Published online Oct 6, 2021. doi: 10.12998/wjcc.v9.i28.8563
Peer-review started: May 27, 2021
First decision: June 15, 2021
Revised: June 28, 2021
Accepted: August 2, 2021
Article in press: August 2, 2021
Published online: October 6, 2021
Processing time: 124 Days and 0.9 Hours
Two or multiple primary malignant neoplasms (MPMNs) rarely occur in the same patient. It has been reported that MPMNs are easily misdiagnosed as the recurrence or metastasis of malignancies in clinical practice, affecting the choice of treatment for the patients, thereby resulting in the delay of optimal diagnosis. Next generation sequencing (NGS) can be used to distinguish between multiple primary lung cancers and intrapulmonary metastasis, and may distinguish the origin of tumours in different sites of the body.
We report the case of 66-year-old woman who suffered from different malignant neoplasms in the rectum and esophageal and gastrointestinal tract. The first neoplasm rectal adenocarcinoma was diagnosed and removed in 2016. The second and third lesions were diagnosed with esophageal squamous-cell carcinoma (ESCC) and gastrointestinal stromal tumour (GIST), respectively, in 2019. Next-generation whole exome sequencing was performed on the tissue specimens of rectal carcinoma, esophageal cancer, GIST, and white blood cells to investigate the relationship between malignancies at different timeframe and determine whether the ESCC and GIST evolved from the rectal adenocarcinoma. Mutations including v-Ki-ras2-Kirsten rat sarcoma viral oncogene homolog, adenomatosis polyposis coli, and mothers against decapentaplegic homolog 4 were detected in rectal adenocarcinoma sample, mast/stem cell growth factor receptor was detected in GIST tissue, and lysine methyltransferase 2D was detected in ESCC specimen. Overall, ESCC and GIST were not genetically evolved from rectal adenocarcinoma, and this patient did not have a trunk driven clone.
NGS is an effective tool to study clonal evolution of tumours and distinguish between MPMNs and intrapulmonary metastasis.
Core Tip: We report a case of multiple primary malignant neoplasms. In this case, next-generation whole exome sequencing confirmed that esophageal squamous-cell carcinoma and gastrointestinal stromal tumour were not genetically evolved from rectal adenocarcinoma that was removed 3 years ago but occurred independently. The results of next-generation whole exome sequencing of the tumour tissues confirmed the specimens’ evolution relationship.
- Citation: Ouyang WW, Li QY, Yang WG, Su SF, Wu LJ, Yang Y, Lu B. Genetic characteristics of a patient with multiple primary cancers: A case report. World J Clin Cases 2021; 9(28): 8563-8570
- URL: https://www.wjgnet.com/2307-8960/full/v9/i28/8563.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i28.8563
Multiple primary malignant neoplasms (MPMNs), also known as multiple primary cancers, refer to the simultaneous occurrence of two or more primary malignancies in one or more organs and tissues of the same patient. To our knowledge, MPMNs are easily misdiagnosed as the recurrence or metastasis of malignant tumours in clinical settings, which affects the choice of treatment for the patients and further delays the optimal diagnosis. As diagnostic techniques and treatments develop, the life of patients with MPMNs is increasing.
Colorectal cancer is a malignant tumour with high morbidity and mortality in the world[1]. In recent decades, the incidence rate of colorectal cancer remains high in developing countries[2]. Esophageal cancer is the fourth most common cause of cancer-related death worldwide, ranking sixth in incidence[3]. There are two main subtypes of this disease, including esophageal squamous-cell cancer (ESCC) and esophageal adenocarcinoma. The incidence of these two types varies by geographic areas, with approximately over half of the esophageal cancer cases detected in China, especially ESCC[4]. MPMNs may occur simultaneously or successively in different sites in an individual with colorectal cancer, accounting for about 4.5%[5-8]. Dual primary tumours are most common in MPMNs, while triple primary tumours are rare[9]. The second most frequently reported cancers since 2011 are renal, uterine, cervical, and lung cancers[9]. Zhou et al[10] reported a case of coexistence of gastrointestinal stromal tumour (GIST) and esophageal and gastric cardia carcinomas in 2013. In the following year, Suzuki et al[11] described a 76-year-old man with a large GIST along with advanced adenocarcinoma in the rectum complicated with prostate carcinoma in 2014. Fan et al[12] also presented a rare case of synchronous occurrence of hereditary gastric adenocarcinoma, gastrointestinal stromal tumour, small cell esophageal carcinoma, and squamous carcinoma in situ in 2017. However, colorectal cancer, esophageal cancer, and GIST have rarely been reported in the same patient and there is no analysis of gene mutations in these rare cases.
