Published online Oct 16, 2021. doi: 10.12998/wjcc.v9.i29.8797
Peer-review started: December 16, 2020
First decision: July 16, 2021
Revised: July 28, 2020
Accepted: September 7, 2021
Article in press: September 7, 2021
Published online: October 16, 2021
Processing time: 296 Days and 7.3 Hours
Identifying a potential single monogenetic disorder in healthy couples is costly due to the Assisted Reproduction facilities' current methodology for screening, which focuses on the detecting multiple genetic disorders at once. Here, we report the successful application of a low-cost and fast preimplantation genetic testing for monogenic/single gene defects (PGT-M) approach for detecting propionic acidemia (PA) in embryos obtained from a confirmed heterozygous propionyl-CoA carboxylase alpha subunit (PCCA) couple.
A fertile 32-years old Mexican couple with denied consanguinity sought antenatal genetic counseling. They were suspected obligate PA carriers due to a previous deceased PA male newborn with an unknown PCCA/propionyl-CoA carboxylase beta subunit (PCCB) genotype. Next-Generation Sequencing revealed a heterozygous genotype for a pathogenic PCCA variant (c.2041-1G>T, ClinVar:RCV
We show that using PGT-M with Whole Genome Amplification templates, coupled with IVF, can reduce the transmission of a pathogenic variant of the PCCA gene.
Core Tip: Propionic acidemia is an uncommon monogenetic disorder resulting in inherited severe complications and death. A heterozygous genotype for a pathogenic variant of the propionyl-Coenzyme A carboxylase alpha subunit gene (PCCA) was located in a fertile Mexican couple. Here we show that a couple can reduce the transmission of a pathogenic variant of a gene using in vitro fertilization, genetic counseling, and sequencing of a whole genome amplification template. Furthermore, after embryo transfer, we report the delivery of a healthy male newborn without propionic acidemia.
- Citation: Neumann A, Alcantara-Ortigoza MA, González-del Angel A, Zarate Díaz NA, Santana JS, Porchia LM, López-Bayghen E. Whole-genome amplification/preimplantation genetic testing for propionic acidemia of successful pregnancy in an obligate carrier Mexican couple: A case report. World J Clin Cases 2021; 9(29): 8797-8803
- URL: https://www.wjgnet.com/2307-8960/full/v9/i29/8797.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i29.8797
For fertile, healthy couples, the diagnosis of genetic disorders occurs postnatally when the disease's characteristics are present. However, for genetic disorders in which the fetus suffers from a detrimental illness, the probability of conceiving and delivering a healthy newborn decrease dramatically. Therefore, many at-risk couples attend Assisted Reproduction facilities for Preimplantation Genetic Testing for Monogenic/single gene defects (PGT-M). However, the cost associated with PGT-M, which includes the assessment of over 200 genetic disorders, can be expensive and, for some patients, preventative. Therefore, there is a need for alternative methods when the cost is an issue or the disorder is not covered under standard PGT-M. Propionic acidemia (PA, MIM#606054) is an autosomal recessive, life-threatening metabolic disorder caused by a deficiency in the mitochondrial enzyme propionyl-CoA carboxylase (PCC). PCC carboxylates propionyl-CoA to methylmalonyl-CoA and reduced/abolished PCC activity results in elevated blood concentrations of levocarnitine ester of propionyl-CoA[1]. PCC is composed of alpha and beta subunits encoded by the PCCA (13q32.3, MIM*232000) and PCCB (3q22.3, MIM*232050) genes, respectively[1]. Shortly after birth, approximately 65% of all PA patients present with symptoms, such as vomiting, lethargy, refuse feeding, hypotonia, and other clinical data[2]. If not treated, PA may progress to a severe illness with neurologic, cardiologic, hematologic, hepatic, and pancreatic complications or death[1,2]. Since 80% of all PA cases worldwide are caused by a single-nucleotide or other small pathogenic changes, which results in loss of PCC activity[1,2], suspected carriers of PCCA or PCCB pathogenic genetic variants would benefit from PGT-M. Here, we report the successful application of a low-cost and fast PGT-M approach for PA in embryos obtained from a confirmed heterozygous PCCA couple, which resulted in a clinical pregnancy and delivery of a healthy newborn male.
A 32-years old healthy Mexican couple with denied consanguinity sought antenatal genetic counseling, after giving birth to PA male.
They were suspected of being obligate PA carriers due to a previous full-term delivered male (3200 g), who passed one month after birth and presented with PA's classical clinical profile. Diagnosis of PA in the deceased newborn was biochemically suspected through an acylcarnitine profile during newborn screening (elevated serum propionyl carnitine) and further confirmed by elevated urine organic acids concentrations (methyl citrate, 3-hydroxypropionate, 2-hydroxyisovaleric, 3-hydroxybutyric, 3-hydroxy propionic).
No significant past medical history associated with PA.
Both parents originated from different states in Mexico. Moreover, they indicated no knowledge of any family history of PA.
Neither parent presented with any symptoms or risk factors for being a PA carrier.
Due to a lack of any previous complication of either parent with respect to reproductive health or otherwise, no laboratory examinations were performed.
No imaging studies were performed.
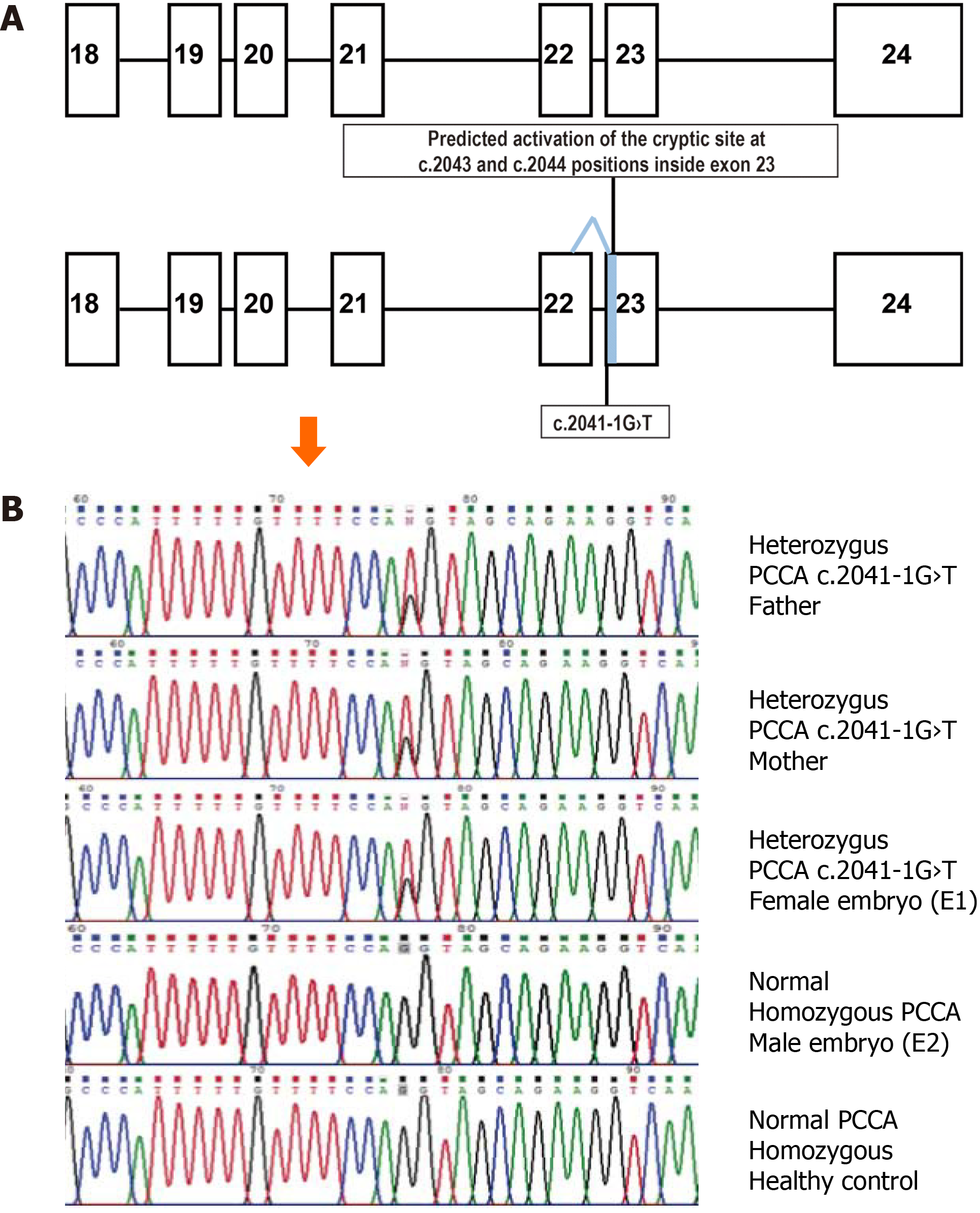
Since the pathogenic PA genotype could not be confirmed for the deceased newborn, both parents were subjected to next-generation sequencing (NGS) for the PCCA and PCCB genes. Quality of the parental genomic DNA, obtained from peripheral blood samples, was evaluated using the Qubit™ Flex Fluorometer (Life Technologies, Singapore). Targets were captured by hybridization to create libraries through Nextera Rapid Capture Exome (Illumina Inc., San Diego, CA, United States) and then sequenced on an Illumina HiSeq2000 2 × 150 platform (San Diego, CA, United States). The bioinformatics pipeline included an overall quality evaluation of the raw output reads with FastQC v0.11.8, trimming of adapters and filtering of low-quality reads using Trimmomatic v0.35, alignment of the filtered reads against the GRCh38 human reference sequence using the Bowtie2 software v2.3.4.1, and calling and annotation of single nucleotide variations and detection of small insertion-deletion with the GATK and snpEff programs, respectively. Only one clinically relevant PCCA gene variant was confirmed by unidirectional Sanger sequencing of an end-point polymerase chain reaction (PCR)-derived 250 bp fragment containing exon 23 of the PCCA gene (Figure 1A). Annotation was carried out using the Alamut Visual 2.14 software (SOPHIA GENETICS, Lausanne, Switzerland).
Both parents were found to be asymptomatic heterozygotes for the same pathogenic PCCA gene variant (Figure 1B), c.2041-1G>T (NM_000282.4; ClinVar:RCV000802701.1; dbSNP:rs1367867218). According to the in silico evaluation by the Splice Site Finder-like, MaxEntScan, NNSPLICE, and GeneSplicer programs, including the Splicing Prediction Module of Alamut Visual 2.14 software, this variant eliminates the natural acceptor splicing site of intron 22 and possible activation of a cryptic acceptor splicing site inside of exon 23 (positions c.2043 and c.2044; Figure 1A).
After the PCCA genotype characterization and post-genetic testing counseling, both parents requested PGT-M for PA, along with PGT for aneuploidies (PGT-A). A single in vitro fertilization (IVF) cycle, embryo biopsy, and PGT were performed according to the standard protocols of the Ingenes Institute, as previously described[3,4]. The mother underwent one standard course of controlled ovarian stimulation with 1650 UI of Gonal (FSH, Merck, Darmstadt, Germany) and 375 UI of Merapur (Hmg Ferring Laboratories, Saint-Prex, Switzerland). After 36 h, 12 oocytes (10 were in Metaphase II) were retrieved with ultrasound guidance and were fertilized by intracytoplasmic sperm injection. Only morphologically optimal embryos were considered for PGT, using the Istanbul Consensus Workshop Criteria on Embryo Assessment[3].
During day 5 of embryonic development, two embryos were determined to be of high-quality and were biopsied. Using micromanipulation, 4-7 cells from the trophectoderm were isolated and placed into a 0.2 mL PCR tube. Afterward, the embryos were cryopreserved using the vitrification technique[4]. The biopsies were subjected to Whole Genome Amplification (WGA, according to the manufacturer's protocol) using the SurePlex DNA Amplification System (Illumina Inc., San Diego, CA, United States). NGS (Veri-seq PGS Library Prep kit, Illumina Inc., San Diego, CA, United States) was performed and embryo ploidy assessment was obtained[4]. To determine the embryos’ PCCA genotype, in triplicate, WGA-DNA samples were subjected to PCR amplifi
Fourteen days after transfer, β-hCG serum levels were 168.0 mUI/mL, confirming a positive pregnancy. After 16 wk, an ultrasound exhibited a single gestational sac. The couple declined to confirm the PCCA genotype of the fetus by invasive diagnostic testing. Duo test was performed during week 11-13, which indicated a low risk of chromosomopathy during the 1st trimester. Also, a TORCH screen was performed, which was negative. Structural ultrasound and five following consultations showed that the baby was healthy throughout the pregnancy. A healthy newborn male was delivered after 38 wk of pregnancy (weight: 4080 g, length: 49 cm, APGAR 9/9). The newborn’s metabolic screening demonstrated a normal acylcarnitine profile. Moreover, none of the typical symptoms for PA were noted in the subsequent days after birth. When this article was submitted, the child was four months of age and has not presented with any symptoms of PA.
Here, we report a couple that underwent preconception counseling, pre- and post-genetic testing counseling, subjected to IVF and PGT-M to avoid the high probability of bearing an affected PA offspring. The preventive reproductive approach was successful and the couple was able to conceive a healthy newborn. As reported with other potentially fatal organic acidemias, the poor clinical prognosis of PA justifies providing preconception genetic counseling and preventive reproductive options, like PGT-M, to at-risk couples[5]. Monogenic disorders have been estimated to occur in about 0.36% of births[6]. These Mendelian diseases can be identified in embryos belonging to at-risk couples through PGT. At present, different technologies are available to directly search for specific mutations for a highly accurate genetic diagnosis of potential embryos. PGT-M has been performed with sequencing verification for 234 pathogenic genetic variants; however, this approach increases the cost and time to complete. As pointed out before, NGS strategies applied to PGT-M allow identifying pathogenic variants located across the genome, including those responsible for “rare disorders”, like organic acidemias[5]. However, this approach is an expensive and time-consuming testing that does not appear to be an affordable option for couples where a specific pathogenic genotype has been previously identified. Therefore, the most straightforward approach is to perform PGT-M to identify the pathogenic genotype from IVF-generated euploid embryos[4]. Indeed, here, our technique presents as an alternative to prenatal diagnosis and avoids termination of a pregnancy in the case of a compromised fetus. Moreover, the cost associated with the methodology presented here can significantly reduce the patient's expenses, making the test accessible.
With embryo biopsies, a significant problem during analysis can arise if there is inadequate DNA or its concentration is very low. The signal can be improved by performing WGA. We have previously shown that using WGA-DNA, obtained by the fragmentation/amplification-based method, yields a suitable DNA template for the generation of small PCR fragments (250 bp aprox). Those fragments are successfully sequenced by the simple automated Sanger method to rapidly determine a specific embryo genotype for a single nucleotide change or other small pathogenic variants[4]. However, such assays must consider a false positive possibility due to the allele drop-out phenomena. As also noted in the recently reported PGT-M procedure performed with the WGA-DNA template obtained by isothermal genome amplification[4], in our triplicate Sanger sequencing assay, we were unable to document the allele drop-out phenomena. It would be necessary to carry out further validation studies to quantify the possibility of allelic amplification imbalances when PGT-M is based on WGA-DNA obtained by a fragmentation/amplification-based method.
The birth prevalence of PA across Asia-Pacific, Europe, and North America is 0.29, 0.33, and 0.33 per 100000 newborns, respectively[7], however, it is still unknown in Mexico. This disorder appears to be very rare for the Mexican population, as determined by at least two expanded neonatal screening reports applying an acylcarnitine profile[8.9], even though PA comprises around 7% of all detected inborn errors of intermediary metabolism in Mexican patients[10]. To date, in the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/), there are around 25 splicing defects responsible for PA. To the best of our knowledge, the c.2041-1G>T variant has not been reported in the literature as associated with PA. However, according to the gnomAD database, it is listed as an extremely infrequent allele, with only single heterozygous individuals identified in the Latino population (allele frequency 0.00002891). This feature contrasts with the consanguinity antecedent denied by the couple. Interestingly, both parents are of Mexican origin. However, they come from different states in Mexico, supporting the state of no endogamy in the couple.
Recently, the c.2041-1G>T variant has been reported in the ClinVar database as a likely pathogenic variant (RCV000802701.1), as it predicts to nullify the recognition of the natural acceptor splice site of PCCA intron 22. Another pathogenic variant, c.2041-2A>G, affects the same acceptor splicing site, conditioning exon 23 skipping in the mRNA but leads to a relatively mild PA phenotype with high residual PCC activity in fibroblasts. This is attributed to generating a small amount of normally spliced transcripts[11]. If the c.2041-1G>T variant also leads to exon 23 skipping, the resulting protein will lose a biotin-binding motif, leading to a non-functional PCC enzyme[11]. Possible residual exon 23 retention and the activation of the in silico predicted out-of-frame cryptic acceptor splice site inside exon 23 must be confirmed by further experimental assays to define the precise pathogenic effect of c.2041-1G>T and its possible genotype-phenotype correlation.
Here, we report the c.2041-1G>T variant of the PCCA gene classified as likely pathogenic and is associated with PA. Moreover, we show that using PGT-M coupled with IVF could be applied to avoid the transmission of PA in patients’ descendances, which should decrease the potential of an unsuccessful pregnancy and increase the possibility of delivering a healthy baby. Lastly, we demonstrate the feasibility of using WGA-DNA templates for a low-cost, rapid, and accurate PGT-M for life-threatening disorders, like PA.
We are grateful to the participants of the study and to Ing. Lucero Cervantes Pozos for her editorial assistance. We also thank the technical support of Semper Genomics SA de CV.
Manuscript source: Unsolicited manuscript
Corresponding Author's Membership in Professional Societies: American Society for Reproductive Medicine; American Society for Neurochemistry, No. 12657; Society for Neuroscience, No. 100008912.
Specialty type: Reproductive biology
Country/Territory of origin: Mexico
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kim HS S-Editor: Zhang H L-Editor: A P-Editor: Li JH
| 1. | Wongkittichote P, Ah Mew N, Chapman KA. Propionyl-CoA carboxylase - A review. Mol Genet Metab. 2017;122:145-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 153] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 2. | Pena L, Franks J, Chapman KA, Gropman A, Ah Mew N, Chakrapani A, Island E, MacLeod E, Matern D, Smith B, Stagni K, Sutton VR, Ueda K, Urv T, Venditti C, Enns GM, Summar ML. Natural history of propionic acidemia. Mol Genet Metab. 2012;105:5-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 110] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 3. | Alpha Scientists in Reproductive Medicine and ESHRE Special Interest Group of Embryology. The Istanbul consensus workshop on embryo assessment: proceedings of an expert meeting. Hum Reprod. 2011;26:1270-1283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 952] [Cited by in RCA: 1296] [Article Influence: 92.6] [Reference Citation Analysis (0)] |
| 4. | Neumann A, Alcántara-Ortigoza MÁ, González-Del Ángel A, Camargo-Diaz F, López-Bayghen E. Diagnosis of Laron syndrome using monoplex-polymerase chain reaction technology with a whole-genome amplification template: A case report. World J Clin Cases. 2019;7:4029-4035. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (1)] |
| 5. | Habibzadeh P, Tabatabaei Z, Farazi Fard MA, Jamali L, Hafizi A, Nikuei P, Salarian L, Nasr Esfahani MH, Anvar Z, Faghihi MA. Pre-implantation genetic diagnosis in an Iranian family with a novel mutation in MUT gene. BMC Med Genet. 2020;21:22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 6. | Verma IC, Puri RD. Global burden of genetic disease and the role of genetic screening. Semin Fetal Neonatal Med. 2015;20:354-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 7. | Almási T, Guey LT, Lukacs C, Csetneki K, Vokó Z, Zelei T. Systematic literature review and meta-analysis on the epidemiology of propionic acidemia. Orphanet J Rare Dis. 2019;14:40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 8. | Torres-Sepúlveda Mdel R, Martínez-de Villarreal LE, Esmer C, González-Alanís R, Ruiz-Herrera C, Sánchez-Peña A, Mendoza-Cruz JA, Villarreal-Pérez JZ. [Expand newborn screening using tandem mass spectrometry: two years' experience in Nuevo León, Mexico]. Salud Publica Mex. 2008;50:200-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 9. | Cantú-Reyna, C, Zepeda, LM, Montemayor, R, Benavides, S, González, HJ, Vázquez-Cantú, M, Cruz-Camino, H. Incidence of inborn errors of metabolism by expanded newborn screening in a Mexican hospital. J Inborn Errors Metab Screen. 2016;4:1-8. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 10. | Ibarra-González I, Fernández-Lainez C, Belmont-Martínez L, Guillén-López S, Monroy-Santoyo S, Vela-Amieva M. [Characterization of inborn errors of intermediary metabolism in mexican patients]. An Pediatr (Barc). 2014;80:310-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 11. | Clavero S, Pérez B, Rincón A, Ugarte M, Desviat LR. Qualitative and quantitative analysis of the effect of splicing mutations in propionic acidemia underlying non-severe phenotypes. Hum Genet. 2004;115:239-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |