Published online Oct 16, 2021. doi: 10.12998/wjcc.v9.i29.8671
Peer-review started: March 25, 2021
First decision: June 14, 2021
Revised: June 17, 2021
Accepted: August 19, 2021
Article in press: August 19, 2021
Published online: October 16, 2021
Processing time: 204 Days and 6.9 Hours
Colon adenocarcinoma (COAD) is one of the most common and fatal malignant tumors, which increases the difficulty of prognostic predictions. Thus, new biomarkers for the diagnosis and prognosis of COAD should be explored. Ferroptosis is a recently identified programmed cell death process that has the characteristics of iron-dependent lipid peroxide accumulation. However, the predictive value of ferroptosis-related genes (FRGs) for COAD still needs to be further clarified.
To identify some critical FRGs and construct a COAD patient prognostic signature for clinical utilization.
The Cancer Genome Atlas database (TCGA) and Gene Expression Omnibus databases were the data sources for mRNA expression and corresponding COAD patient clinical information. Differentially expressed FRGs were recognized using R and Perl software. We constructed a multi-FRG signature of the TCGA-COAD cohort by performing a univariate Cox regression and least absolute shrinkage and selection operator Cox regression analysis. COAD patients from the Gene Expression Omnibus cohort were utilized for verification.
Our research showed that most of the FRGs (85%) were differentially expressed between the corresponding adjacent normal tissues and cancer tissues in the TCGA-COAD cohort. Seven FRGs were related to overall survival (OS) in the univariate Cox analysis (all P < 0.05). A model with five FRGs (AKR1C1, AKR1C3, ALOX12, CRYAB, and FDFT1) was constructed to divide patients into high- and low-risk groups. The OS of patients in the high-risk group was significantly lower than that of the low-risk group (all P < 0.01 in the TCGA and Gene Expression Omnibus cohorts). The risk score was an independent prognosticator of OS in the multivariate Cox analysis (hazard ratio > 1, P < 0.01). The predictive capacity of the model was verified by a receiver operating characteristic curve analysis. In addition, a nomogram based on the expression of five hub FRGs and risk score can precisely predict the OS of individual COAD cancer patients. Immune correlation analysis and functional enrichment analysis results revealed that immunology-related pathways were abundant, and the immune states of the high-risk group and the low-risk group were different.
In conclusion, a novel five FRG model can be utilized for predicting prognosis in COAD. Targeting ferroptosis may be a treatment option for COAD.
Core Tip: Colon adenocarcinoma is one of the most ordinary and fatal malignant tumors. Ferroptosis is a recently appeared type of programmed cell death process, which has the characteristics of iron-dependent lipid peroxide accumulation. In this work, we excavated five ferroptosis-related genes of colon adenocarcinoma from The Cancer Genome Atlas database. Then, we constructed a very accurate prognostic model based on the five genes (AKR1C1, AKR1C3, ALOX12, CRYAB, FDFT1). Our study also highlights the crucial roles of the risk score was an independent prognostic biomarker for colorectal cancer patients, and we validated these genes in the outside data access (GSE39582).
- Citation: Miao YD, Kou ZY, Wang JT, Mi DH. Prognostic implications of ferroptosis-associated gene signature in colon adenocarcinoma. World J Clin Cases 2021; 9(29): 8671-8693
- URL: https://www.wjgnet.com/2307-8960/full/v9/i29/8671.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i29.8671
According to Global Cancer Statistics 2020[1], colon cancer is one of the most fatal cancers, with approximately 1148515 new cases and 576858 deaths from 36 cancers in 185 countries. The incidence and mortality are both ranked fifth worldwide in 2020. It is the fifth most commonly diagnosed cancer in men and the fourth most frequently diagnosed cancer in women. The incidence rates of colon cancer vary by approximately 9-fold in different regions of the world, with the highest rates in Europe, Australia/New Zealand and North America, with Hungary and Norway ranking first for men and women, respectively. The development of colon cancer is a complex biological process caused by a variety of factors, including obesity, lifestyle, eating habits, and environmental influences[2]. A systematic review and meta-analysis of 43 randomized controlled trials or cohorts indicated that dietary fiber led to a reduced risk of colorectal cancer (CRC), while red and preserved meats led to an increased risk and was rated “convincing,” which is the strongest grade assigned[3-5]. Despite significant advances in diagnosis and treatment, the average 5-year overall survival (OS) rate for CRC is 48%, and the 5-year OS rate for patients with metastatic colon cancer is only 10%[6,7]. The complex biological factors and high heterogeneity associated with colon cancer increase the difficulty of predicting prognosis. In addition, given the limited treatment strategies for colon cancer, it is imperative to develop new prognostic models. The application of second-generation DNA sequencing technology by whole-transcriptome, whole-genome, and whole-exome methods can lead to remarkable advances in cancer genomics[8], and the Cancer Genome Atlas (TCGA) has led to unprecedented opportunities for the study of clinically significant cancer biology[9].
In 2012, Dixon first proposed that ferroptosis was a new form of cell death[10]. Unlike apoptosis, autophagy, and necrosis in terms of biochemistry, morphology, and genetics, ferroptosis is a unique reactive oxygen species-reliant and iron-dependent form of nonapoptotic cell death in which cytological changes are the main features[11]. The iron-dependent accumulation of lipid hydroperoxides to lethal levels is characterized by ferroptosis-regulated cell death[12-15]. Previous studies have shown that ferroptosis was originally defined in RAS-mutated cancer cells. Sensitivity to the induction of ferroptosis is mainly found in numerous diverse types of cancer cells with RAS mutations[16,17]. Lanperisone facilitates the production of reactive oxygen species to kill embryonic fibroblasts of K-Ras-mutant mice by ferroptosis methods, and in a mouse model, it induced ferroptotic death in lung cancer cells by inhibiting Cys2 uptake[18]. The induction of ferroptosis has become an encouraging treatment option for inducing cancer cell death, particularly for malignancies resistant to conventional therapies[19-21].
In addition to ferroptosis inducers, many genes have also been identified as markers or modulators of ferroptosis, such as iron metabolism-related genes, including transferrin receptor 1, transferrin, ferritin heavy chain 1, and ferritin light chain[19]. Previous studies have shown that ferroptosis plays an essential role in several cancers and several genes, such as Nrf2, which promotes cancer progression by negatively regulating ferroptosis. of tumor suppressor P53 (TP53), as a transcriptional repressor of SLC7A11, participates in ferroptosis, impairs cysteine input, and promotes the initiation of ferroptosis[22-24]. MUC1-C can upregulate the expression of glutathione by forming a complex with CD44v, which can lead to ferroptosis in breast cancer cells[25]. The coinhibition of NFS1 and Cys transport can cause hypertrophy in vitro and inhibit lung cancer cell growth[26]. However, whether these ferroptosis-related genes (FRGs) are related to colon adenocarcinoma (COAD) patient prognosis is still largely unknown.
In the current research, we obtained mRNA expression data and corresponding clinical information on COAD patients from two public databases. In addition, we constructed a prognostic model based on five differentially expressed FRGs from the TCGA-COAD cohort and validated it in the Gene Expression Omnibus (GEO)-GSE39582 cohort. Furthermore, we performed an immune correlation analysis and functional enrichment analysis to identify the potential mechanisms.
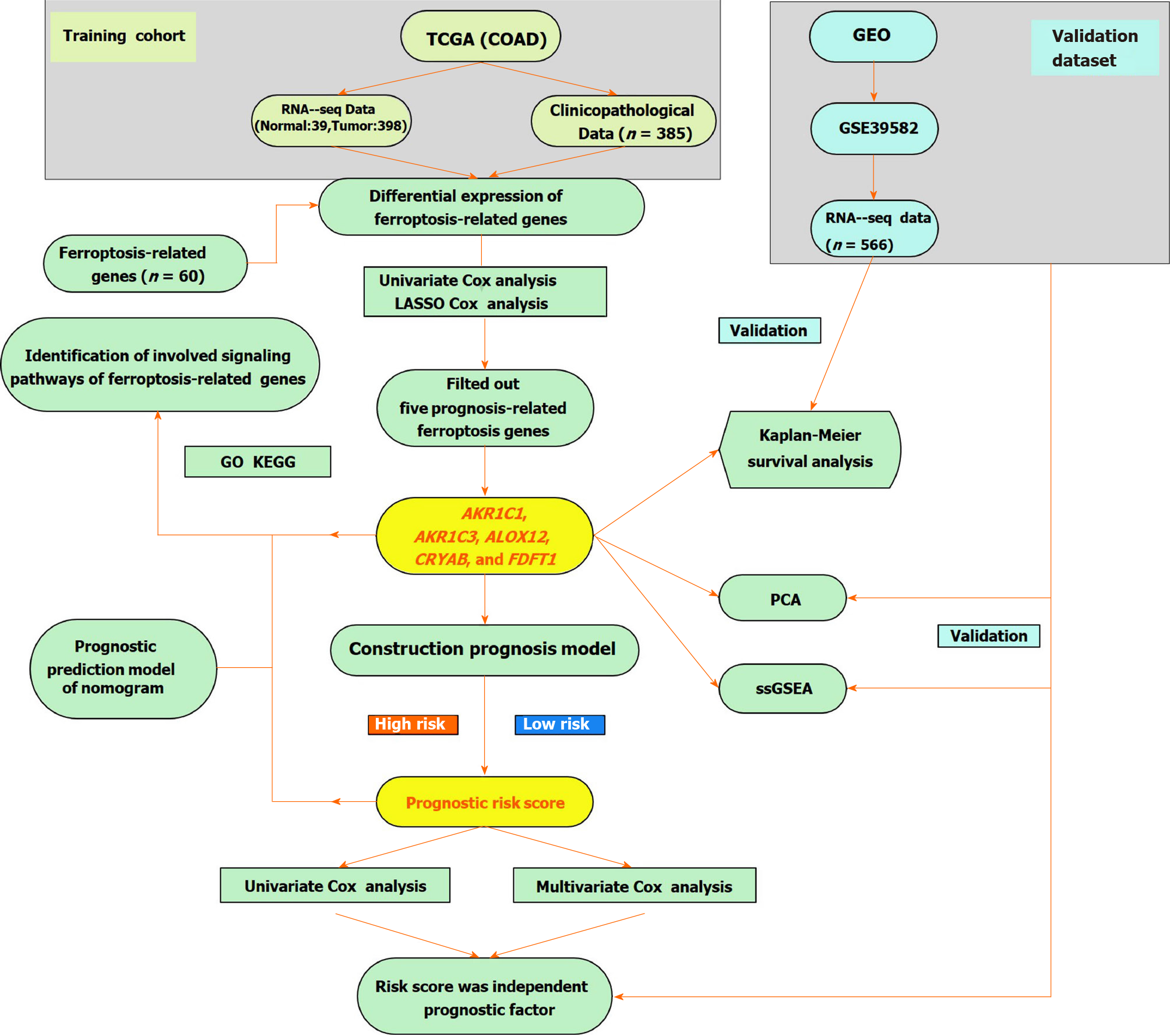
TCGA-COAD cohort and GSE39582 cohort: We obtained level 3 RNA-sequencing (RNA-seq) information and corresponding clinical data of COAD from the TCGA database (Data Release 27.0, October 29, 2020, https://tcga-data.nci.nih.gov/tcga/). Detailed data selection criteria for mRNA sequencing include the following: (1) The keywords for cases were [Primary Site] “colon,” [Program] “TCGA,” [Project] “TCGA-COAD,” and [Disease Type] “Adenomas and Adenocarcinomas;” (2) The keywords for files are [Data Category] “Transcriptome Profiling,” [Data Category] “Gene Expression Quantification,” [Experimental Strategy] “RNA-Seq,’” and [Workflow Type] “HTSeq - FPKM;” and (3) The filter criteria of clinical data are [Data Category] “Clinical” and [Data Format] “BCR XML”. We obtained 437 mRNA expression profiles for COAD. Among them, 39 (8.9%) samples were normal and 398 samples (91.1%) were cancerous. The data from the GSE39582 cohort were obtained from the GEO database (https:// www.ncbi.nlm.nih.gov/geo/) and used for verification, and they consisted of 585 samples (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE39582) that contained 19 colon mucosa samples and 566 stage I-IV colon adenocarcinoma samples. We normalized the raw data of the gene chip by the RMA algorithm provided by the R package “limma”[27]. The patients’ clinicopathological features are listed in Table 1. The mRNA expression and clinical data are both available from public databases (TCGA and GEO). Therefore, the current research was exempt from approval by the local ethics committee. The present study follows the data access strategy and publication guidelines of the TCGA and GEO databases. Then, 60 FRGs were obtained from the previous literature[12,20,28] and were shown in Supplementary Table 1.