In the present study, we report a patient who developed ESCC and GIST in 2019, with a history of surgery for rectal adenocarcinoma 3 years ago. Whole exome sequencing (WES) was used to investigate the patient's genetic characteristics, as well as the relevance of different tumours in this patient.
In February 2019, a 66-year-old woman was admitted to Guizhou Cancer Hospital with swallowing and choking difficulties for more than 5 mo, and her symptoms worsened in the past 1 mo.
In March 2016, the patient was hospitalized at a local hospital due to anal distention and changes in bowel habits. On admission, the colonoscopy and biopsy examinations revealed a rectal carcinoma with high-grade epithelioid neoplasia. Meanwhile, digital rectal examination showed the presence of an ulcerous neoplasm that was palpable 4-5 cm from the lateral wall of the rectum and invasive for about half a week. Besides, the neoplasm was about 3 cm × 3 cm in size, hard in texture, and inactive. In the same month, the case received laparoscopic radical resection of rectal carcinoma and rectal sigmoid anastomosis, whereas preoperative neoadjuvant therapy was not performed at the patient's request. Finally, the patient was diagnosed with stage IIIB (T3N1cM0) mucinous adenocarcinoma of the rectum with a tumour size of 4.5 cm × 3.5 cm. After discharge, the patient did not return for further treatments such as chemotherapy.
The patient had no family history of similar lesions.
Karnofsky Performance Status Scale (KPS) has been widely adopted to quantify the functional status of cancer patients. In this study, the KPS of the case was 80.
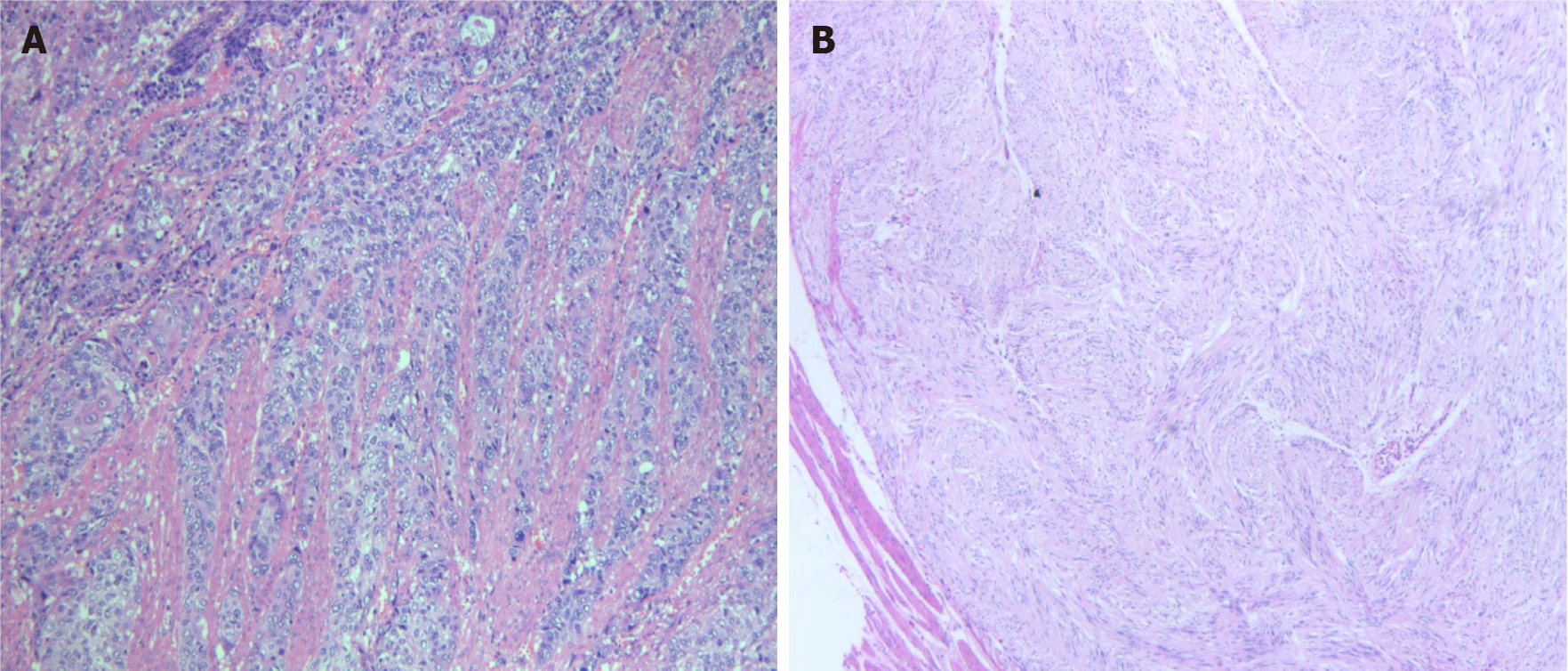
Postoperative pathology exhibited moderately to highly differentiated ESCC (4 cm × 2 cm) (Figure 1A), with tumour invasion of the whole esophageal wall, vascular invasion (+), and nerve invasion (+/-), without the two cutting edges of the samples. The results of hematoxylin-eosin stained tissue morphology and immunohistochemical markers revealed four spindle cell nodules in the lesser curvature and cardia, including one leiomyoma and three GISTs (Figure 1B) (nuclear fission: 0-1/5 mm2, tumour size: 1 cm × 0.8 cm × 0.2 cm, and extreme low risk for invasion). Immunohistochemical staining showed CD117+, Calponin-, MSA-, Des-, SMA-, CK-, Vim+, Ki67+ (2%), CD34+ and Dog+ in GIST cells, and MSA+, Des+, SMA+ and Ki67+ (2%) in smooth muscle cells.
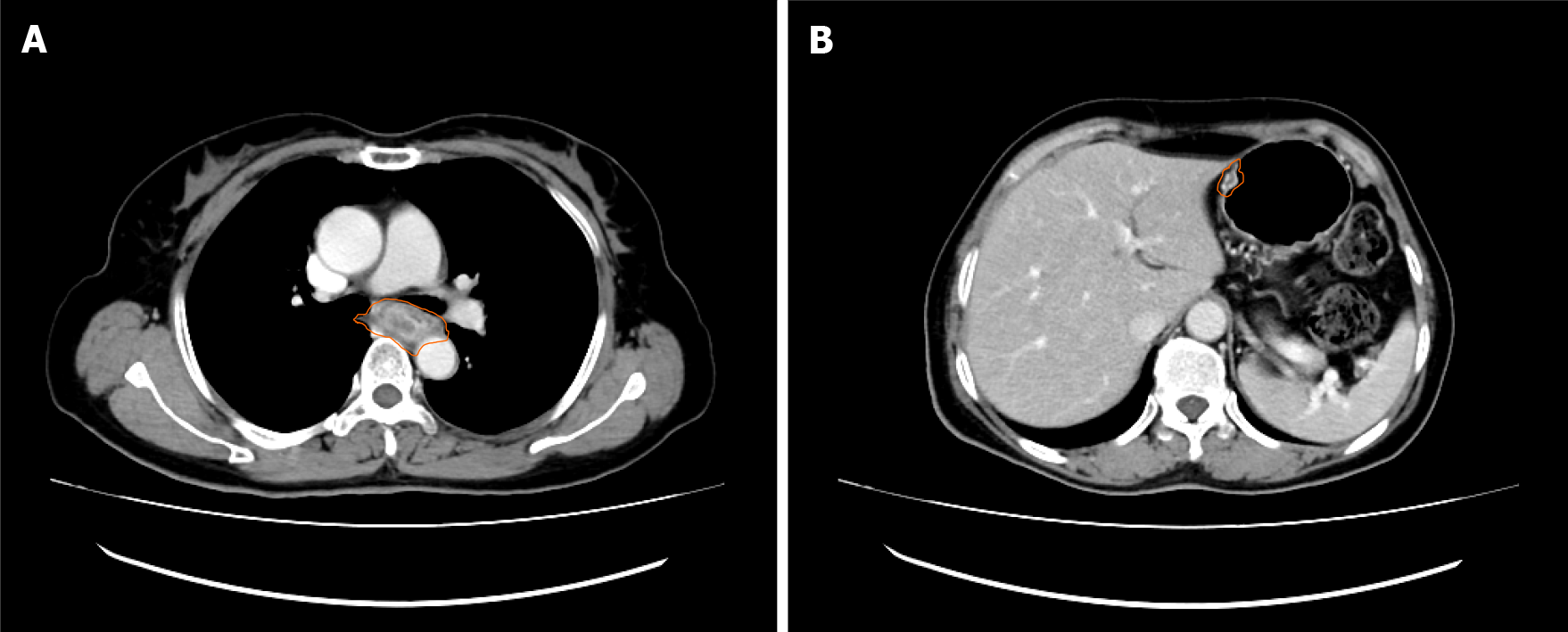
Enhanced computed tomography (CT) scans of the chest showed a thickened esophageal wall and enlarged peripheral lymph nodes, suggesting esophageal cancer with peripheral lymph node metastasis (Figure 2A). In addition, the increased density of the lower lobe of both lungs might be caused by inflammation. Subsequently, esophagoscopy examination showed esophageal neoplasia with stenosis, indicating progressive esophageal cancer. Pathological results revealed that ESCC was about 25 cm away from the incisors. The CT image of nodules of the lesser curvature of the stomach is shown in Figure 2B.
The patient was diagnosed as having postoperative stage III middle thoracic ESCC (pT4N0M0), and spindle cell nodules of the lesser curvature and cardia (1 leiomyoma and 3 GISTs) after rectal adenocarcinoma.
The patient underwent thoracoscopic radical resection of esophageal cancer, reconstruction of the digestive tract, and systematic lymph node dissection. After surgery, she received both adjuvant chemotherapy and targeted therapy (5-fluorouracil, cisplatin, and docetaxel therapy and nimotuzumab). On day 1, the patient was administered intravenously with nimotuzumab (100 mg) for more than 60 min, followed by intravenous infusion of cisplatin (75 mg/m2, 100 mg) on days 2 and 3. At the same time, she also received pump injection of fluorouracil (500 mg/m2/d, 3400 mg) on days 1 to 5. Besides, the patient was treated by antiemesis, acid suppression, gastric protection, hydration, dexamethasone allergy prevention and nutritional support for 21-18 d/cycle, followed by re-examination after two cycles of chemo
The observation indexes were non-target lesion and the occurrence of new lesion. The efficacy and the toxic and side effects were evaluated according to the RECIST standard and CTC4.0 standard, respectively. After the completion of chemotherapy and targeted therapy in August 2019, the patient was in good physical condition and routine follow-up showed no abnormalities.
To study the patient's genetic characteristics, informed consent was obtained from the patient. Whole exome sequencing (WES) was carried out on the tissue specimens of rectal carcinoma, esophageal cancer, GIST, and white blood cells. For sequencing, the genomic DNA was first sheared into 150-200 bp fragments using Covaris M220 Focused-ultrasonicator Instrument (Covaris, MA, United States). Next, the fragmented DNA libraries were constructed via NimbleGen SeqCap EZ Exome Library Kit (Roche, WI, United States) according to manufacturer’s instructions. Subsequently, the captured DNA fragments were sequenced using Illumina Novaseq 6000 sequencing system at least 50× for blood cells and 100× for tumour tissue samples (Genecast, Wuxi, Jiangsu Province, China). After filtering out low quality reads, the clean reads were aligned with the human reference genome (Hg19, NCBI Build 37.5) via Burrows-Wheeler Aligner (version 0.7.17)[13]. Picard toolkit (version 2.1.0)[14] was used to make duplicates, while Genome Analysis ToolKit (version 3.7)[15] was adopted for realignment. VarDict (version 1.5.1)[16] was introduced to single nucleotide variations calling, while compound heterozygous mutations were merged with FreeBayes (version 1.2.0)[17]. All mutations were annotated through ANNOVAR[18]. Finally, the remaining mutations were annotated with pathway information[19]. Copy number variations (CNVs) were paired via software CONTRA (version 2.0.8)[20] with copy number threshold 3 for CNV gain and 1.2 for CNV loss.
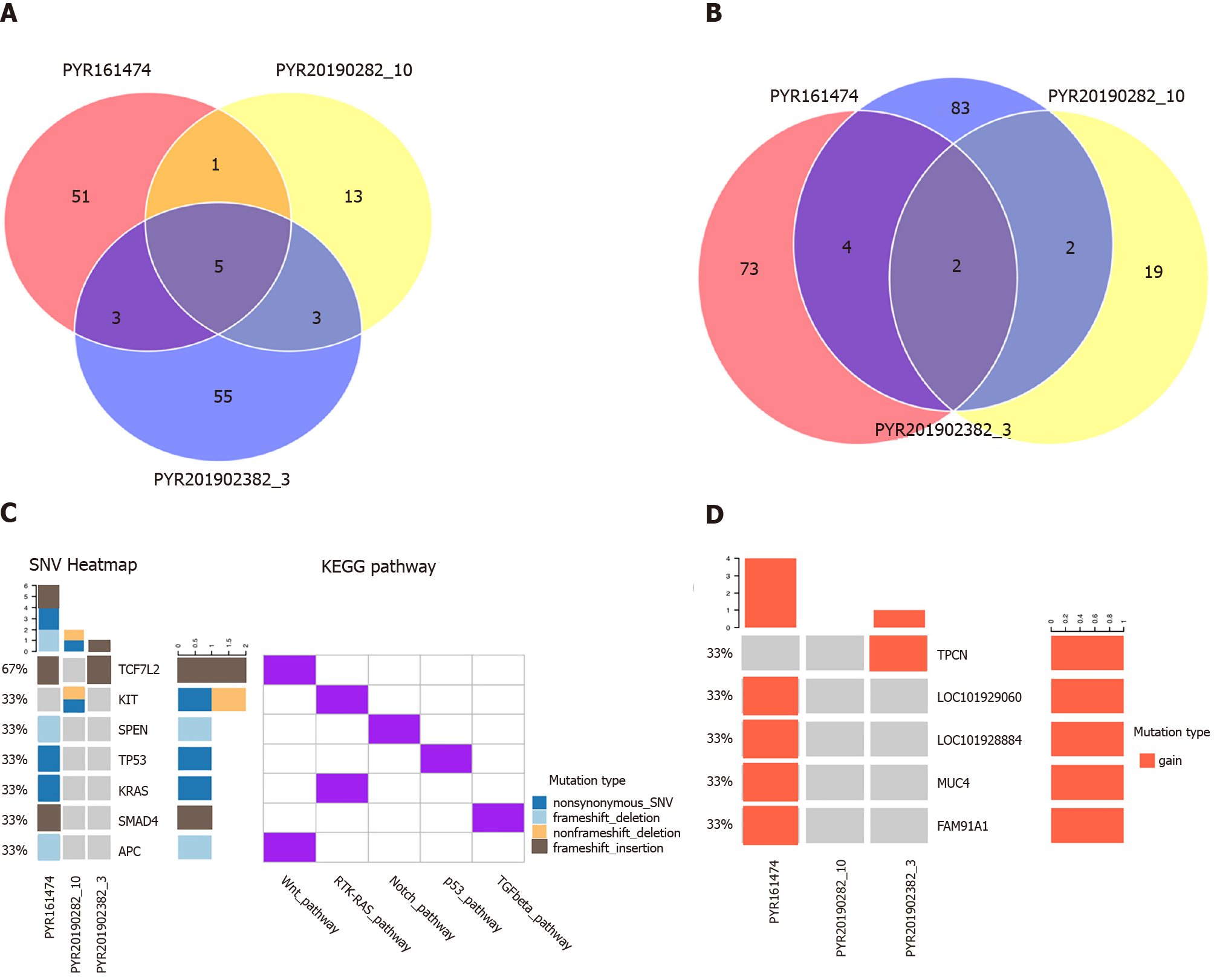
The results of genetic characteristics showed that carboxyl ester lipase, mucin-16, mucin-3A (MUC3A), mucin-4 (MUC4), and zinc finger protein 88 were common mutated genes in the three lesions (Supplementary Figure 1). There were 51 unique mutated genes in intestinal tumour tissues, 13 in gastric tumour tissues, and 55 in esophageal tumour tissues (Figure 3A), among which the common mutation sites were MUC3A p.S697N and MUC3A p.T463P. Additionally, the numbers of unique mutated gene loci in intestinal tumour tissues, gastric tumour tissues, and esophageal tumour tissues were 73, 19, and 83, respectively (Figure 3B), indicating a strong heterogeneity of the three lesions, with few mutated genes and loci in common. According to gene analysis, v-Ki-ras2-Kirsten rat sarcoma viral oncogene homolog (KRAS), adenomatosis polyposis coli (APC), and mothers against decapentaplegic homolog 4 (SMAD4) gene mutations were detected in rectal adenocarcinoma sample, but not in esophageal and gastric tumour samples (Figure 3C). Furthermore, the mast/stem cell growth factor receptor (KIT) mutation was detected in GIST tissue, but not observed in intestinal and esophageal tumour specimens (Figure 3C); lysine methyltransferase 2D (KMT2D) was noticed in ESCC specimen, but not in intestinal and GIST specimens (Supplementary Figure 1). The results of CNV revealed that the three lesions were inconsistent, and the copy numbers of MUC4, family with sequence similarity 91 member A1, LOC101928884, and LOC101929060 in the rectal adenocarcinoma and two pore segment channel 2 in the ESCC samples were significantly increased, while there was no significance between gene copy numbers in the GIST tissue (Figure 3D). There
Next generation sequencing (NGS) is considered a genetic testing that can be used to distinguish between multiple primary lung cancers and intrapulmonary metastasis. In a NGS analysis of 60 patients with multifocal tumours, researchers at Memorial Sloan-Kettering Cancer Center found that prospective histological predictions were incon
MPMNs are sometimes confused with multiple metastases from the same cancer. Genetic testing is an effective tool for studying these problems; however, the application of genetic testing to distinguish colorectal cancer, esophageal cancer, and GIST in the same patient has rarely been reported. Compared with NGS panel se
KIT p.V559D mutation was detected in GIST tissue, but not in intestinal and esophageal tumour specimens, which is located in exon 11 of KIT gene. Nearly 90% of GISTs harbour activating mutations in the KIT gene, and approximately 80% of patients with metastatic GISTs show at least some clinical response to the targeted small molecule KIT inhibitor imatinib[28]. In addition, KMT2D mutation was detected in ESCC specimen, which was not found in intestinal and GIST specimens. The CNV results revealed that the three lesions were inconsistent.
From the results of genetic testing, there was no dominant mutation clone in the rectal adenocarcinoma, GIST, and esophageal cancer lesions in this patient. The two lesions in 2019 did not evolve from the main clone of the lesion in 2016, and this result was supported by the favourable prognosis of the patient. After that, no pathogenic germ-line mutation was identified by WES and the patient also reported no family history of similar lesions, suggesting that the MPMNs were not triggered by genetic factors. Therefore, the cause of MPMNs in this case remains unclear.
In addition to studying the origin and evolution of tumours, the results of WES could also guide the medication for patients. The patient in the case with KRAS p.G13D should not be administrated with cetuximab. The KIT p.V559D mutation indicated that the patients might be treated with KIT inhibitor such as imatinib.
Due to limited data, this research only provides a reference for the identification of multiple primary tumours and metastatic tumours at the genetic level. For large-scale clinical application, more prospective studies with large sample size are needed to guide clinical decision-making, and the cost of detection and the economic cost of patients are also important factors that must be considered.
Genetic testing provides important information for the study of MPMNs. In this case, next-generation WES confirmed that ESCC and GIST did not genetically evolve from rectal adenocarcinoma resected 3 years ago, but occurred independently. Besides, the subject of this study did not have a trunk driven clone. In summary, NGS is a useful tool to study clonal evolution of tumours.
Manuscript source: Unsolicited manuscript
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C, C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Ali SM, Elpek GO, Ho CM, Li X S-Editor: Yan JP L-Editor: Wang TQ P-Editor: Wu RR
| 1. | Siegel RL, Fedewa SA, Miller KD, Goding-Sauer A, Pinheiro PS, Martinez-Tyson D, Jemal A. Cancer statistics for Hispanics/Latinos, 2015. CA Cancer J Clin. 2015;65:457-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 323] [Cited by in RCA: 351] [Article Influence: 35.1] [Reference Citation Analysis (0)] |
| 2. | Kersemaekers AM, Fleuren GJ, Kenter GG, Van den Broek LJ, Uljee SM, Hermans J, Van de Vijver MJ. Oncogene alterations in carcinomas of the uterine cervix: overexpression of the epidermal growth factor receptor is associated with poor prognosis. Clin Cancer Res. 1999;5:577-586. [PubMed] |
| 3. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23762] [Cited by in RCA: 25543] [Article Influence: 1824.5] [Reference Citation Analysis (7)] |
| 4. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11065] [Cited by in RCA: 12187] [Article Influence: 1523.4] [Reference Citation Analysis (3)] |
| 5. | Cai H, Dong RZ, Wu JH, Zhu HY, Wang YN, Shi YQ, Mo SJ. [Clinical analysis of 168 cases of multiple primary colorectal carcinoma]. Zhonghua Wai Ke Za Zhi. 2008;46:370-374. [PubMed] |
| 6. | Wang W, Zhou ZW, Wan DS, Lu ZH, Chen G, Pan ZZ, Li LR, Wu XJ, Ding PR. [Clinical analyses of 70 cases of multiple primary colorectal carcinoma]. Ai Zheng. 2008;27:505-509. [PubMed] |
| 7. | Fu J, Huang Z, Lin Y, Xiao B. [Clinical analysis of 39 cases of multiple primary colorectal carcinoma]. Nan Fang Yi Ke Da Xue Xue Bao. 2013;33:578-581. [PubMed] |
| 8. | Wu A, He S, Li J, Liu L, Liu C, Wang Q, Peng X, Zhou J, Cao PG, Cao K. Colorectal cancer in cases of multiple primary cancers: Clinical features of 59 cases and point mutation analyses. Oncol Lett. 2017;13:4720-4726. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 9. | Seegobin K, Staggs E, Khawaja R, Maharaj S, Gautam S, Smotherman C, Rana F. Pilot study on the occurrence of multiple cancers following cancer-related therapy at the University of Florida, Jacksonville (2011-2016). J Investig Med. 2018;66:1050-1054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 10. | Zhou Y, Wu XD, Shi Q, Jia J. Coexistence of gastrointestinal stromal tumor, esophageal and gastric cardia carcinomas. World J Gastroenterol. 2013;19:2005-2008. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 11] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 11. | Suzuki T, Suwa K, Hanyu K, Okamoto T, Fujita T, Yanaga K. Large gastrointestinal stromal tumor and advanced adenocarcinoma in the rectum coexistent with an incidental prostate carcinoma: A case report. Int J Surg Case Rep. 2014;5:640-642. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 12. | Fan H, Lu P, Xu L, Qin Y, Li J. Synchronous occurrence of hereditary gastric adenocarcinoma, gastrointestinal stromal tumor, and esophageal small cell and squamous carcinoma in situ: an extremely rare case report. BMC Cancer. 2017;17:720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 13. | Li H. Aligning sequence reads, clone sequences and assembly contigs with bwa-mem. 2013 Preprint. Available from: arXiv:1303.3997. |
| 14. | Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078-2079. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51763] [Cited by in RCA: 41767] [Article Influence: 2610.4] [Reference Citation Analysis (0)] |
| 15. | DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491-498. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9842] [Cited by in RCA: 8302] [Article Influence: 593.0] [Reference Citation Analysis (0)] |
| 16. | Lai Z, Markovets A, Ahdesmaki M, Chapman B, Hofmann O, McEwen R, Johnson J, Dougherty B, Barrett JC, Dry JR. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44:e108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 448] [Cited by in RCA: 625] [Article Influence: 69.4] [Reference Citation Analysis (0)] |
| 17. | Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. 2012 Preprint. Available from: arXiv:1207.3907. |
| 18. | Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7976] [Cited by in RCA: 10465] [Article Influence: 697.7] [Reference Citation Analysis (0)] |
| 19. | Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, Dimitriadoy S, Liu DL, Kantheti HS, Saghafinia S, Chakravarty D, Daian F, Gao Q, Bailey MH, Liang WW, Foltz SM, Shmulevich I, Ding L, Heins Z, Ochoa A, Gross B, Gao J, Zhang H, Kundra R, Kandoth C, Bahceci I, Dervishi L, Dogrusoz U, Zhou W, Shen H, Laird PW, Way GP, Greene CS, Liang H, Xiao Y, Wang C, Iavarone A, Berger AH, Bivona TG, Lazar AJ, Hammer GD, Giordano T, Kwong LN, McArthur G, Huang C, Tward AD, Frederick MJ, McCormick F, Meyerson M; Cancer Genome Atlas Research Network, Van Allen EM, Cherniack AD, Ciriello G, Sander C, Schultz N. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173:321-337.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2333] [Cited by in RCA: 2120] [Article Influence: 302.9] [Reference Citation Analysis (0)] |
| 20. | Li J, Lupat R, Amarasinghe KC, Thompson ER, Doyle MA, Ryland GL, Tothill RW, Halgamuge SK, Campbell IG, Gorringe KL. CONTRA: copy number analysis for targeted resequencing. Bioinformatics. 2012;28:1307-1313. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 239] [Cited by in RCA: 271] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 21. | Chang JC, Alex D, Bott M, Tan KS, Seshan V, Golden A, Sauter JL, Buonocore DJ, Vanderbilt CM, Gupta S, Desmeules P, Bodd FM, Riely GJ, Rusch VW, Jones DR, Arcila ME, Travis WD, Ladanyi M, Rekhtman N. Comprehensive Next-Generation Sequencing Unambiguously Distinguishes Separate Primary Lung Carcinomas From Intrapulmonary Metastases: Comparison with Standard Histopathologic Approach. Clin Cancer Res. 2019;25:7113-7125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 75] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 22. | Murphy SJ, Harris FR, Kosari F, Barreto Siqueira Parrilha Terra S, Nasir A, Johnson SH, Serla V, Smadbeck JB, Halling GC, Karagouga G, Sukov WR, Leventakos K, Yang P, Peikert T, Mansfield AS, Wigle DA, Yi ES, Kipp BR, Vasmatzis G, Aubry MC. Using Genomics to Differentiate Multiple Primaries From Metastatic Lung Cancer. J Thorac Oncol. 2019;14:1567-1582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 23. | Jiang T, Fang Z, Tang S, Cheng R, Li Y, Ren S, Su C, Min W, Guo X, Zhu W, Zhang H, Hou L, Pan Y, Zhou Z, Zhang J, Zhang G, Yue Z, Chen L, Zhou C. Mutational Landscape and Evolutionary Pattern of Liver and Brain Metastasis in Lung Adenocarcinoma. J Thorac Oncol. 2021;16:237-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 24. | Wei Q, Ye Z, Zhong X, Li L, Wang C, Myers RE, Palazzo JP, Fortuna D, Yan A, Waldman SA, Chen X, Posey JA, Basu-Mallick A, Jiang BH, Hou L, Shu J, Sun Y, Xing J, Li B, Yang H. Multiregion whole-exome sequencing of matched primary and metastatic tumors revealed genomic heterogeneity and suggested polyclonal seeding in colorectal cancer metastasis. Ann Oncol. 2017;28:2135-2141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 90] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 25. | Naxerova K, Reiter JG, Brachtel E, Lennerz JK, van de Wetering M, Rowan A, Cai T, Clevers H, Swanton C, Nowak MA, Elledge SJ, Jain RK. Origins of lymphatic and distant metastases in human colorectal cancer. Science. 2017;357:55-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 375] [Article Influence: 46.9] [Reference Citation Analysis (0)] |
| 26. | Sorich MJ, Wiese MD, Rowland A, Kichenadasse G, McKinnon RA, Karapetis CS. Extended RAS mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: a meta-analysis of randomized, controlled trials. Ann Oncol. 2015;26:13-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 382] [Cited by in RCA: 404] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 27. | Ryan MB, Corcoran RB. Therapeutic strategies to target RAS-mutant cancers. Nat Rev Clin Oncol. 2018;15:709-720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 255] [Article Influence: 42.5] [Reference Citation Analysis (0)] |
| 28. | Hornick JL, Fletcher CD. The role of KIT in the management of patients with gastrointestinal stromal tumors. Hum Pathol. 2007;38:679-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 127] [Article Influence: 7.1] [Reference Citation Analysis (0)] |