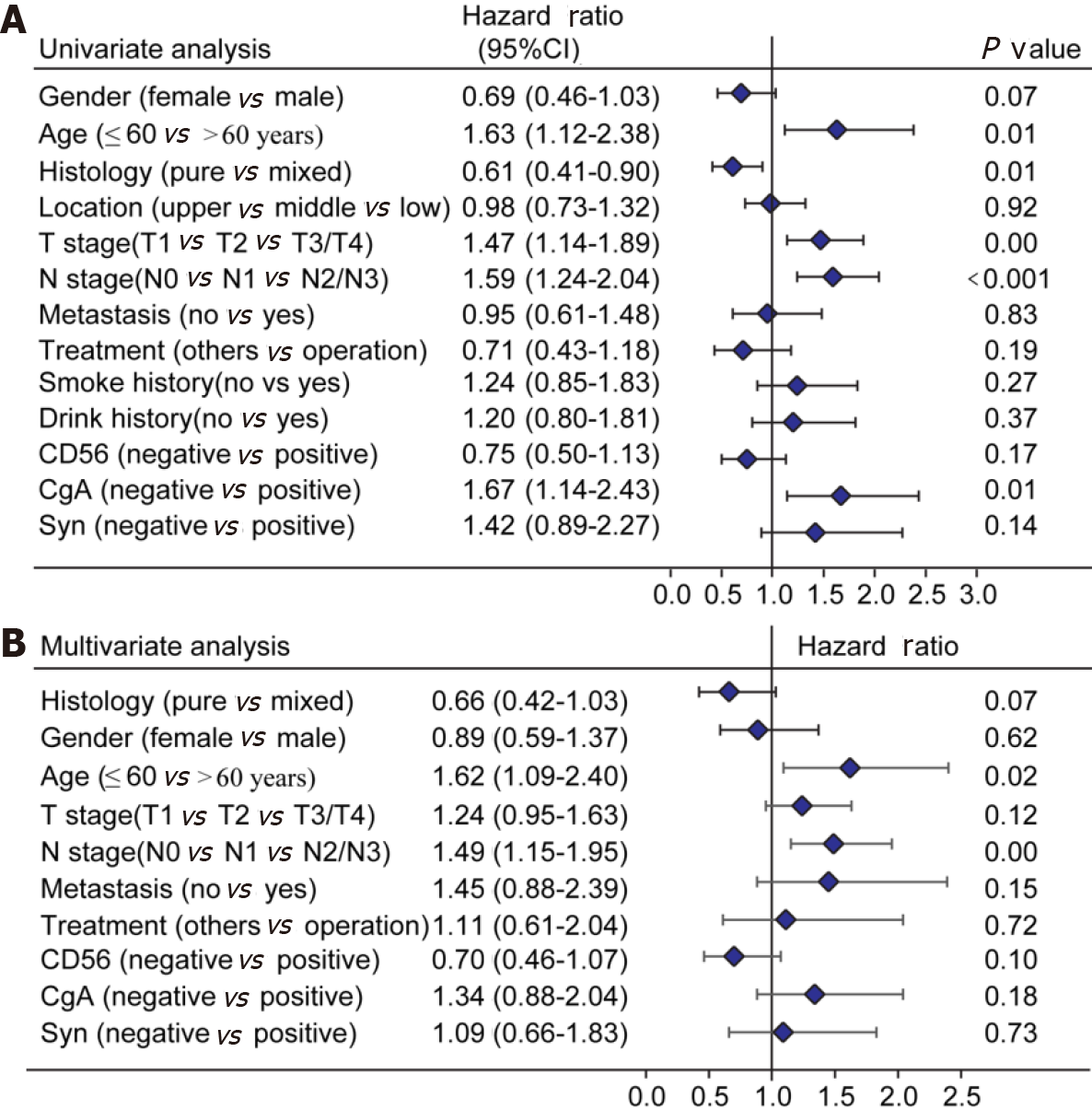
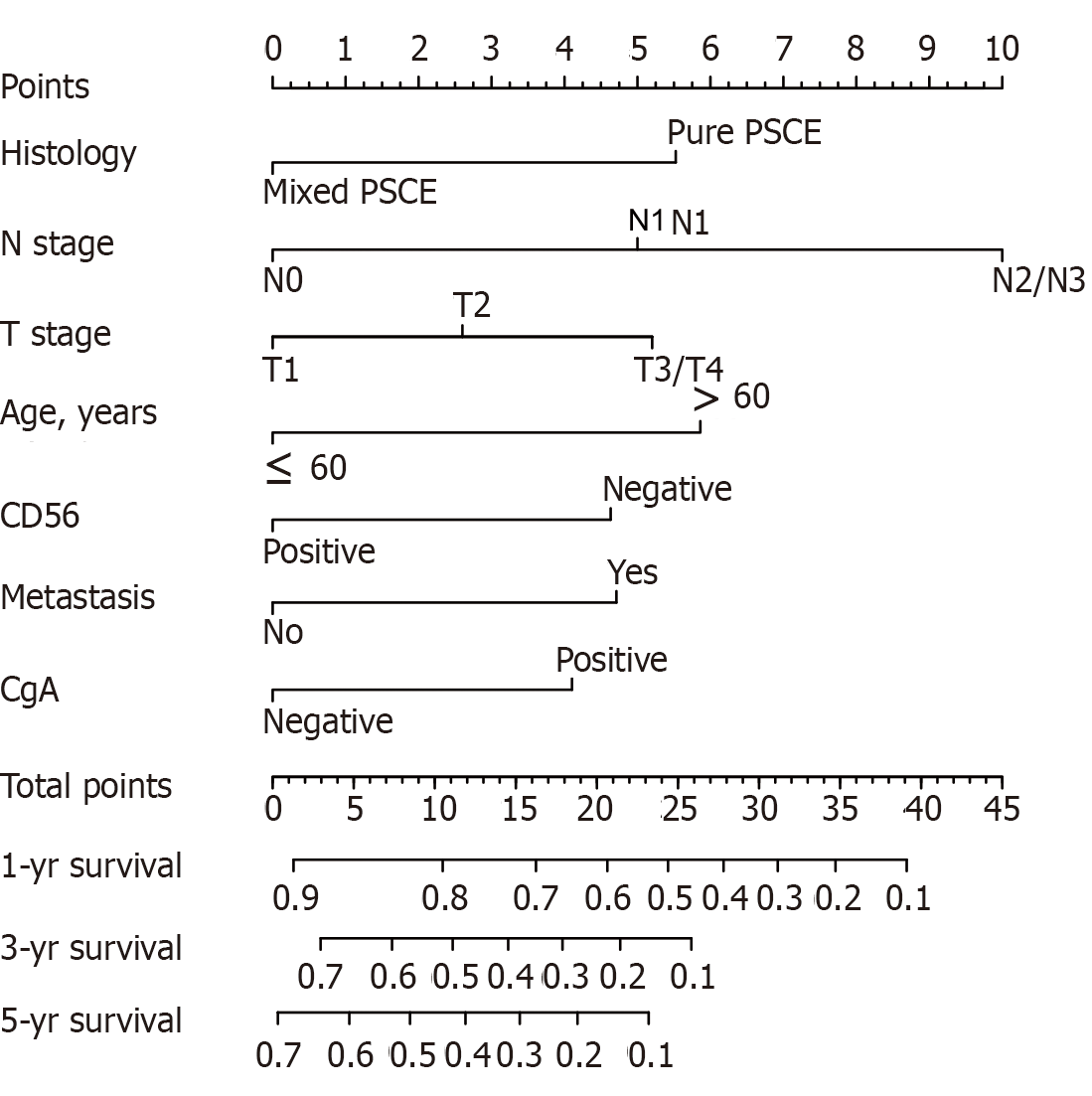
Of the initial 13 variables, tumor location, alcohol consumption, and smoking history were excluded from the Cox proportional hazards regression analysis due to their weak and non-significant OS correlation in the univariate analysis (P > 0.2) (Figure 1). Based on the results of univariate analysis and clinical parameters, histology type, gender, age, T stage, N stage, M, operation, Syn, CgA, and CD56 were included in the Cox proportional hazards regression model. Finally, the most suitable nomogram model was determined using the backward step selection process with the smallest AIC, which included histology type, age, T stage, N stage, M, CgA, and CD56. The model suggested that N stage had the greatest influence on patient prognosis, followed by histological type and age. T stage, M, CD56, and CgA had moderate effects on OS. Each subtype of the variables was assigned a score on the scoring table. By adding the total score and positioning it in relation to the total subscale, a straight line was drawn to determine the estimated probability of survival at each time point. Figure 2 shows the nomogram model for predicting 1-, 3-, and 5-year OS in the training cohort, indicating that the higher the score, the worse the prognosis.
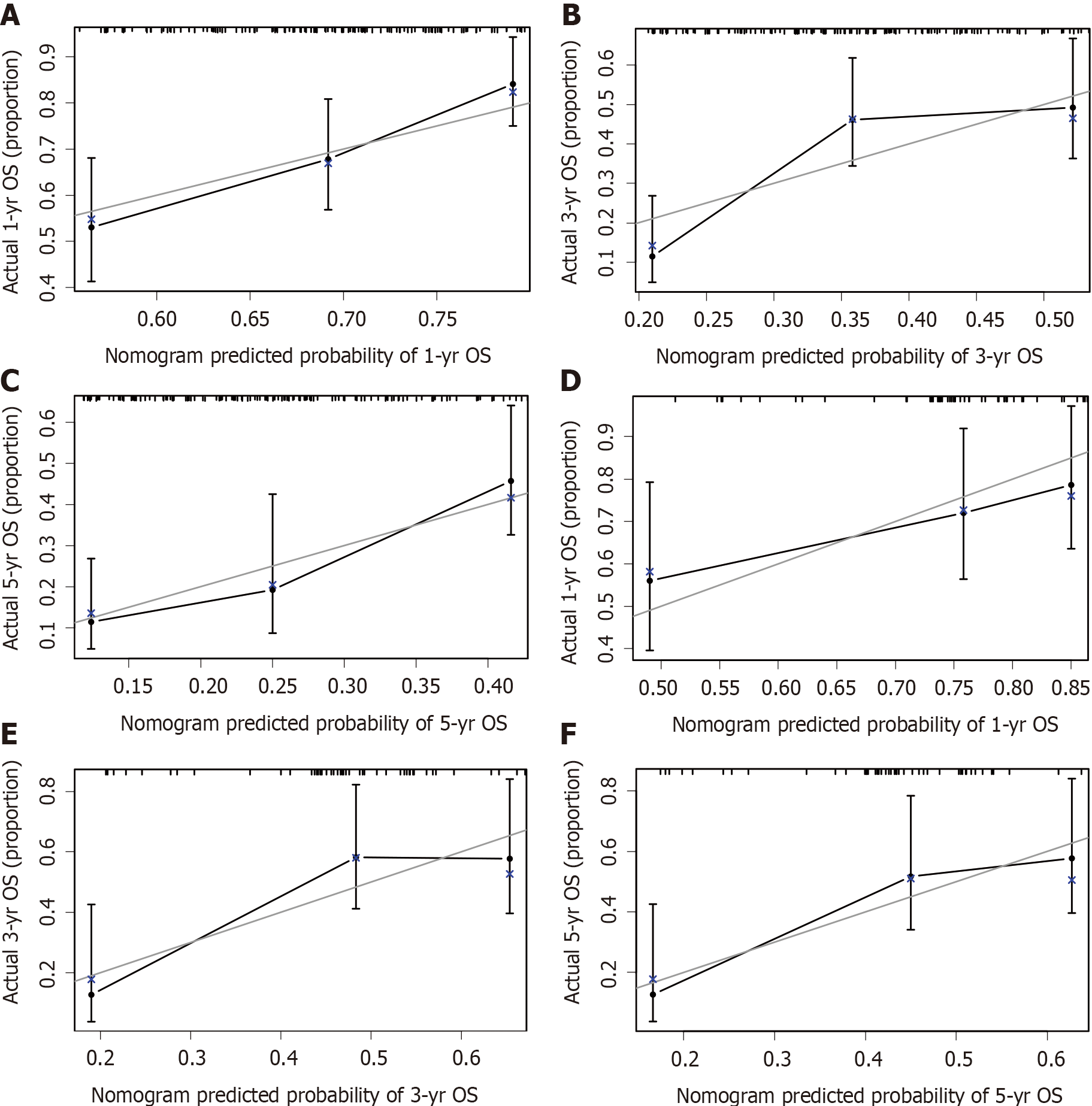
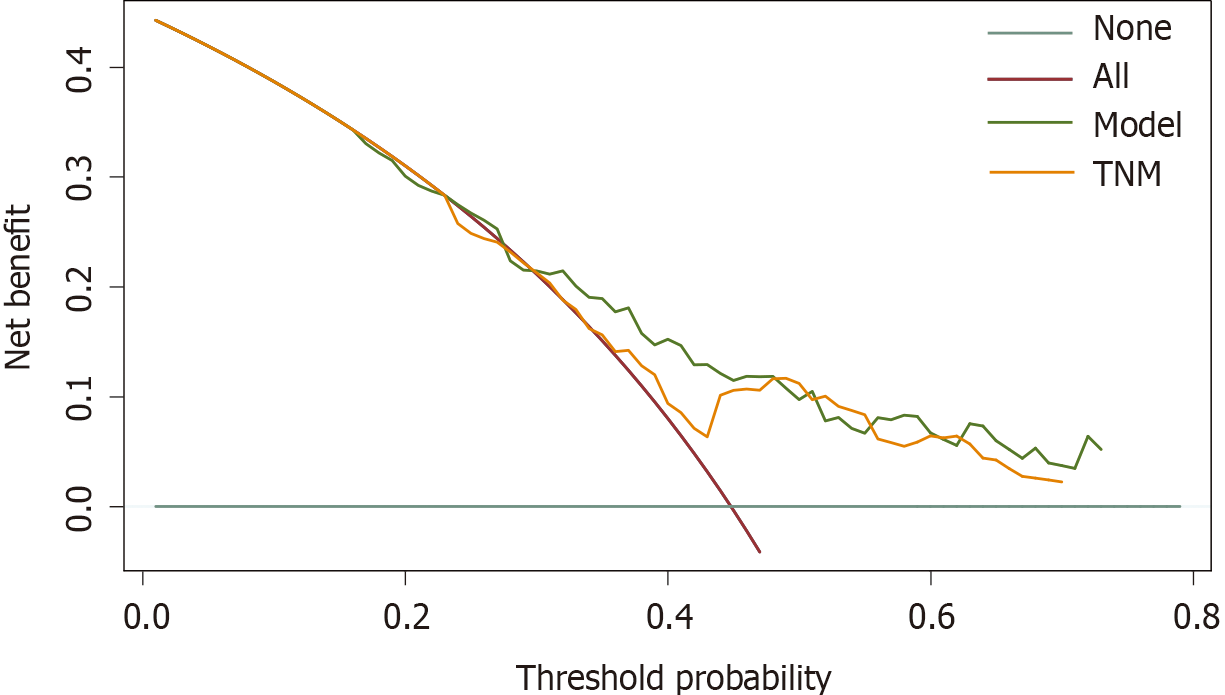
The calibration plots showed that the nomogram prediction was in good agreement with the actual observation of 1-, 3-, and 5-year OS in both cohorts (Figure 3). The accuracy of prognosis prediction was also compared between the nomogram model and the 7th TNM staging using the C-index (Table 2). In the training cohort, the C-index for OS prediction was 0.659 (95%CI: 0.607-0.712), which was significantly higher than that of the 7th TNM staging (0.591, 95%CI: 0.517-0.666, P = 0.033). This superior tendency in OS prediction was also verified using external validation in the validation cohort, with the C-index of the former (0.700, 95%CI: 0.622-0.778) also being higher than that of the latter (0.605, 95%CI: 0.490-0.721, P = 0.041). Figure 4 shows the results of DCA at 18 mo for PSCE, which indicated that our nomogram model had a higher overall net benefit than the 7th TNM staging within a wide range of threshold probabilities.
| Variable | Primary cohort | Validation cohort | ||
| C-index (95%CI) | P value | C-index (95%CI) | P value | |
| Nomogram model | 0.659 (0.607-0.712) | 0.033 | 0.700 (0.622-0.778) | 0.041 |
| 7th TNM staging | 0.591 (0.517-0.666) | 0.605 (0.490-0.721) | ||
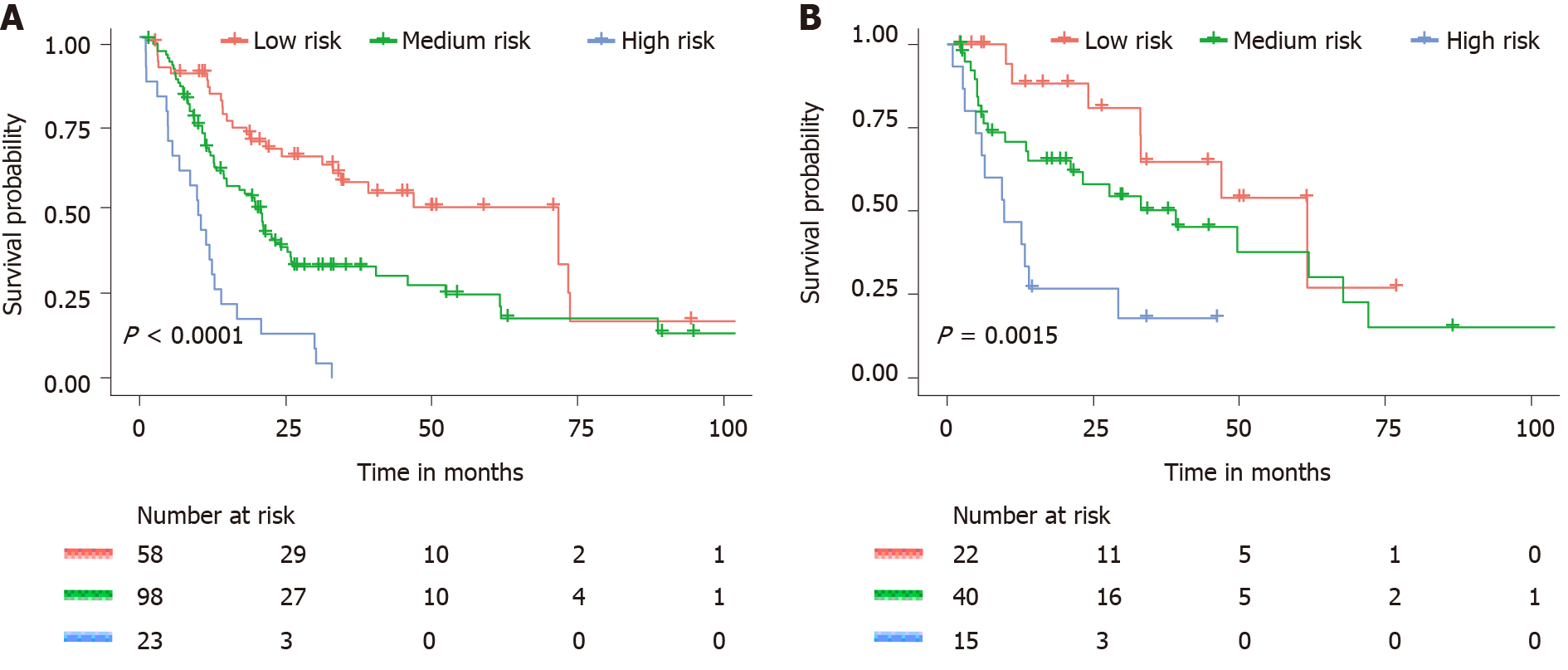
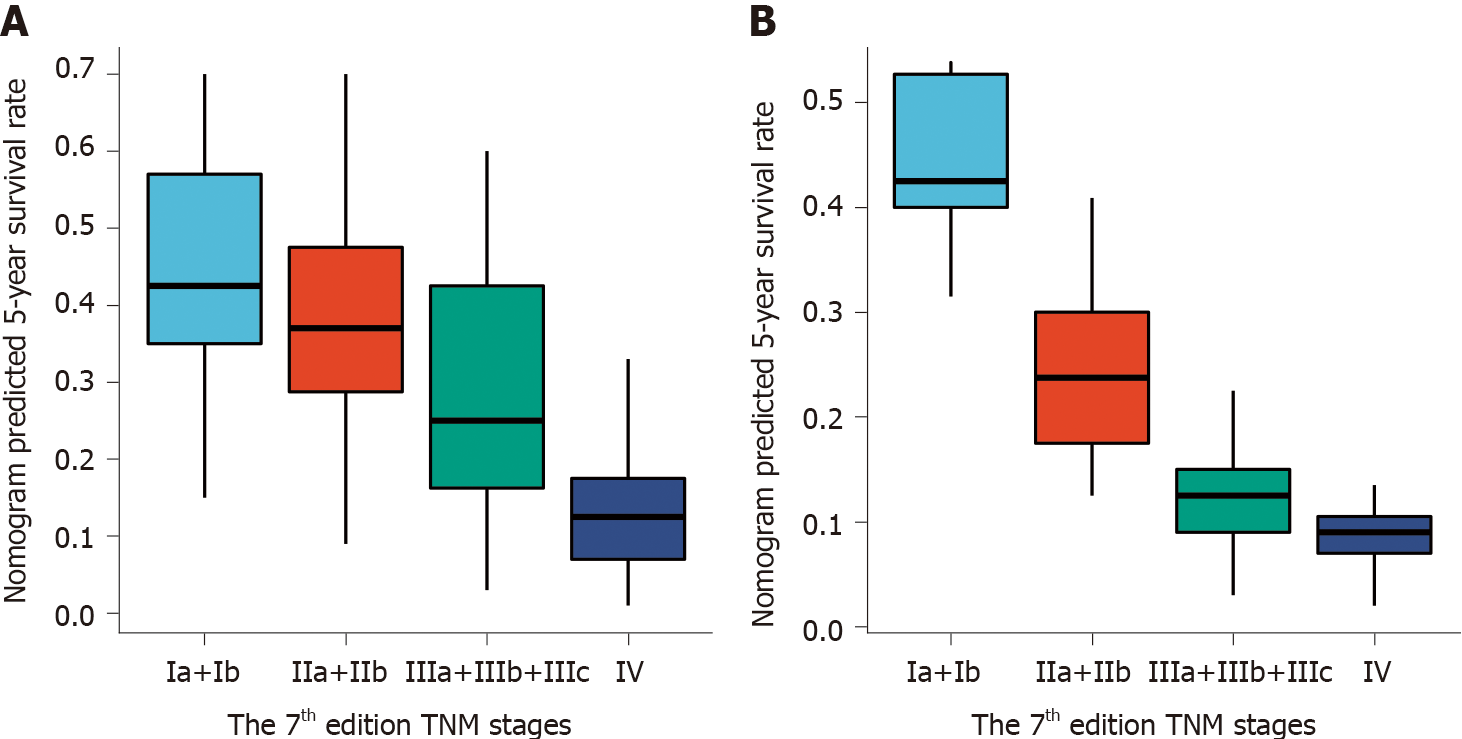
As predicted by the nomogram model, the X-tile program was used to categorize the primary cohort into three groups: Low risk prognosis (TPS ≤ 13.2, 58 patients), medium risk prognosis (13.2 < TPS ≤ 22.8, 98 patients), and high risk prognosis (TPS > 22.8, 23 patients). The survival rate was highest in the low risk group (83.41%, 57.53%, and 33.40% for 1-, 3-, and 5-year survival rates), followed by the medium risk group (66.41, 31.71, and 21.14%) and the high risk group (34.78%, 4.35%, and 0%; Table 3). We then used cutoff values to plot K-M curves for both cohorts (Figure 5), which were also significantly correlated with OS (P < 0.01). Figure 6 shows the distribution of 5-year survival predicted by the nomogram for each stage of the 7th TNM staging in the primary and validation cohorts. And the results revealed that the higher the stage, the lower the survival rate.
| Groups | OS mean | 1-yr (%) | 3-yr (%) | 5-yr (%) | Sig | HR (95%CI) |
| Low risk | 71.7 | 83.4 | 57.5 | 33.4 | - | - |
| Medium risk | 19.8 | 66.4 | 31.7 | 21.4 | 0.004 | 1.93 (1.23-3.03) |
| High risk | 10.03 | 34.8 | 4.4 | - | 0.000 | 5.47 (3.08-9.73) |
A nomogram can be used to establish a statistical prognostic model to estimate the prognosis of cancer patients[10]. No studies have determined a prognostic nomogram for PSCE patients due to the rarity of the disease. In this study, we successfully constructed an effective nomogram model to predict PSCE prognosis. Combined with clinical correlations, we identified that age, histology type, T stage, N stage, M, CD56, and CgA were prognostic predictors of PSCE. However, tumor location and grade in the 7th TNM staging were not independent prognostic factors in this study. A previous study also failed to find a significant correlation of tumor location and grade with OS in Chinese PSCE patients[6]. In addition, the analysis of four non-TNM risk factors, including age, histology type, CgA, and CD56, showed significant increases in risk scores for patients over 60 years old (6.0 points), pure PSCE (6.0 points), positive CgA (4.0 points), and negative CD56 (5.0 points). The nomogram model is beneficial in the development of a personalized scoring system for patients[23].
Histopathologically, some PSCEs are mainly composed of neuroendocrine tumor cells and may have other cancerous components, such as adenocarcinoma or squamous cell carcinoma[24]. PSCE has been reported to be a highly metastatic disease with a poor prognosis. This estimate corresponds well with our study finding that 24.1% of patients presented with stage IV and 40.8% of patients had coexisting squamous cell carcinoma. Mixed PSCE may be derived from multipotential stem cells of the esophageal mucosa[25]. With regard to survival, mixed PSCE patients had a superior outcome with a median OS of 23.21 mo and a 5-year OS of 31.2%. Pure PSCE patients were associated with a median OS of 16.67 mo and a 5-year OS of 20.3%. CD56 is a type of neurocellular adhesion molecule associated with the prognosis of multiple myeloma[26]. CgA is an acidic glycoprotein, which belongs to a class of regulated secretory proteins[27] and is a prognosis factor for several types of small cell carcinomas[28,29].
In addition, several previous studies have shown that surgery can enhance the prognosis of some PSCE patients[30,31]. In our study, compared with patients who received other treatments, patients in the surgery group showed an increasing trend in clinical outcome, with a median OS of 21.11 vs 12.39 mo and a 5-year OS of 28.3% vs 15.8%, although these differences were not statistically significant. A meta-analysis by Raja et al[32] showed that surgery or radiotherapy combined with chemotherapy can significantly improve patient prognosis. However, since only 14 patients received surgery plus radiotherapy and 10 underwent surgery and chemotherapy in our training cohort, we were unable to conduct further survival analysis to determine any differences. Therefore, treatment is not considered to be a potential prognostic factor in our nomogram model.
As far as we know, this is the first nomogram model of PSCE based on seven important prognostic factors. The discrimination and calibration were evaluated by internal and external validation[33]. In our study, the C-index of the nomogram was higher than that of the 7th TNM staging , indicating higher discrimination of the model. We then used the optimal cut-off analyses to successfully classify the patients in the training cohort into three risk groups. Subgroup stratification of patients with different risk levels is beneficial for clinicians to carry out individualized treatment[34]. The calibration curves indicated that there was good consistency between the predicted and actual values for 1-, 3-, and 5-year OS. DCA also suggested that the prognostic prediction model had greater potential for clinical application than the 7th TNM staging .
However, this study also had several limitations. First, due to the retrospective nature of our research, potential biases were inevitable. Second, due to the low incidence of PSCE, other relevant clinical parameters (e.g., incidence area, family history, body mass index, and reproductive history) were unavailable. These confounding factors might affect treatment choice or survival, which should be considered in future related studies.
We have developed, for the first time, a nomogram model for predicting OS in Chinese patients with PSCE. The novel nomogram classifies patients into different risk subgroups and showed superiority in predicting survival compared with the 7th TNM staging. Therefore, our nomogram may help clinicians make individualized prognostic predictions and better treatment recommendations for PSCE patients in the future.
Primary small cell carcinoma of the esophagus (PSCE) is a rare tumor, accounting for 0.05% to 3.1% of all esophageal malignancies and approximately 2% of extrapulmonary small cell carcinomas. PSCE patients seem to have earlier metastasis and a worse prognosis than those with esophageal squamous cell carcinoma, which requires a more accurate prognostic prediction model.
Several previous studies have reported the prognostic factors for PSCE with controversial results, partly due to their small sample size. To date, only one nomogram has been used to predict the overall survival probability for PSCE patients in the United States. In addition, the model did not include relevant neuroendocrine markers.
The present study aimed to build a prognostic predictive nomogram model including clinicopathological factors and neuroendocrine biomarkers for Chinese PSCE patients. It was also determined whether the nomogram model can predict overall survival (OS) more accurately than the 7th tumor-node-metastasis (TNM) staging system.
The nomogram was based on a retrospective study of 256 PSCE patients, derived from our esophageal and gastric cardia carcinoma database including 500000 cases (1973-2015), established by State Key Laboratory of Esophageal Cancer Prevention and Treatment and Henan Key Laboratory for Esophageal Cancer Research of the First Affiliated Hospital of Zhengzhou University in Henan, China. The predictive accuracy and discriminative ability of the nomogram were determined by the concordance index (C-index), calibration plot, and decision curve analysis (DCA), and the results were also compared with the 7th TNM staging.
The final nomogram model included histology type, age, tumor invasion depth, lymph node invasion, distant metastases, chromogranin A, and neuronal cell adhesion molecule 56. The C-index of the model had a prognostic superiority over the 7th edition TNM staging system in both the primary cohort [0.659 (95%CI: 0.607-0.712) vs 0.591 (95%CI: 0.517-0.666), P = 0.033] and the validation cohort [0.700 (95%CI: 0.622-0.778) vs 0.605 (95%CI: 0.490-0.721), P = 0.041]. Good calibration curves were observed for the prediction probabilities of 1-, 3- and 5-year OS in the primary and validation cohorts. DCA analysis showed that our nomogram model had a higher overall net benefit than the 7th TNM staging.
We have developed and validated a nomogram for predicting 1-year, 3-year, and 5-year OS in Chinese PSCE patients. The new nomogram classifies patients into different risk subgroups and shows a superiority of survival prediction over the 7th TNM staging.
The nomogram model can be used to predict the survival probability of PSCE patients, which might help clinicians to make individualized prognosis predictions and give better treatment recommendations for PSCE patients in China.
We thank Professor Shi XZ (Department of Epidemiology and Biostatistics, College of Public Health in Zhengzhou University) for help in statistical analysis.
Manuscript source: Unsolicited manuscript
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Narumiya K, Oda M S-Editor: Wang LL L-Editor: Wang TQ P-Editor: Wang LYT
| 1. | Vos B, Rozema T, Miller RC, Hendlisz A, Van Laethem JL, Khanfir K, Weber DC, El Nakadi I, Van Houtte P. Small cell carcinoma of the esophagus: a multicentre Rare Cancer Network study. Dis Esophagus. 2011;24:258-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 2. | Li AF, Li AC, Hsu CY, Li WY, Hsu HS, Chen JY. Small cell carcinomas in gastrointestinal tract: immunohistochemical and clinicopathological features. J Clin Pathol. 2010;63:620-625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 3. | Deng HY, Ni PZ, Wang YC, Wang WP, Chen LQ. Neuroendocrine carcinoma of the esophagus: clinical characteristics and prognostic evaluation of 49 cases with surgical resection. J Thorac Dis. 2016;8:1250-1256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 51] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 4. | Rice TW, Rusch VW, Ishwaran H, Blackstone EH; Worldwide Esophageal Cancer Collaboration. Cancer of the esophagus and esophagogastric junction: data-driven staging for the seventh edition of the American Joint Committee on Cancer/International Union Against Cancer Cancer Staging Manuals. Cancer. 2010;116:3763-3773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 315] [Cited by in RCA: 357] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 5. | Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471-1474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5537] [Cited by in RCA: 6460] [Article Influence: 430.7] [Reference Citation Analysis (0)] |
| 6. | Ku JW, Zhang DY, Song X, Li XM, Zhao XK, Lv S, Hu SJ, Cheng R, Zhou FY, Wu HF, Wang LD. Characterization of tissue chromogranin A (CgA) immunostaining and clinicohistopathological changes for the 125 Chinese patients with primary small cell carcinoma of the esophagus. Dis Esophagus. 2017;30:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 7. | Hu Y, Hu C, Zhang H, Ping Y, Chen LQ. How does the number of resected lymph nodes influence TNM staging and prognosis for esophageal carcinoma? Ann Surg Oncol. 2010;17:784-790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 68] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 8. | Ishida H, Kasajima A, Kamei T, Miura T, Oka N, Yazdani S, Ozawa Y, Fujishima F, Sakurada A, Nakamura Y, Tanaka Y, Kurosumi M, Ishikawa Y, Okada Y, Ohuchi N, Sasano H. SOX2 and Rb1 in esophageal small-cell carcinoma: their possible involvement in pathogenesis. Mod Pathol. 2017;30:660-671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 9. | Zhu Y, Qiu B, Liu H, Li Q, Xiao W, Hu Y, Liu M. Primary small cell carcinoma of the esophagus: review of 64 cases from a single institution. Dis Esophagus. 2014;27:152-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 10. | Karakiewicz PI, Briganti A, Chun FK, Trinh QD, Perrotte P, Ficarra V, Cindolo L, De la Taille A, Tostain J, Mulders PF, Salomon L, Zigeuner R, Prayer-Galetti T, Chautard D, Valeri A, Lechevallier E, Descotes JL, Lang H, Mejean A, Patard JJ. Multi-institutional validation of a new renal cancer-specific survival nomogram. J Clin Oncol. 2007;25:1316-1322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 411] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 11. | Rudloff U, Jacks LM, Goldberg JI, Wynveen CA, Brogi E, Patil S, Van Zee KJ. Nomogram for predicting the risk of local recurrence after breast-conserving surgery for ductal carcinoma in situ. J Clin Oncol. 2010;28:3762-3769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 244] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 12. | Qie S, Wang XF, Ran YG, Liu ML, Cui GM, Shi HY. Nomogram for predicting the survival of patients with small cell carcinoma of the esophagus: A population study based on the surveillance, epidemiology, and end results database. Medicine (Baltimore). 2021;100:e25427. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 13. | van der Gaag NA, Kloek JJ, de Bakker JK, Musters B, Geskus RB, Busch ORC, Bosma A, Gouma DJ, van Gulik TM. Survival analysis and prognostic nomogram for patients undergoing resection of extrahepatic cholangiocarcinoma. Ann Oncol. 2012;23:2642-2649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 77] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 14. | Ji LF, Fan ZM, Wu MJ, Wang R, Kong GQ, Meng H, Zhou YF, Liu TJ, Liu ZC, Fu WT, Wu Y, Cheng R, Wang LD. Clinicpathopathologic features and survival impact factors of 286 patients with esophagus spindle cell carcinoma. Zhengzhou Daxue Xuebao (Yixueban). 2016;51:565-568. [DOI] [Full Text] |
| 15. | Sun K, Huang SH, Wong DS, Jang SS. Design and Application of a Variable Selection Method for Multilayer Perceptron Neural Network With LASSO. IEEE Trans Neural Netw Learn Syst. 2017;28:1386-1396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 16. | Kim SY, Yoon MJ, Park YI, Kim MJ, Nam BH, Park SR. Nomograms predicting survival of patients with unresectable or metastatic gastric cancer who receive combination cytotoxic chemotherapy as first-line treatment. Gastric Cancer. 2018;21:453-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 17. | Li S, Zhao J, Zhu L, Su F, Chen K. Development and validation of a nomogram predicting the overall survival of stage IV breast cancer patients. Cancer Med. 2017;6:2586-2594. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 18. | Yu SC, Qi X, Hu YH, Zheng WJ, Wang QQ, Yao HY. [Overview of multivariate regression model analysis and application]. Zhonghua Yu Fang Yi Xue Za Zhi. 2019;53:334-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 19. | Barton JB, Langdale LA, Cummins JS, Stelzner M, Lynge DC, Mock CN, Nason KS, Billingsley KG. The utility of routine preoperative computed tomography scanning in the management of veterans with colon cancer. Am J Surg. 2002;183:499-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 29] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 20. | Kramer AA, Zimmerman JE. Assessing the calibration of mortality benchmarks in critical care: The Hosmer-Lemeshow test revisited. Crit Care Med. 2007;35:2052-2056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 476] [Cited by in RCA: 637] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 21. | Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26:565-574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3515] [Cited by in RCA: 3479] [Article Influence: 183.1] [Reference Citation Analysis (1)] |
| 22. | Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252-7259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1947] [Cited by in RCA: 2936] [Article Influence: 146.8] [Reference Citation Analysis (0)] |
| 23. | Long J, Wang A, Bai Y, Lin J, Yang X, Wang D, Jiang Y, Zhao H. Development and validation of a TP53-associated immune prognostic model for hepatocellular carcinoma. EBioMedicine. 2019;42:363-374. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 257] [Article Influence: 42.8] [Reference Citation Analysis (0)] |
| 24. | Yun JP, Zhang MF, Hou JH, Tian QH, Fu J, Liang XM, Wu QL, Rong TH. Primary small cell carcinoma of the esophagus: clinicopathological and immunohistochemical features of 21 cases. BMC Cancer. 2007;7:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 86] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 25. | Walenkamp AM, Sonke GS, Sleijfer DT. Clinical and therapeutic aspects of extrapulmonary small cell carcinoma. Cancer Treat Rev. 2009;35:228-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 147] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 26. | Skerget M, Skopec B, Zadnik V, Zontar D, Podgornik H, Rebersek K, Furlan T, Cernelc P. CD56 Expression Is an Important Prognostic Factor in Multiple Myeloma Even with Bortezomib Induction. Acta Haematol. 2018;139:228-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 27. | Modlin IM, Gustafsson BI, Moss SF, Pavel M, Tsolakis AV, Kidd M. Chromogranin A--biological function and clinical utility in neuro endocrine tumor disease. Ann Surg Oncol. 2010;17:2427-2443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 261] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 28. | Liao LM, Zhang X, Ren YF, Sun XY, Di N, Zhou N, Pan RK, Ma SH, Zhou LX. Chromogranin A (CgA) as poor prognostic factor in patients with small cell carcinoma of the cervix: results of a retrospective study of 293 patients. PLoS One. 2012;7:e33674. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 29. | Berruti A, Bollito E, Cracco CM, Volante M, Ciccone G, Porpiglia F, Papotti M, Scarpa RM, Dogliotti L. The prognostic role of immunohistochemical chromogranin a expression in prostate cancer patients is significantly modified by androgen-deprivation therapy. Prostate. 2010;70:718-726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 41] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 30. | Xu L, Li Y, Liu X, Sun H, Zhang R, Zhang J, Zheng Y, Wang Z, Liu S, Chen X. Treatment Strategies and Prognostic Factors of Limited-Stage Primary Small Cell Carcinoma of the Esophagus. J Thorac Oncol. 2017;12:1834-1844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 31. | Situ D, Lin Y, Long H, Zhang L, Lin P, Zheng Y, Jiang L, Tan Z, Meng Y, Ma G. Surgical treatment for limited-stage primary small cell cancer of the esophagus. Ann Thorac Surg. 2013;95:1057-1062. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 32. | Raja S, Rice TW, Rajeswaran J, Zhong J, Blackstone EH. Esophageal small-cell cancer: study of a rare disease. Dis Esophagus. 2013;26:690-695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 33. | Iasonos A, Schrag D, Raj GV, Panageas KS. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol. 2008;26:1364-1370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1306] [Cited by in RCA: 2305] [Article Influence: 135.6] [Reference Citation Analysis (0)] |
| 34. | Liang W, Zhang L, Jiang G, Wang Q, Liu L, Liu D, Wang Z, Zhu Z, Deng Q, Xiong X, Shao W, Shi X, He J. Development and validation of a nomogram for predicting survival in patients with resected non-small-cell lung cancer. J Clin Oncol. 2015;33:861-869. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 467] [Article Influence: 46.7] [Reference Citation Analysis (0)] |