| TCGA-COAD | GSE39582 | |
| Number of patients | 385 | 566 |
| Survival status | ||
| Alive | 306 (79.5) | 371 (65.5) |
| Dead | 79 (20.5) | 191 (33.7) |
| Unknown | 4 (0.8) | |
| Survival time in d | ||
| Range for OS days | 0-4502 | 0-2412 |
| OS days, median | 670 | 612 |
| Age in yr | 67.00 ± 12.76 | 66.87 ± 13.28 |
| Range | 31-90 | 22-97 |
| Median | 69 | 68 |
| Sex | ||
| Female | 180 (46.7) | 256 (45.2) |
| Male | 205 (53.2) | 310 (54.8) |
| Stage | ||
| I | 66 (17.1) | 33 (5.8) |
| II | 151 (39.2) | 264 (46.6) |
| III | 103 (26.8) | 205 (36.2) |
| IV | 54 (14.0) | 60 (10.6) |
The Wilcox test in the R package “limma” was utilized to explore the differentially expressed FRGs (DEFRGs) between corresponding adjacent non-neoplastic tissues and cancer tissues in the TCGA-COAD cohort (false discovery rate < 0.05). FRGs with prognostic values for OS were screened out by a univariate Cox analysis (P < 0.05). The interacting genes between FRGs with prognostic values and DEFRGs were visualized through the R package “Venn”[29]. The R packages “igraph” and “reshape2” were used to perform the correlation analysis of interacting genes.
To minimize the risk of overfitting, a least absolute shrinkage and selection operator Cox regression analysis[30] was used to construct the optimal model of FRGs using the R-package “glmnet.” The response variables were the status and OS of TCGA-COAD patients, and the independent variable in the regression analysis was the standardized expression data of candidate prognostic DEFRGs. The risk score for each patient was computed based on the normalized expression level of each FRG and their corresponding regression coefficients. The formula was defined as follows: Risk score = ∑nj=1Coefficient (n) × gene expression (n), where Coefficient (n) denotes the coefficient and indicates the relative expression levels of each ferroptosis-associated gene in the prognostic risk score model. The median risk score was chosen as the cut-off value for TCGA-COAD cohort dichotomy, and the COAD patients were categorized into high- and low-risk groups. Survival information was obtained from the clinical data of the aforementioned samples. We used the R packages “survminer” and “survival” to draw the survival curve according to the high- and low-risk values. Based on the expression of ferroptosis-associated genes in the model, a principal component analysis was carried out with the R package “ggplot2” and t-distributed stochastic neighbor embedding was used to identify the distribution of diverse groups using the R package “Rtsne.” Receiver operating characteristic (ROC) curves were utilized to examine the sensitivity and specificity of survival prediction by the ferroptosis-associated risk score and were drawn using the R package “survival ROC.” The area under the curve (AUC) value is an accurate prognostic indicator. The risk curve, heatmap, and survival state map were drawn according to the diverse risk scores of the patients. The independent prognostic FRGs were identified by univariate and multivariate Cox hazard regression analyses. A total of 566 COAD patient samples with prognostic information from the GSE39582 dataset were used as verification cohorts to validate the predictive ability of the ferroptosis-associated gene prognostic model based on TCGA-COAD data.
A nomogram was drawn according to the expression of five FRGs by using the R packages “Hmisc,” “Formula,” “lattice,” “foreign,” “rms,” and “ggplot2.” In addition, a calibration curve was utilized to evaluate the consistency between actual and predicted survival rates. Furthermore, the consistency index was computed to evaluate the prognostic capability of the model. Values of 0.5 and 1.0 represent a random probability and an excellent performance for predicting survival with the model, respectively.
We applied the “FUNCTIONS-Expression analysis- Expression DIY-Box Plot” component of the webserver: Gene expression profiling interactive analysis[31] (GEPIA2, http://gepia2.cancer-pku.cn/#analysis) to acquire box plots of different expression between corresponding normal tissues from the Genotype-tissue expression database and tumor tissues using the following parameters: log2FC (Fold change) cutoff = 2, P value cutoff = 0.05, and “Match TCGA normal and Genotype-tissue expression data.” In addition, we acquired a violin illustration of the expression levels of five FRGs in COAD and the corresponding normal tissues of Genotype-tissue expression and TCGA. We drew box and violin plots by using log2 [TPM (transcripts per million) +1] converted expression data.
We used the Human Protein Atlas (http://www.proteinatlas.org/) database to explore the expression level of the core FRGs in the normal and tumor samples. For most of the protein-coding genes, we performed an immunohistochemistry analysis according to the tissue microarray analysis of each protein and the corresponding cancer type of patients. Immunohistochemistry staining was conducted based on prior published literature. The staining index scores were distributed as follows: staining intensity (negative: 0; weak: 1; moderate: 2; and strong: 3) and positive staining (0: < 5%; 1: 5-25%; 2: 26-50%; 3: 51-75%; 4: > 75%). The staining index score was computed by multiplying the staining intensity score by the positive staining score, which ranged from 0 to 12[32].
Based on the DEFRGs between the high- and low-risk groups, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses using the R packages “clusterProfiler,” “enrichplot,” “org.Hs.eg.db,” and “ggplot2.” Then, we calculated the activity of 13 immune-related pathways and the infiltration score of 16 immune cells via a single-sample gene set enrichment analysis using the R packages “GSVA” and “GSEABase.”
We compared the differential expression of FRGs between corresponding adjacent nonneoplastic tissues and COAD tissues by the Wilcoxon test. The OS between diverse groups was contradistinguished based on a Kaplan-Meier analysis with the log-rank test. We explored the independent predictors of OS by using univariate and multivariate Cox regression analyses. The Mann-Whitney test with adjusted P values through the Benjamini and Hochberg method was used to contradistinguish the single-sample gene set enrichment analysis scores of immune cells or pathways between the high- and low-risk groups. All statistical analyses were performed with R software (Version-4.0.3). P < 0.05 indicated statistical significance and all P values were two-tailed.
We enrolled 385 COAD patients from the TCGA-COAD cohort and 566 COAD patients from the GEO (GSE39582) dataset. The detailed clinical features of these patients are shown in Table 1. The flow chart of this current study is shown in Figure 1.
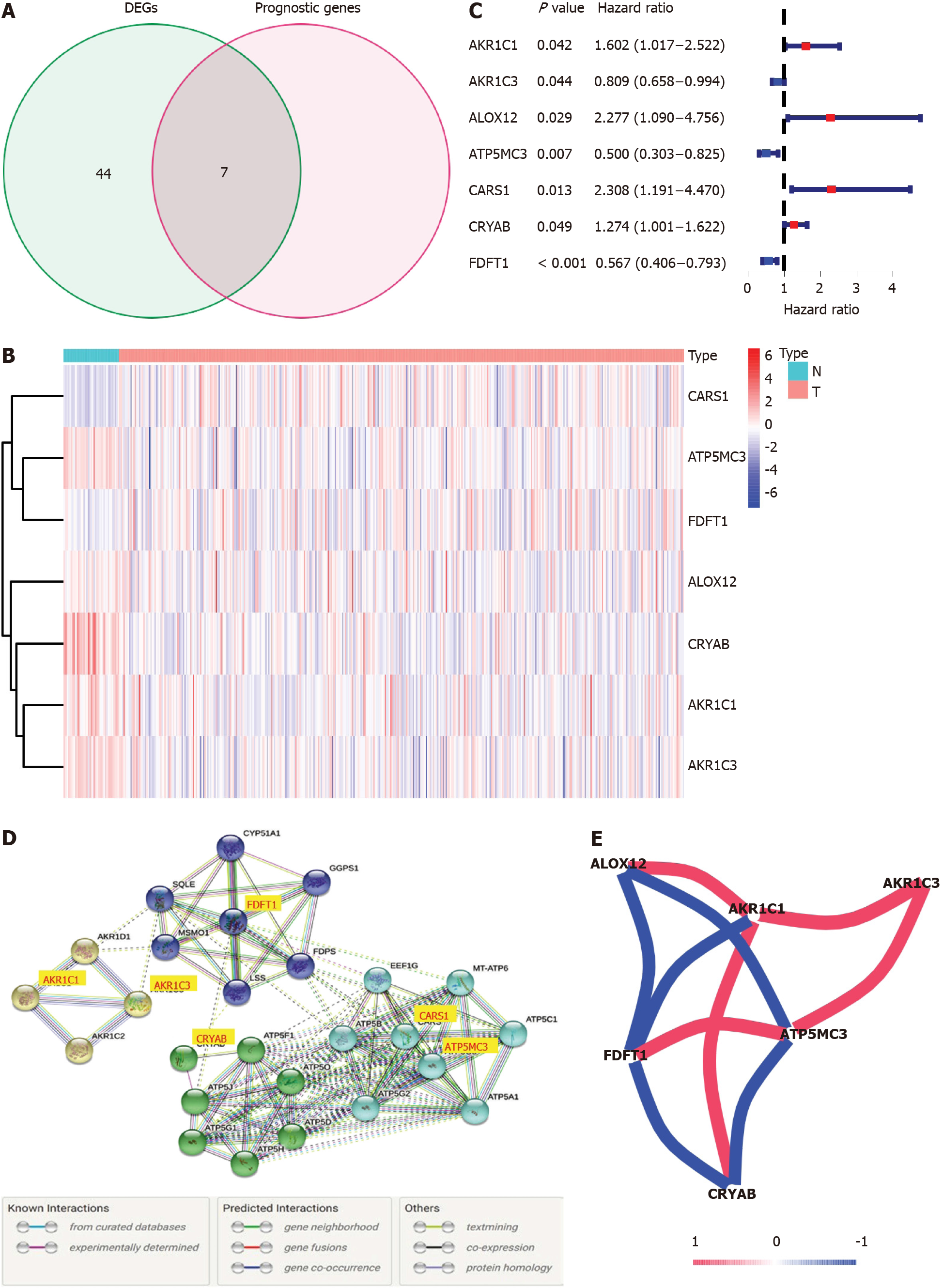
Most of the FRGs (51/60, 85%) were differentially expressed between corresponding adjacent non-neoplastic tissues and COAD tissues, with 31 upregulated FRGs and 20 downregulated FRGs in the COAD tissues (Figure 2A). A total of seven DEFRGs were related to OS in the univariate Cox regression analysis. AKR1C3, ATP5MC3 and farnesyl-diphosphate farnesyltransferase 1 (FDFT1) were low-risk DEFRGs [hazard ratio (HR) < 1, P < 0.05]; and AKR1C1, ALOX12, CARS1, and crystallin alpha B (CRYAB) were high-risk DEFRGs (HR > 1, P < 0.05) (Figure 2B and C). The interaction network among these genes demonstrated that AKR1C1, AKR1C3, ATP5MC3, CARS1, CRYAB, and FDFT1 are the core genes (Figure 2D). ALOX12 positively correlated with AKR1C1 while negatively correlated with FDFT1 and ATP5MC3. AKR1C1 has a positive correlation with ALOX12, AKR1C3, and CRYAB while negatively correlated with FDFT1. AKR1C3 positively correlated with AKR1C1 and ATP5MC3. FDFT1 has a positive correlation with ATP5MC3 while negatively correlated with ALOX12, AKR1C1, and CRYAB. ATP5MC3 positively correlated with AKR1C3 and FDFT1 while negatively correlated with ALOX12 and CRYAB. The correlation between these genes is shown in Figure 2E.
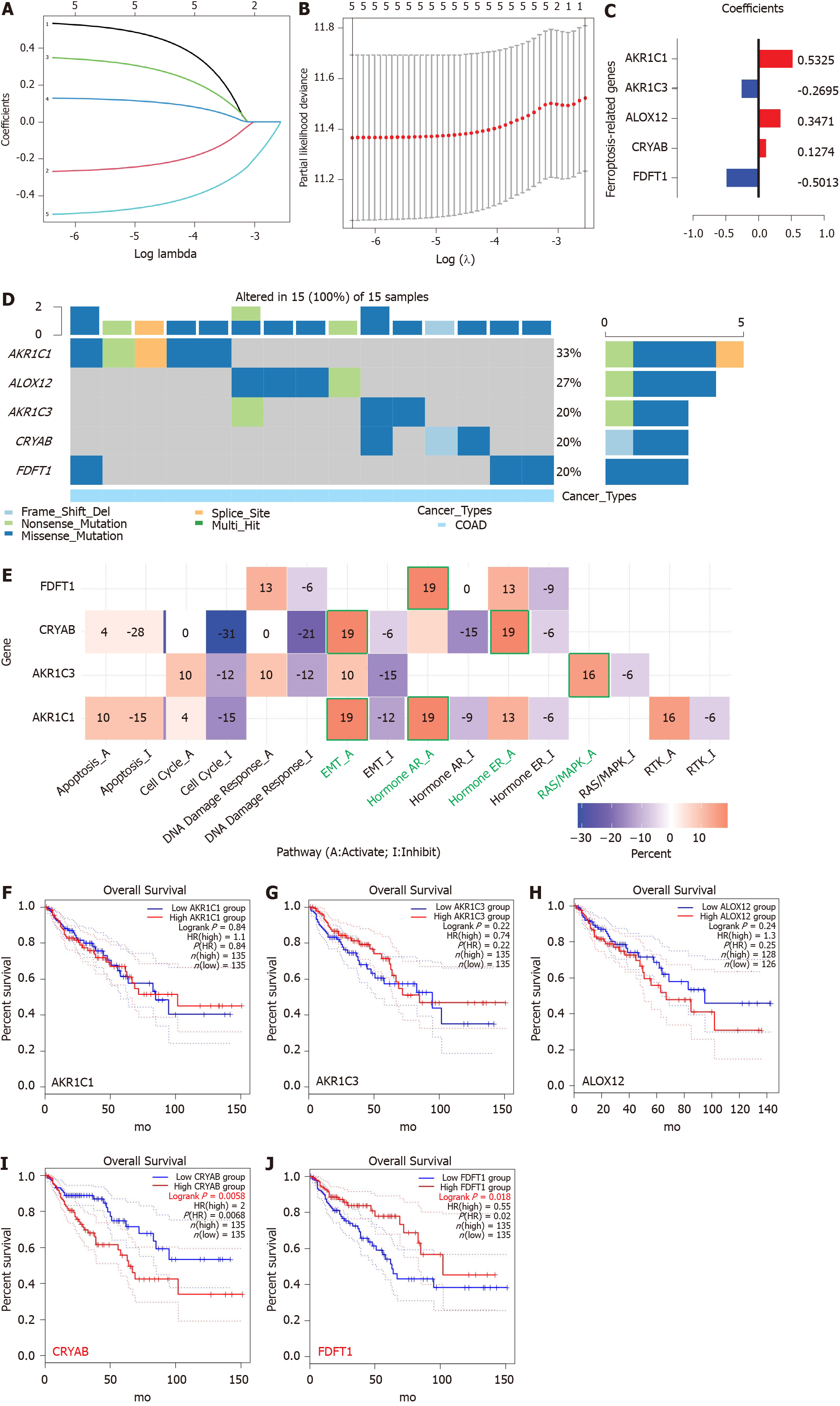
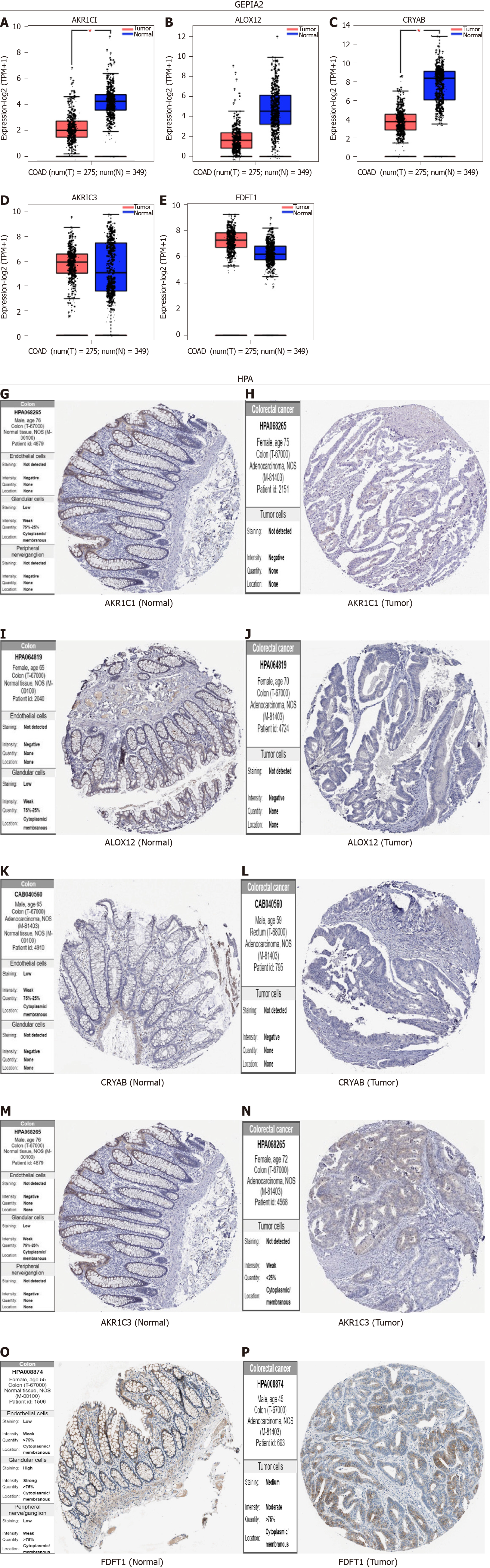
Five FRGs were screened out by the least absolute shrinkage and selection operator Cox regression analysis, and the coefficients were calculated. The model produced the best data when five FRGs were included (Figure 3A and B). The coefficient of each FRG is shown in Figure 3C. The prognosis signature was constructed based on five FRGs (AKR1C1, AKR1C3, ALOX12, CRYAB, and FDFT1). We investigated the genetic alterations of these five FRGs in COAD using the Gene Set Cancer Analysis Lite (http://bioinfo.life.hust.edu.cn/web/GSCALite/). The interesting genes in COAD were altered in 15 of 15 queried samples (100%) (Figure 3D). The frequent genetic alterations demonstrated the essential roles of these FRGs in the development of COAD. In addition, AKR1C1 mainly participated in the regulation of epithelial-mesenchymal transition (EMT) and hormone androgen receptor activity. AKR1C3 is mainly involved in the regulation of RAS/MAPK activity, and CRYAB is mainly associated with the regulation of EMT and hormone estrogen receptor activity. FDFT1 mainly participated in the regulation of hormone androgen receptor activity (Figure 3E). To further explore the association between the expression of each gene and COAD, we applied the “FUNCTIONS-Expression Analysis-Survival Analysis” module of the webserver GEPIA2 to obtain the significance profile data between these five FRGs and OS in COAD. We used the values of cutoff-high (50%) and cutoff-low (50%) as the expression thresholds to divide the patients into high- and low-expression groups. The log-rank test was utilized in the hypothesis test, and survival curves were also acquired via the “Survival Analysis” module of the web database GEPIA2. The results indicated that the expression of AKR1C1, AKR1C3, and ALOX12 did not have a significant correlation with the OS of COAD (all P > 0.05, Figure 3F-H). High expression of CRYAB was correlated with poor OS in COAD (P < 0.05, Figure 3I), while high expression of FDFT1 was associated with better OS in COAD (P < 0.05, Figure 3J).
A prognostic model of five FRGs was explored according to the optimal value of λ (Figure 3A and B). The risk score for each patient was calculated based on the following formula: Risk score = 0.5325 × expression level of AKR1C1 + (-0.2695) × expression level of AKR1C3 + 0.3471 × expression level of ALOX12 + 0.1274 × expression level of CRYAB + (-0.5013) × expression of FDFT1.
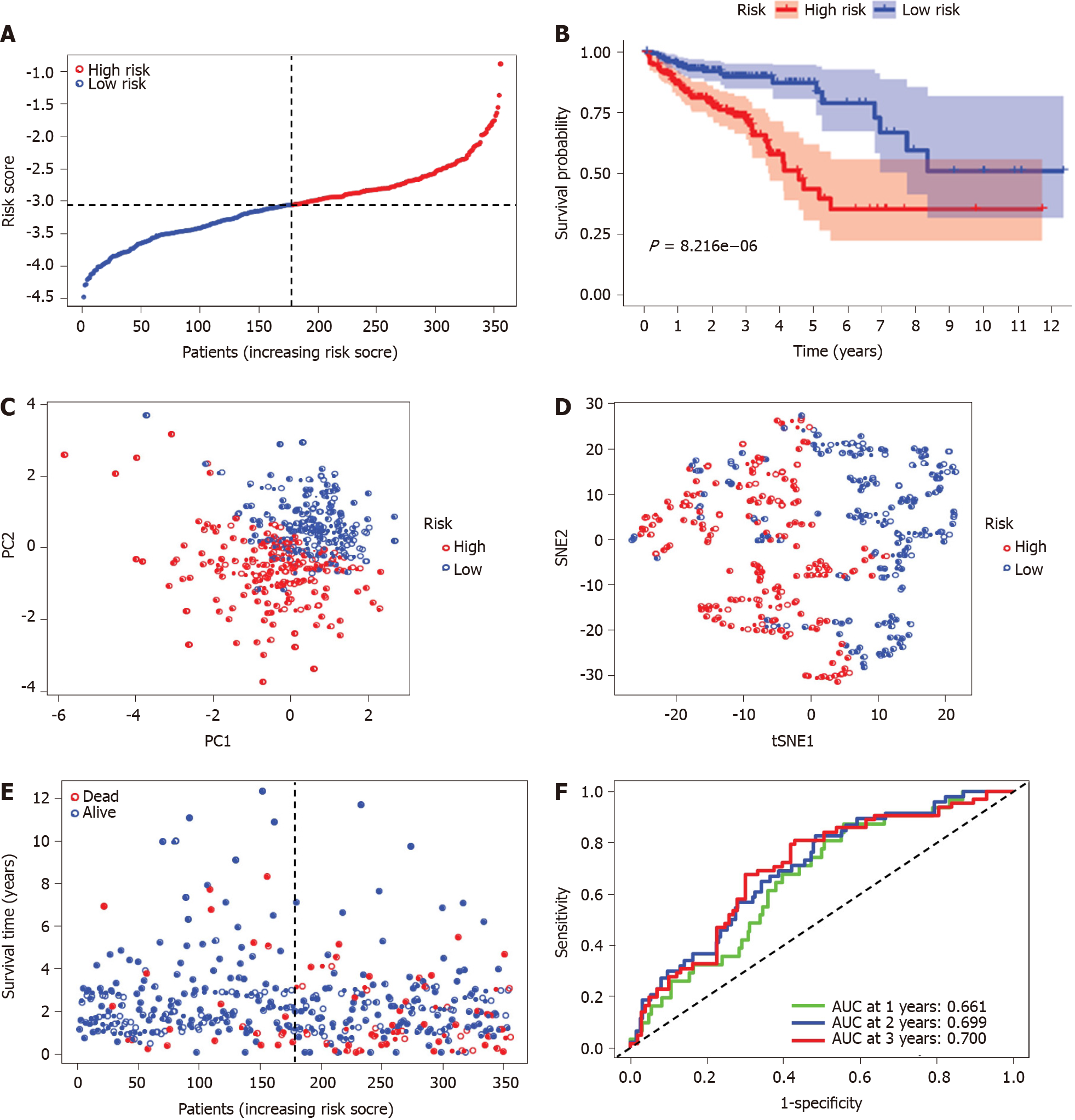
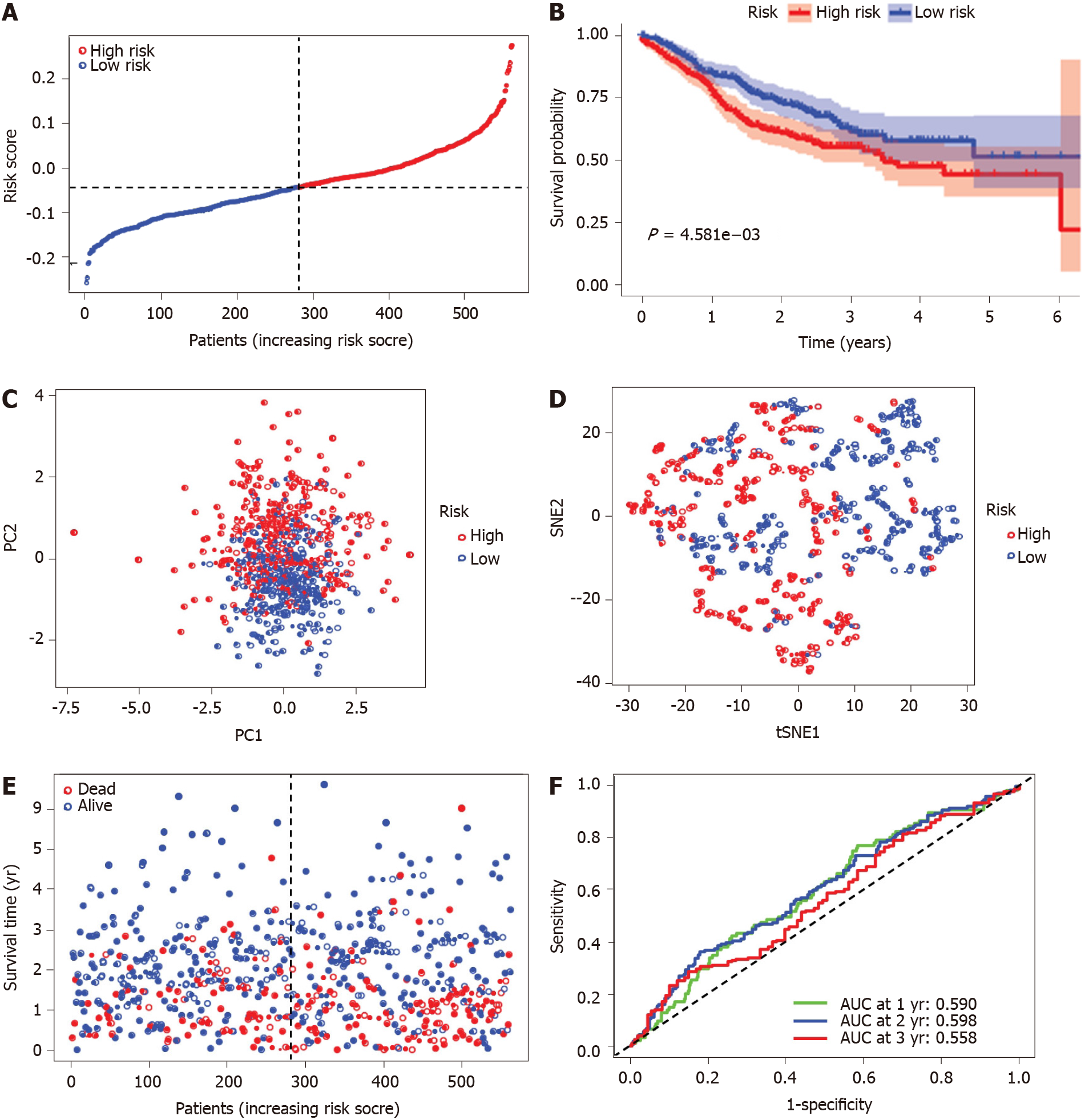
The risk score was utilized to predict prognosis. Using the median cut-off value, patients were divided into a high-risk group (n = 177) and a low-risk group (n = 178) (Figure 4A). Kaplan-Meier cumulative curves showed that patients in the high-risk group had a significantly poorer OS than those in the low-risk group (P < 0.001, Figure 4B). The patients in the high-risk group were significantly correlated to advanced TNM stage in the TCGA-COAD cohort (Table 2). The principal component analysis and t-distributed stochastic neighbor embedding analysis demonstrated that the patients in diverse risk groups were distributed in two directions (Figure 4C and D). The survival time diminished as the risk score increased, and a higher risk score corresponded to more deaths in TCGA-COAD (Figure 4E). A time-dependent ROC analysis was used to further assess the prognostic accuracy of the prognostic model. The AUC value was 0.661 at 1 year, 0.699 at 2 years, and 0.700 at 3 years, which showed a moderate diagnostic ability (Figure 4F).
| Features | TCGA-COAD cohort | GEO-GSE39582 cohort | ||||
| High-risk | Low-risk | P value | High-risk | Low-risk | P value | |
| Age | 0.795 | 0.862 | ||||
| < 65 yr | 70 (39.5) | 68 (38.2) | 105 (37.4) | 107 (38.1) | ||
| ≥ 65 yr | 107 (60.5) | 110 (61.8) | 176 (62.6) | 174 (61.9) | ||
| Sex | 0.265 | 0.351 | ||||
| Female | 86 (48.6) | 76 (42.7) | 132 (47.0) | 121 (43.1) | ||
| Male | 91 (51.4) | 102 (57.3) | 149 (53.0) | 160 (56.9) | ||
| TNM-stage | P < 0.001 | P = 0.002 | ||||
| I and II | 83 (46.9) | 114 (64.0) | 134 (47.7) | 170 (60.5) | ||
| III and IV | 89 (50.3) | 58 (32.6) | 145 (51.6) | 109 (38.8) | ||
| Unknown | 5 (2.8) | 6 (3.4) | 2 (0.7%) | 2 (0.7%) | ||
To verify the stability of the signature determined by the TCGA-COAD cohort, patients from the GSE39582 database were also classified as high or low risk, with the median value computed using the same formula as that from the TCGA-COAD dataset (Figure 5A). Similar to the results obtained from the TCGA-COAD database, the OS of patients in the high-risk group was significantly worse than that of patients in the low-risk group (P < 0.01, Figure 5B). The high-risk group was also associated with advanced TNM stage in the GEO-GSE39582 cohort (Table 2). The principal component analysis and t-distributed stochastic neighbor embedding analysis showed that patients in the high-risk and low-risk groups were also distributed in discrete directions (Figure 5C and D). Similarly, an elevated risk score was correlated with a decreased survival time and an increase in mortality (Figure 5E). In addition, the AUC value of the five FRG signatures was 0.590 at 1 year, 0.598 at 2 years, and 0.558 at 3 years in the GSE39582 cohort (Figure 5F).
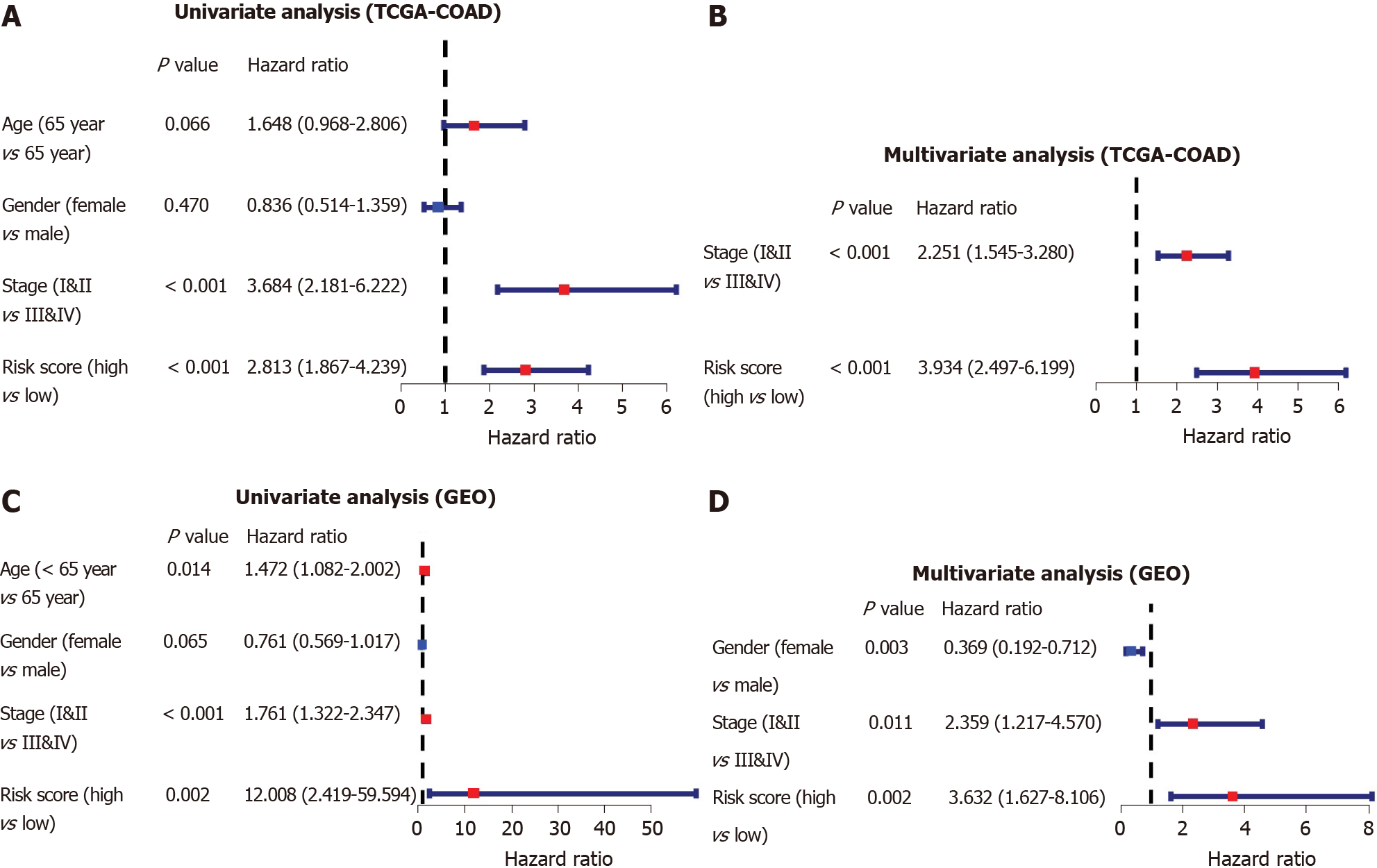
To identify whether the risk score was an independent prognostic factor for OS, we performed univariate and multivariate Cox regression analyses among the available variables. The risk score was remarkably related to OS in both the TCGA and GSE39582 cohorts in the univariate Cox regression analysis (HR = 2.813, P < 0.001; HR = 12.008, P = 0.002, respectively, Figure 6A-C). After adjustment for other farrago elements, the risk score was confirmed as an independent predictor of OS in the multivariate Cox analysis (TCGA-COAD cohort: HR = 3.934, P < 0.001; GSE39582 cohort: HR = 3.632, P = 0.002; Figure 6B-D).
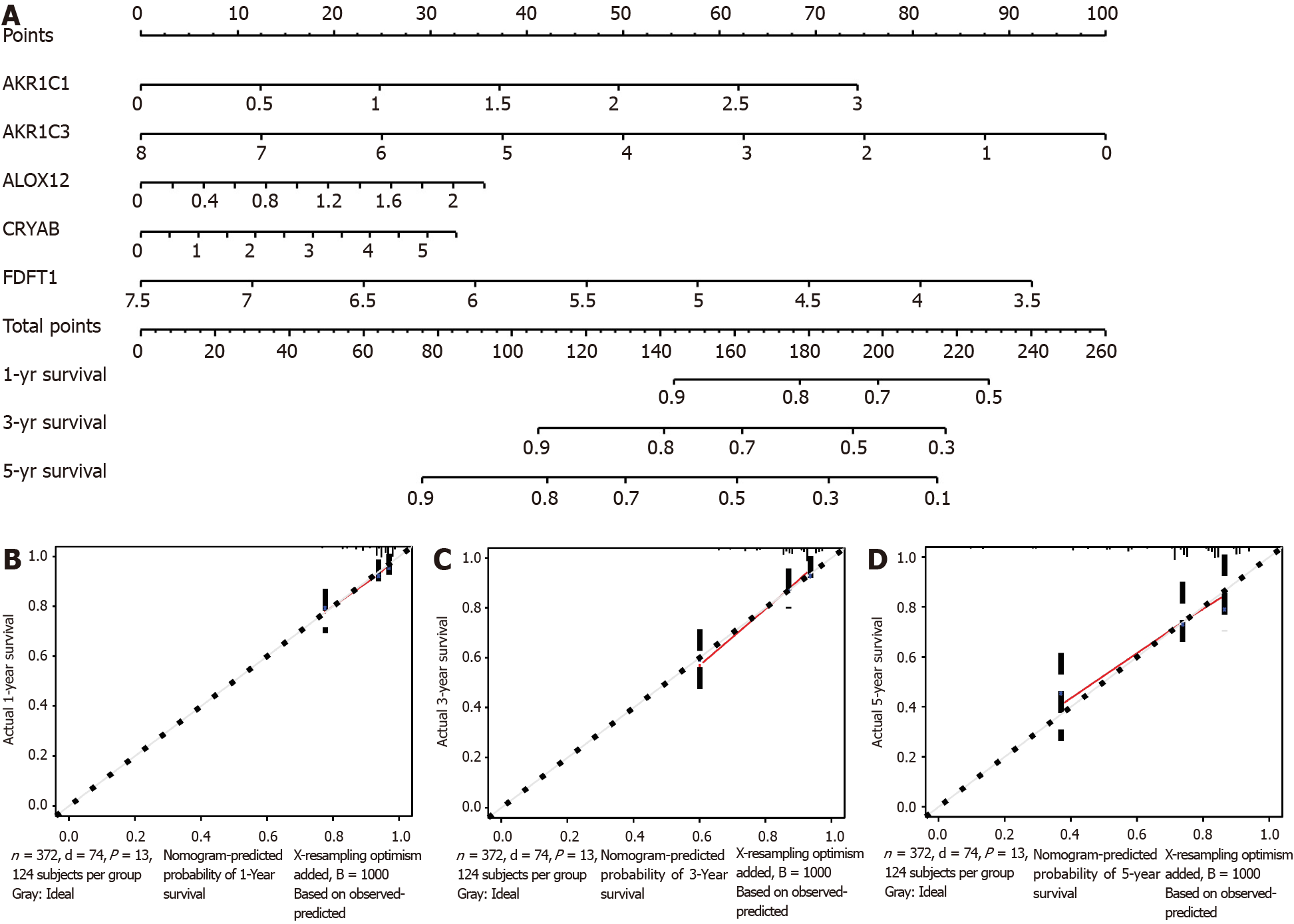
A nomogram can quantitatively identify an individual’s clinical risk by combining multiple risk factors[33]. We used the nomogram to predict the probability of 1-, 3-, and 5-year OS by merging the five FRG signatures for TCGA-COAD (Figure 7A). The distribution point of each FRG was proportional to its risk contribution to survival and was normalized to the distribution of 0 to 100. We calculated each patient’s total points by summarizing the number of points for all FRGs and estimated the survival rates by drawing a vertical line between each prognosis axis and the total point axis. This tool may help practitioners make clinical decisions for COAD patients. The calibration curve demonstrated that the actual survival rate matched the predicted 1-, 3-, and 5-year survival rate (Figure 7B-D). The consistency index was 0.764 in the TCGA-COAD cohort.
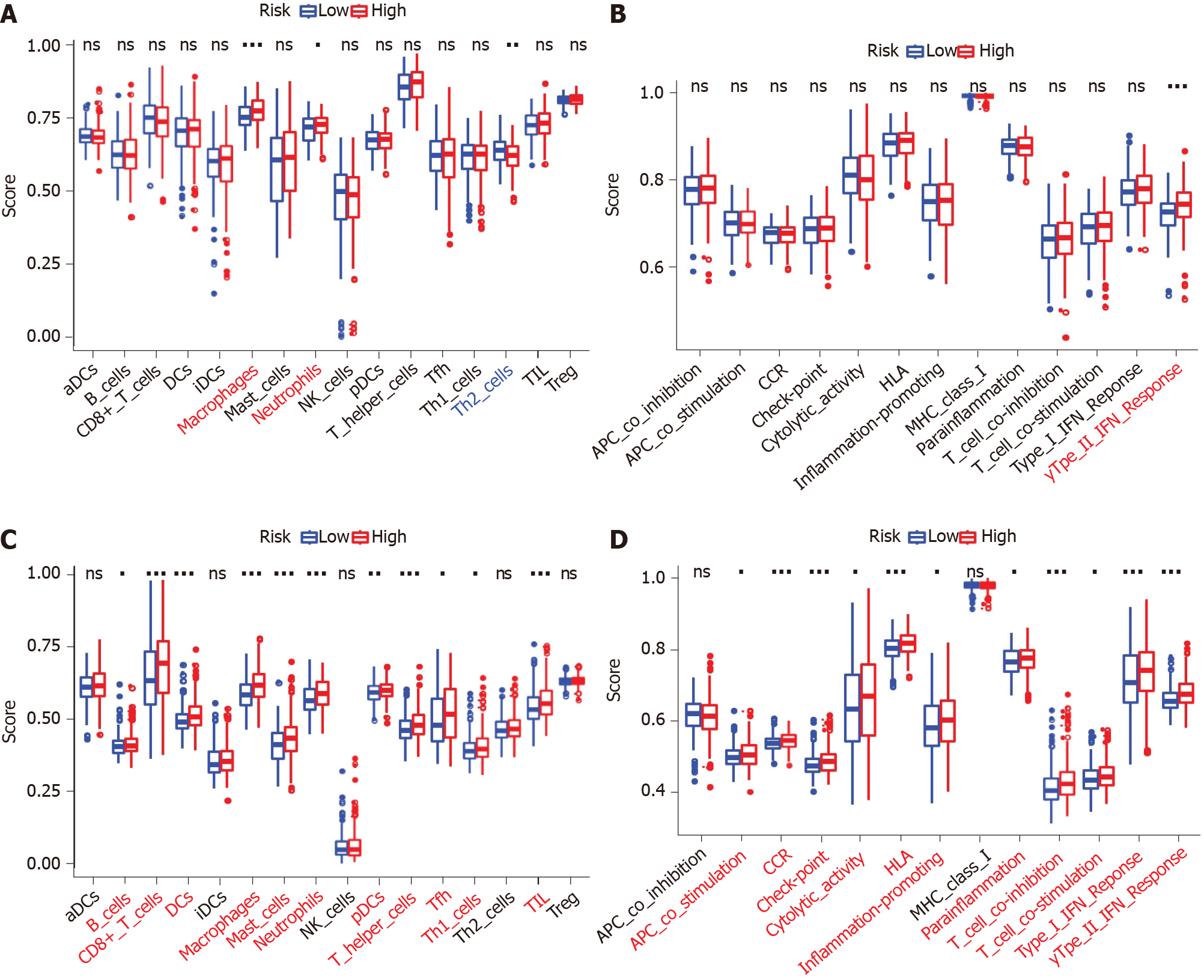
The scores of macrophages, neutrophils, and Th2 cells were considerably different between the high- and low-risk groups in the TCGA-COAD dataset. Macrophages and neutrophils scored higher in the high-risk group, while Th2 cells scored higher in the low-risk group (P < 0.05, Figure 8A). The Type II interferon (IFN) Response score was significantly higher in the high-risk group than the low-risk group (adjusted P < 0.05, Figure 8B). Comparisons in the GSE39582 cohort showed that the scores of B cells, CD8+ T cells, dendritic cells, macrophages, mast cells, neutrophils, plasmacytoid dendritic cells, T helper cells, Thf cells, Th1 cells, and tumor infiltrating lymphocytes were significantly higher in the high-risk group than the low-risk group (adjusted P < 0.05, Figure 8C). The scores of antigen presenting cell coinhibition, CC chemokine receptor, checkpoint, cytolytic activity, human leukocyte antigen, inflammation promotion, parainflammation, T cell coinhibition, T cell costimulation, type I IFN response, and type II IFN response were significantly different between the two risk groups (P < 0.05, Figure 8D). In particular, the scores of macrophages, neutrophils, and type II IFN response were significantly different between the two risk groups in both the TCGA-COAD and GSE39582 cohorts.
The GEPIA2 analysis indicated that the expression levels of AKR1C1, ALOX12, and CRYAB in the tissues of COAD were lower than those in the matching normal tissues (Figure 9A-C). In contrast, the expression levels of AKR1C3 and FDFT1 in the tumor tissues of COAD were higher than those in the corresponding normal tissues (Figure 9D-F).
The immunohistochemistry results of the Human Protein Atlas database were used to explore the expression of the five hub FRGs in COAD, and the results showed that AKR1C1, ALOX12, and CRYAB were highly expressed in normal tissues compared with matching tumor tissues (Figure 9G-L). Furthermore, the protein expression of AKR1C3 was not significantly different between normal and COAD tissues (Figure 9M and N). FDFT1 was highly expressed in both normal and COAD tissues (Figure 9O and P).
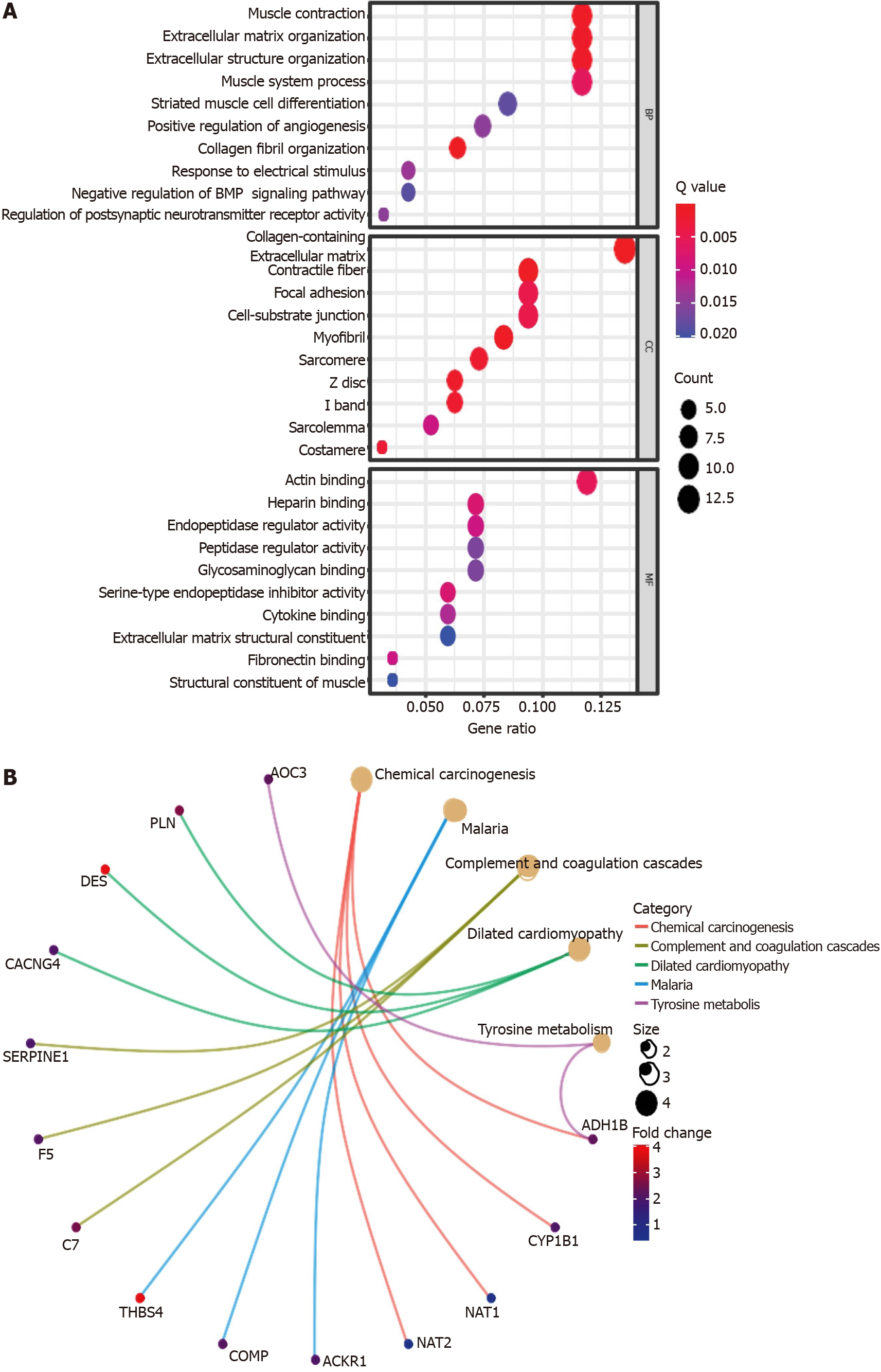
GO enrichment and KEGG pathway analyses were performed to identify the biological functions and pathways related to the risk score and DEFRGs between the high-risk and low-risk groups. The GO enrichment analysis showed that these genes might be involved in the regulation of several essential biological processes, including muscle contraction, extracellular matrix organization, extracellular structure organization, muscle system process, and regulation of vasculature development. In addition, these genes also participated in the regulation of several molecular functions, such as actin binding, endopeptidase regulator activity, and peptidase regulator activity (Figure 10A). The KEGG pathway analysis indicated that the DEFRGs were mainly involved in the regulation of chemical carcinogenesis and complement and coagulation cascade pathways (Figure 10B).
Ferroptosis is a new type of cell death with unique capabilities and identifies functions related to cancers[19]. Recent research shows that ferroptosis has been consistently accepted as a recognized function to eliminate malignant cells, thereby demonstrating its key role in inhibiting tumorigenesis by eliminating cells from the environment that lack key nutrients[34]. More clues about the role of ferroptosis can be obtained from the latest studies of TP53. Xie et al[35] found that CRC was resistant to ferroptosis, leading to suppression of dipeptidyl-peptidase-4 activity via TP53 as a transcription-independent method. TP53 loss increases the dipeptidyl-peptidase-4 concentration at the plasma membrane and thus enhances dipeptidyl-peptidase-4-dependent lipid peroxidation, ultimately leading to ferroptotic cell death. In the current research, we comprehensively investigated the expression of 60 FRGs in COAD tumor tissues and their relationship to OS. A new prognostic signature based on five FRGs was constructed and validated in an outside dataset.
Although a few previous studies[36-38] have demonstrated that certain genes may be associated with drug-induced ferroptosis in COAD, the association with the OS of COAD patients is still largely unknown. Impressively, most of the FRGs (85%) were differentially expressed between corresponding adjacent non-neoplastic tissues and COAD tissues. These results demonstrated the underlying role of ferroptosis in COAD and the possibility of constructing a prognostic signature based on these FRGs.
First, we attempted to construct a prognostic model for COAD patients based on five FRGs (AKR1C1, AKR1C3, ALOX12, CRYAB, and FDFT1) screened by a least absolute shrinkage and selection operator Cox regression analysis and identified the risk score. The prognostic signature is precise and has an accurate predictive advantage. We found that the risk score was an independent prognostic element. AKR1C1 (aldo-keto reductase family 1 member C1) encodes a member of the aldo/keto reductase superfamily. Concurrent STK11/KEAP1 mutations led to considerable upregulation of ferroptosis-protective genes (AKR1C1/2/3) and resistance to pharmacologically induced ferroptosis in lung cancer. In the pathway analysis, we found that AKR1C1 mainly participated in the regulation of EMT. Previous research has shown that aldo-keto reductases protect metastatic melanoma from estrogen receptor stress-independent ferroptosis due to the EMT-related gene expression reprogramming process[39].
ALOX12, an arachidonate 12-lipoxygenase-encoding enzyme, acts on diverse polyunsaturated fatty acid substrates to produce bioactive lipid mediators containing lipoxins and eicosanoids. Loss of one ALOX12 allele is sufficient to expedite tumorigenesis in Eμ-Myc lymphoma models. ALOX12 is not necessary for ferroptosis induced by GPX4 or erastin inhibitors, whereas ACSL4 is necessary for ferroptosis upon GPX4 inhibition but not necessary for TP53-mediated ferroptosis[40]. Inhibition of ALOX12 in SW620 cells or blood endothelial cells attenuated circular chemorepellent-induced defects[41]. In addition, genetic variation in ALOX12 could influence the risk of rectal cancer and an association may occur between nonsteroidal anti-inflammatory drug use and variants in ALOX12 on CRC risk[42].
CRYAB is a member of the small heat shock protein family and interacts with TP53 to isolate its translocation to mitochondria, thereby indirectly constraining its proapoptotic influence on apoptotic Bcl-2 molecules[43]. Previous reports indicated that CRC patients with high expression of CRYAB and positive distant metastasis showed significantly reduced OS. High CRYAB expression could promote tumor cell proliferation, invasion, and metastasis of CRC by EMT[44,45]. These results are consistent with our study, in which CRYAB, as a high-risk gene, is mainly associated with the regulation of EMT activity and higher expression of CRYAB is related to poor OS in COAD.
FDFT1 encodes a membrane-associated enzyme located at a branch point in the mevalonate pathway. Weng et al[46] reported that the downregulation of FDFT1 is related to malignant progression and poor prognosis in CRC and that FDFT1 acts as an essential cancer suppressor in CRC, which is consistent with our result showing that high FDFT1 expression is associated with better OS in COAD.
In summary, three of the genes (AKR1C1, AKR1C3, and ALOX12) in the prognostic signature have been clarified to protect cells from ferroptosis, whereas the relationship between the two surplus genes (CRYAB and FDFT1) and ferroptosis has not been reported. Whether these genes play a role in the prognosis of COAD patients by affecting the course of ferroptosis remains to be clarified because few associated studies on these genes (except for AKR1C1 and AKR1C3) have been reported.
Whether the expression level of FRGs can be applied as a prognostic marker is an essential subject for study. Our COAD prognostic signature based on five FRGs was discovered to be valuable, and the scores of 1-, 3-, and 5-year risk and the AUC value of the ROC curve were consistent with previous studies[47-50]. The prognoses were divided into high-risk and low-risk groups, and the five FRGs we selected may be ideal prognostic markers. In the high-risk group, the 5-year OS was approximately 43% and 85% in the low-risk group, which is consistent with previous research[51]. In addition, we constructed a nomogram to predict the clinical prognosis of individual COAD patients. A nomogram is a reliable and steady tool to quantify the risk of individual patients by integrating and describing risk elements, and this tool has been applied for tumor prognosis, including CRC[52,53]. Nomograms produce a graphical statistical forecasting model that can assign scores to every element. Risk scores based on genetic markers can be included in a predictive nomogram model to predict clinical prognosis. A combination of metabolism-related gene markers and a prognostic element has better prognostic value than an individual marker for CRC[52]. We were able to show that a nomogram, including the five FRGs signature, could better predict 1-, 3-, and 5-year survival possibility of COAD patients. These results showed that the five FRG prognostic model is of value in determining the treatment plan of COAD patients.
In recent years, the mechanisms underlying tumor susceptibility to ferroptosis have been a hot area of study; however, the potential regulatory role between tumor immunity and ferroptosis has not been clarified. Macrophages and neutrophils scored higher in the high-risk group, while Th2 cells scored higher in the low-risk group. Previous studies have indicated that tumor-associated macrophages correlate with shorter survival in colon cancer[54] because of their role in immune invasion. Neutrophils also promote the spread of tumor cells by using neutrophilic extracellular traps to capture circulating CRC cells and promote their distant migration[55]. Epithelial notch signaling recruits neutrophils to drive colon cancer metastasis[56]. In addition, higher risk scores related to impaired antitumor immunity included the activity of the type II IFN response. In addition, the type II IFN response is involved in the recurrence of lower grade glioma[57]. Therefore, weakened antitumor immunity in high-risk patients may be one of the reasons for their poor prognosis.
Based on the differentially expressed FRGs between the high- and low-risk groups, we performed a GO analysis and surprisingly found that several EMT-associated biological processes and pathways were enriched. Therefore, a rational assumption is that ferroptosis is closely related to EMT. Interestingly, there was a significant difference in the content of the antigen presentation process between the high-risk and low-risk groups in this research.
Although a prognostic model based on the five FRGs was constructed and the risk score was an independent prognostic marker for OS in COAD, our research still has some shortcomings. First, this research mainly focused on performing a bioinformatic analysis of retrospective data from public databases. In the future, more prospective real-world data are needed to confirm the clinical utility of the findings. Second, although FRG expression was discovered to be associated with the tumor immune cell infiltration level and patient survival, we were unable to identify whether FRGs affect patient survival through immune cell infiltration. Future prospective studies should focus on the expression of these FRGs and infiltration of immune cells in COAD, which may help provide more mechanistic insights associated with this problem.
In summary, our research defined a new prognostic signature with five FRGs in COAD. The FRG signature proved to be independently related to OS in both the training and validation datasets, thus providing insights into prognostic predictions for COAD. The potential mechanisms between FRGs and tumor immunity in COAD are still poorly understood and need to be further investigated. These results provided a comprehensive perspective for further studies on the role of hub FRGs in the pathogenesis of COAD and identified underlying molecular markers for the diagnosis and treatment of COAD.
Colon adenocarcinoma (COAD) is one of the most common and fatal malignant tumors, which increases the difficulty of prognostic predictions. Ferroptosis is a recently identified programmed cell death process that has the characteristics of iron-dependent lipid peroxide accumulation. However, the predictive value of ferroptosis-related genes (FRGs) for COAD still needs to be further clarified. Therefore, the screening of novel therapeutic targets or prognostic biomarkers based on FRGs could be an essential work for COAD therapy, which could contribute to the improvement of treatment schemes.
This article aims to identify critical FRGs and construct a COAD patient prognostic signature for clinical utilization.
This research addressed the question of the novel FRG gene biomarkers and potential mechanism involved in the development of COAD.
The Cancer Genome Atlas database (TCGA) and Gene Expression Omnibus databases were the data sources for mRNA expression and corresponding COAD patient clinical information. Differentially expressed FRGs were recognized using R and Perl software. We constructed a multi-FRG signature of the TCGA-COAD cohort by performing a univariate Cox regression and least absolute shrinkage and selection operator Cox regression analysis. COAD patients from the Gene Expression Omnibus cohort were utilized for verification.
Most of the FRGs (85%) were differentially expressed between the corresponding adjacent normal tissues and cancer tissues in the TCGA-COAD cohort. Seven FRGs were related to overall survival (OS) in the univariate Cox analysis (all P < 0.05). A model with five FRGs (AKR1C1, AKR1C3, ALOX12, CRYAB, and FDFT1) was con
Our research constructed a new prognostic signature with five FRGs in COAD. The FRG signature proved to be independently related to OS in both the training and validation datasets, thus providing insights into prognostic predictions for COAD. Targeting ferroptosis may be a treatment option for COAD.
In this study, we identified five FRGs signatures in COAD, and this signature proved to be independently related to OS, suggesting that the five FRG signatures might be novel therapeutic targets for COAD. In our subsequent work, the potential mechanisms between FRGs and tumor immunity in COAD will be investigated, which might contribute to the improvement of treatment schemes.
We acknowledge the Cancer Genome Atlas and Gene Expression Omnibus database for providing their platforms and contributors for uploading their meaningful datasets. The authors would like to express their gratitude to AJE (www.aje.com) for the expert linguistic services provided. In addition, Yan-Dong Miao would like to give special thanks to Wu-Xia Quan for her patience, care, and support over the past years.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Abdellateif MS S-Editor: Wang LL L-Editor: Filipodia P-Editor: Liu JH
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64104] [Article Influence: 16026.0] [Reference Citation Analysis (174)] |
| 2. | Murphy N, Jenab M, Gunter MJ. Adiposity and gastrointestinal cancers: epidemiology, mechanisms and future directions. Nat Rev Gastroenterol Hepatol. 2018;15:659-670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 127] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 3. | Aune D, Chan DS, Lau R, Vieira R, Greenwood DC, Kampman E, Norat T. Dietary fibre, whole grains, and risk of colorectal cancer: systematic review and dose-response meta-analysis of prospective studies. BMJ. 2011;343:d6617. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 830] [Cited by in RCA: 720] [Article Influence: 51.4] [Reference Citation Analysis (0)] |
| 4. | Magalhães B, Peleteiro B, Lunet N. Dietary patterns and colorectal cancer: systematic review and meta-analysis. Eur J Cancer Prev. 2012;21:15-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 168] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 5. | O'Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol. 2016;13:691-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 521] [Cited by in RCA: 767] [Article Influence: 85.2] [Reference Citation Analysis (0)] |
| 6. | Sharma R. An examination of colorectal cancer burden by socioeconomic status: evidence from GLOBOCAN 2018. EPMA J. 2020;11:95-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 62] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 7. | Lafitte M, Sirvent A, Roche S. Collagen Kinase Receptors as Potential Therapeutic Targets in Metastatic Colon Cancer. Front Oncol. 2020;10:125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 8. | Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11:685-696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 844] [Cited by in RCA: 781] [Article Influence: 52.1] [Reference Citation Analysis (0)] |
| 9. | Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V; Cancer Genome Atlas Research Network, Hu H. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell. 2018;173:400-416.e11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2534] [Cited by in RCA: 2329] [Article Influence: 332.7] [Reference Citation Analysis (0)] |
| 10. | Dixon SJ. Ferroptosis: bug or feature? Immunol Rev. 2017;277:150-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 232] [Article Influence: 33.1] [Reference Citation Analysis (0)] |
| 11. | Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, Morrison B 3rd, Stockwell BR. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060-1072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4711] [Cited by in RCA: 11531] [Article Influence: 887.0] [Reference Citation Analysis (1)] |
| 12. | Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascón S, Hatzios SK, Kagan VE, Noel K, Jiang X, Linkermann A, Murphy ME, Overholtzer M, Oyagi A, Pagnussat GC, Park J, Ran Q, Rosenfeld CS, Salnikow K, Tang D, Torti FM, Torti SV, Toyokuni S, Woerpel KA, Zhang DD. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell. 2017;171:273-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4608] [Cited by in RCA: 4901] [Article Influence: 612.6] [Reference Citation Analysis (0)] |
| 13. | Yagoda N, von Rechenberg M, Zaganjor E, Bauer AJ, Yang WS, Fridman DJ, Wolpaw AJ, Smukste I, Peltier JM, Boniface JJ, Smith R, Lessnick SL, Sahasrabudhe S, Stockwell BR. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447:864-868. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1311] [Cited by in RCA: 1224] [Article Influence: 68.0] [Reference Citation Analysis (0)] |
| 14. | Cao JY, Dixon SJ. Mechanisms of ferroptosis. Cell Mol Life Sci. 2016;73:2195-2209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1001] [Cited by in RCA: 1112] [Article Influence: 123.6] [Reference Citation Analysis (0)] |
| 15. | Xie Y, Hou W, Song X, Yu Y, Huang J, Sun X, Kang R, Tang D. Ferroptosis: process and function. Cell Death Differ. 2016;23:369-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1553] [Cited by in RCA: 2593] [Article Influence: 288.1] [Reference Citation Analysis (0)] |
| 16. | Xu T, Ding W, Ji X, Ao X, Liu Y, Yu W, Wang J. Molecular mechanisms of ferroptosis and its role in cancer therapy. J Cell Mol Med. 2019;23:4900-4912. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 185] [Cited by in RCA: 454] [Article Influence: 75.7] [Reference Citation Analysis (0)] |
| 17. | Schott C, Graab U, Cuvelier N, Hahn H, Fulda S. Oncogenic RAS Mutants Confer Resistance of RMS13 Rhabdomyosarcoma Cells to Oxidative Stress-Induced Ferroptotic Cell Death. Front Oncol. 2015;5:131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 75] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 18. | Shaw AT, Winslow MM, Magendantz M, Ouyang C, Dowdle J, Subramanian A, Lewis TA, Maglathin RL, Tolliday N, Jacks T. Selective killing of K-ras mutant cancer cells by small molecule inducers of oxidative stress. Proc Natl Acad Sci U S A. 2011;108:8773-8778. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 207] [Cited by in RCA: 199] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 19. | Mou Y, Wang J, Wu J, He D, Zhang C, Duan C, Li B. Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J Hematol Oncol. 2019;12:34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 392] [Cited by in RCA: 1227] [Article Influence: 204.5] [Reference Citation Analysis (0)] |
| 20. | Hassannia B, Vandenabeele P, Vanden Berghe T. Targeting Ferroptosis to Iron Out Cancer. Cancer Cell. 2019;35:830-849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 900] [Cited by in RCA: 1610] [Article Influence: 268.3] [Reference Citation Analysis (0)] |
| 21. | Liang C, Zhang X, Yang M, Dong X. Recent Progress in Ferroptosis Inducers for Cancer Therapy. Adv Mater. 2019;31:e1904197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 687] [Cited by in RCA: 1042] [Article Influence: 173.7] [Reference Citation Analysis (0)] |
| 22. | Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H, Baer R, Gu W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1173] [Cited by in RCA: 2376] [Article Influence: 237.6] [Reference Citation Analysis (0)] |
| 23. | Galluzzi L, Bravo-San Pedro JM, Kroemer G. Ferroptosis in p53-dependent oncosuppression and organismal homeostasis. Cell Death Differ. 2015;22:1237-1238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 24. | Liu DS, Duong CP, Haupt S, Montgomery KG, House CM, Azar WJ, Pearson HB, Fisher OM, Read M, Guerra GR, Haupt Y, Cullinane C, Wiman KG, Abrahmsen L, Phillips WA, Clemons NJ. Inhibiting the system xC-/glutathione axis selectively targets cancers with mutant-p53 accumulation. Nat Commun. 2017;8:14844. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 244] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 25. | Hasegawa M, Takahashi H, Rajabi H, Alam M, Suzuki Y, Yin L, Tagde A, Maeda T, Hiraki M, Sukhatme VP, Kufe D. Functional interactions of the cystine/glutamate antiporter, CD44v and MUC1-C oncoprotein in triple-negative breast cancer cells. Oncotarget. 2016;7:11756-11769. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 156] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 26. | Alvarez SW, Sviderskiy VO, Terzi EM, Papagiannakopoulos T, Moreira AL, Adams S, Sabatini DM, Birsoy K, Possemato R. NFS1 undergoes positive selection in lung tumours and protects cells from ferroptosis. Nature. 2017;551:639-643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 517] [Cited by in RCA: 560] [Article Influence: 70.0] [Reference Citation Analysis (0)] |
| 27. | Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16184] [Cited by in RCA: 25417] [Article Influence: 2541.7] [Reference Citation Analysis (0)] |
| 28. | Doll S, Freitas FP, Shah R, Aldrovandi M, da Silva MC, Ingold I, Goya Grocin A, Xavier da Silva TN, Panzilius E, Scheel CH, Mourão A, Buday K, Sato M, Wanninger J, Vignane T, Mohana V, Rehberg M, Flatley A, Schepers A, Kurz A, White D, Sauer M, Sattler M, Tate EW, Schmitz W, Schulze A, O'Donnell V, Proneth B, Popowicz GM, Pratt DA, Angeli JPF, Conrad M. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575:693-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1492] [Cited by in RCA: 2100] [Article Influence: 350.0] [Reference Citation Analysis (0)] |
| 29. | Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 2011;12:35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1253] [Cited by in RCA: 1819] [Article Influence: 129.9] [Reference Citation Analysis (0)] |
| 30. | Frost HR, Amos CI. Gene set selection via LASSO penalized regression (SLPR). Nucleic Acids Res. 2017;45:e114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 31. | Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556-W560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1991] [Cited by in RCA: 3029] [Article Influence: 504.8] [Reference Citation Analysis (0)] |
| 32. | Zhang Z, Wang L, Wang Q, Zhang M, Wang B, Jiang K, Ye Y, Wang S, Shen Z. Molecular Characterization and Clinical Relevance of RNA Binding Proteins in Colorectal Cancer. Front Genet. 2020;11:580149. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 33. | Liu J, Huang X, Yang W, Li C, Li Z, Zhang C, Chen S, Wu G, Xie W, Wei C, Tian C, Huang L, Jeen F, Mo X, Tang W. Nomogram for predicting overall survival in stage II-III colorectal cancer. Cancer Med. 2020;9:2363-2371. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 34. | Fearnhead HO, Vandenabeele P, Vanden Berghe T. How do we fit ferroptosis in the family of regulated cell death? Cell Death Differ. 2017;24:1991-1998. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 118] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 35. | Xie Y, Zhu S, Song X, Sun X, Fan Y, Liu J, Zhong M, Yuan H, Zhang L, Billiar TR, Lotze MT, Zeh HJ 3rd, Kang R, Kroemer G, Tang D. The Tumor Suppressor p53 Limits Ferroptosis by Blocking DPP4 Activity. Cell Rep. 2017;20:1692-1704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 681] [Article Influence: 97.3] [Reference Citation Analysis (0)] |
| 36. | Malfa GA, Tomasello B, Acquaviva R, Genovese C, La Mantia A, Cammarata FP, Ragusa M, Renis M, Di Giacomo C. Betula etnensis Raf. (Betulaceae) Extract Induced HO-1 Expression and Ferroptosis Cell Death in Human Colon Cancer Cells. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 79] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 37. | Li Y, Chen W, Qi Y, Wang S, Li L, Li W, Xie T, Zhu H, Tang Z, Zhou M. H2 S-Scavenged and Activated Iron Oxide-Hydroxide Nanospindles for MRI-Guided Photothermal Therapy and Ferroptosis in Colon Cancer. Small. 2020;16:e2001356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 38. | Wei G, Sun J, Hou Z, Luan W, Wang S, Cui S, Cheng M, Liu Y. Novel antitumor compound optimized from natural saponin Albiziabioside A induced caspase-dependent apoptosis and ferroptosis as a p53 activator through the mitochondrial pathway. Eur J Med Chem. 2018;157:759-772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 39. | Gagliardi M, Cotella D, Santoro C, Corà D, Barlev NA, Piacentini M, Corazzari M. Aldo-keto reductases protect metastatic melanoma from ER stress-independent ferroptosis. Cell Death Dis. 2019;10:902. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 120] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 40. | Chu B, Kon N, Chen D, Li T, Liu T, Jiang L, Song S, Tavana O, Gu W. ALOX12 is required for p53-mediated tumour suppression through a distinct ferroptosis pathway. Nat Cell Biol. 2019;21:579-591. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 246] [Cited by in RCA: 643] [Article Influence: 107.2] [Reference Citation Analysis (0)] |
| 41. | Holzner S, Brenner S, Atanasov AG, Senfter D, Stadler S, Nguyen CH, Fristiohady A, Milovanovic D, Huttary N, Krieger S, Bago-Horvath Z, de Wever O, Tentes I, Özmen A, Jäger W, Dolznig H, Dirsch VM, Mader RM, Krenn L, Krupitza G. Intravasation of SW620 colon cancer cell spheroids through the blood endothelial barrier is inhibited by clinical drugs and flavonoids in vitro. Food Chem Toxicol. 2018;111:114-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 42. | Resler AJ, Makar KW, Heath L, Whitton J, Potter JD, Poole EM, Habermann N, Scherer D, Duggan D, Wang H, Lindor NM, Passarelli MN, Baron JA, Newcomb PA, Le Marchand L, Ulrich CM. Genetic variation in prostaglandin synthesis and related pathways, NSAID use and colorectal cancer risk in the Colon Cancer Family Registry. Carcinogenesis. 2014;35:2121-2126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 43. | Liu S, Yan B, Lai W, Chen L, Xiao D, Xi S, Jiang Y, Dong X, An J, Chen X, Cao Y, Tao Y. As a novel p53 direct target, bidirectional gene HspB2/αB-crystallin regulates the ROS level and Warburg effect. Biochim Biophys Acta. 2014;1839:592-603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 44. | Li Q, Wang Y, Lai Y, Xu P, Yang Z. HspB5 correlates with poor prognosis in colorectal cancer and prompts epithelial-mesenchymal transition through ERK signaling. PLoS One. 2017;12:e0182588. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 45. | Shi C, Yang X, Bu X, Hou N, Chen P. Alpha B-crystallin promotes the invasion and metastasis of colorectal cancer via epithelial-mesenchymal transition. Biochem Biophys Res Commun. 2017;489:369-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 46. | Weng ML, Chen WK, Chen XY, Lu H, Sun ZR, Yu Q, Sun PF, Xu YJ, Zhu MM, Jiang N, Zhang J, Zhang JP, Song YL, Ma D, Zhang XP, Miao CH. Fasting inhibits aerobic glycolysis and proliferation in colorectal cancer via the Fdft1-mediated AKT/mTOR/HIF1α pathway suppression. Nat Commun. 2020;11:1869. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 171] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 47. | Liu Y, Guo F, Guo W, Wang Y, Song W, Fu T. Ferroptosis-related genes are potential prognostic molecular markers for patients with colorectal cancer. Clin Exp Med. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 48. | Zhu L, Yang F, Wang L, Dong L, Huang Z, Wang G, Chen G, Li Q. Identification the ferroptosis-related gene signature in patients with esophageal adenocarcinoma. Cancer Cell Int. 2021;21:124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 76] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 49. | Zhuo S, Chen Z, Yang Y, Zhang J, Tang J, Yang K. Clinical and Biological Significances of a Ferroptosis-Related Gene Signature in Glioma. Front Oncol. 2020;10:590861. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 102] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 50. | Liang JY, Wang DS, Lin HC, Chen XX, Yang H, Zheng Y, Li YH. A Novel Ferroptosis-related Gene Signature for Overall Survival Prediction in Patients with Hepatocellular Carcinoma. Int J Biol Sci. 2020;16:2430-2441. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 213] [Cited by in RCA: 411] [Article Influence: 82.2] [Reference Citation Analysis (0)] |
| 51. | Dueland S, Foss A, Solheim JM, Hagness M, Line PD. Survival following liver transplantation for liver-only colorectal metastases compared with hepatocellular carcinoma. Br J Surg. 2018;105:736-742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 57] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 52. | Miao Y, Li Q, Wang J, Quan W, Li C, Yang Y, Mi D. Prognostic implications of metabolism-associated gene signatures in colorectal cancer. PeerJ. 2020;8:e9847. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 53. | Miao YD, Wang JT, Yang Y, Ma XP, Mi DH. Identification of prognosis-associated immune genes and exploration of immune cell infiltration in colorectal cancer. Biomark Med. 2020;14:1353-1369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 54. | Rigo A, Gottardi M, Zamò A, Mauri P, Bonifacio M, Krampera M, Damiani E, Pizzolo G, Vinante F. Macrophages may promote cancer growth via a GM-CSF/HB-EGF paracrine loop that is enhanced by CXCL12. Mol Cancer. 2010;9:273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 95] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 55. | Mizuno R, Kawada K, Itatani Y, Ogawa R, Kiyasu Y, Sakai Y. The Role of Tumor-Associated Neutrophils in Colorectal Cancer. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 218] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 56. | Ruland J. Colon Cancer: Epithelial Notch Signaling Recruits Neutrophils to Drive Metastasis. Cancer Cell. 2019;36:213-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 57. | Huang R, Li Z, Zhu X, Yan P, Song D, Yin H, Hu P, Lin R, Wu S, Meng T, Zhang J, Huang Z. Collagen Type III Alpha 1 chain regulated by GATA-Binding Protein 6 affects Type II IFN response and propanoate metabolism in the recurrence of lower grade glioma. J Cell Mol Med. 2020;24:10803-10815. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |