Published online Sep 16, 2021. doi: 10.12998/wjcc.v9.i26.7653
Peer-review started: December 24, 2020
First decision: May 8, 2021
Revised: May 28, 2021
Accepted: July 7, 2021
Article in press: July 7, 2021
Published online: September 16, 2021
Processing time: 260 Days and 0.9 Hours
Ulcerative colitis (UC) is a refractory intestinal disease with alternating onset and remission and a long disease course, which seriously affects the health and quality of life of patients. The goal of treatment is to control clinical symptoms, induce and maintain remission, promote mucosal healing, and reduce recurrence. Clinical trials have shown unsatisfactory clinical response rates. As a supplementary alternative medicine, traditional Chinese medicine has a rich history and has shown good results in the treatment of UC. Because of the quality of herbal medicine and other factors, the curative effect of traditional Chinese medicine is not stable enough. The mechanism underlying the effect of Jianpi Qingchang Huashi Recipe (JPQCHSR) on inducing UC mucosal healing is not clear.
To investigate the potential mechanism of JPQCHSR for the treatment of UC based on network pharmacology and molecular docking.
Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform was used to extract the active components and action targets of JPQCHSR, and the target names were standardized and corrected through UniProt database. The related targets of UC were obtained through GeneCards database, and the intersection targets of drugs and diseases were screened by jvenn online analysis tool. The visual regulatory network of "Traditional Chinese medicine-active components-target-disease" was constructed using Cytoscape software, the protein interaction network was constructed using STRING database, and enrichment analysis of gene ontology and Kyoto Encyclopedia of Genes and Genomes pathways was conducted through R software. At last, the active components were docked with the core target through SYBYL-X 2.1.1 software.
Through database analysis, a total of 181 active components, 302 targets and 205 therapeutic targets were obtained for JPQCHSR. The key compounds include quercetin, luteolin, kaempferol, etc. The core targets involved STAT3, AKT1, TP53, MAPK1, MAPK3, JUN, TNF, etc. A total of 2861 items were obtained by GO enrichment analysis, and 171 items were obtained by KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis. The results of molecular docking showed that the key active components in JPQCHSR had certain affinity with the core target.
The treatment of UC with JPQCHSR is a complex process of multi-component, multi-target and multi-pathway regulation. The mechanism of this Recipe in the treatment of UC can be predicted through network pharmacology and molecular docking, so as to provide theoretical reference for it to better play its therapeutic role.
Core Tip: Ulcerative colitis (UC) is a chronic non-specific inflammatory disease that can cause varying degrees of mucosal inflammation from the rectum to the proximal colon. The mechanism of this Recipe in the treatment of UC can be predicted through network pharmacology and molecular docking, so as to provide theoretical reference for it to better play its therapeutic role.
- Citation: Zheng L, Wen XL, Dai YC. Mechanism of Jianpi Qingchang Huashi Recipe in treating ulcerative colitis: A study based on network pharmacology and molecular docking. World J Clin Cases 2021; 9(26): 7653-7670
- URL: https://www.wjgnet.com/2307-8960/full/v9/i26/7653.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i26.7653
Ulcerative colitis (UC) is a chronic non-specific inflammatory disease that can cause varying degrees of mucosal inflammation from the rectum to the proximal colon[1]. Its typical clinical manifestations include diarrhea, mucus and blood in stools, and abdominal pain[2]. The disease can affect the intestinal mucosa and muscularis mucosa and is featured by a recurrent and prolonged course. Its pathogenesis involves genetic susceptibility, epithelial barrier defects, dysregulated immune response, and environmental factors[3]. As one of the common refractory diseases in the digestive system, UC has unclear and complex pathogenic mechanisms. Western medicine-based treatments for UC rely mostly on drug therapies. Although these drugs can achieve quick remission in most patients, there is no radical cure. Furthermore, patients receiving Western medicine-based therapies often suffer from problems including hormone dependence, drug resistance, and treatment-related toxicities[4]. Therefore, many patients with UC as well as physicians and researchers are increasingly considering complementary and alternative medicine (CAM) option[5]. In North American and European studies, the rate of current or past use of CAM for the treatment of UC is 21%-60%[6,7], of which herbal medicine, especially CAM intervention, is the first choice[8]. As a supplementary alternative medicine, traditional Chinese medicine (TCM) has a long history and has shown good results in the treatment of UC. Through multi-component, multi-target holistic therapies, TCM can effectively improve intestinal inflammation.
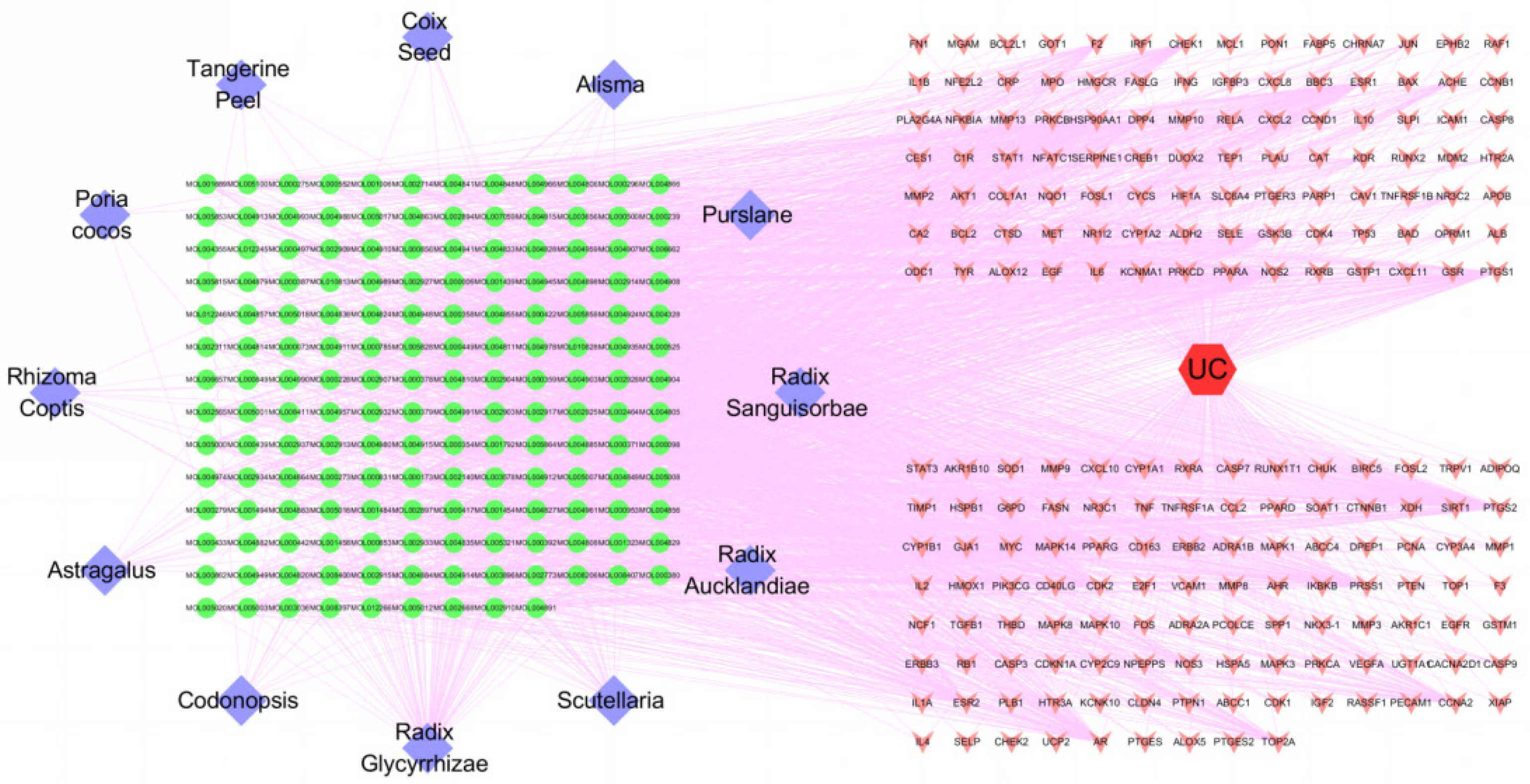
Jianpi Qingchang Recipe (JPQCR, or spleen-invigorating and intestine-clearing recipe) is modified from Shenling Baizhu Powder, which was recorded in Taiping Huimin Heji Ju Fang (Prescriptions of the Bureau of Taiping People's Welfare Pharmacy), and Diyu Powder, which was described in Sheng Ji Zonglu (General Records of Holy Universal Relief). Previous studies from our group showed that oral Chinese medicine compounds were effective for the treatment of UC, inducing and maintaining remission, although the time until the effect was evident was relatively slow. Many studies have shown that JPQCR can markedly improve dextran sulphate sodium (DSS)-induced UC in mouse models[9], which may be explained that JPQCR can inhibit the activation of nuclear factor-kappa B (NF-κB), down-regulate inflammatory mediators [e.g., MPO, interleukin (IL)-1β, IL-8, and tumor necrosis factor alpha (TNF-α)], and improve the barrier function of the intestinal epithelium. In addition, JPQCR can regulate DSS-induced abnormal intestinal motility in UC mice by inhibiting the cascade of intestinal inflammation and reducing autophagy in interstitial cells of Cajal[10,11]. Based on JPQCR, the Jianpi Qingchang Huashi Recipe (JPQCHSR) has newly added TCM drugs including Poria, Tangerine Peel, Coix Seed, Alisma, and Radix Scutellariae. The prescription is composed of Codonopsis, Astragalus, Rhizoma Coptis, Poria cocos, Tangerine Peel, Coix Seed, Alisma, Purslane, Radix Sanguisorbae, Radix Aucklandiae, Scutellaria, and Radix Glycyrrhizae and is effective for clinical treatment of UC.
TCM drugs are characterized by multi-component, multi-target, and multi-pathway. The TCM formulas are prepared in accordance with the principles of "Sovereign, Minister, Assistant, and Courier". Under the guidance of a patient-centered holistic view, TCM formulas start from the integration between “whole” and “parts” and systematically regulate human body through the in vivo metabolism of the active components in the drugs. However, it is often difficult to identify the mechanisms of action of a specific formula. Compared with western medicine[12], traditional Chinese medicine formula may not quickly induce UC remission and control clinical symptoms. In addition, because of the quality of herbal medicine and other factors, the curative effect of TCM is not stable enough. The patients reported a poor taste of traditional Chinese medicine formula and had poor follow-up compliance. TCM drugs exert their effects in both independent and synergistic manners. The “independence” is reflected in the direct or indirect influence or action of a certain drug component on a target, whereas the “synergy” refers that there are synergistic or antagonistic effects among multiple components of a TCM drug and they treat diseases as a whole. In recent years, network pharmacology has been proposed for overall, multi-level, and multi-grade research on TCM formulas. Based on systems biology and computer networks, network pharmacology is the study of the interactions among related nodes, so as to explain the mechanisms of action of drugs in treating diseases. The core concept of "multi-component, multi-target, and multi-pathway" of network pharmacology is consistent with the "holistic view" of TCM theory, and thus network pharmacology may enable the accurate prediction and analysis of the mechanisms of action of TCM compound prescriptions[13]. Molecular docking is a receptor-based virtual screening technology that mainly studies the interaction among molecules and predicts the binding mode and affinity between ligands and receptors[14,15]. In recent years, molecular docking has become an important technology in computer-aided drug design[16,17]. This study aims to explore the mechanisms of action of JPQCHSR in the treatment of UC through network pharmacology and molecular docking, in an attempt to further promote the development and use of JPQCHSR and provide new insights into the treatment of UC.
Data were searched from databases including Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) (https://tcmspw.com/tcmsp.php)[18], UniProt (https://www.uniprot.org/), GeneCards (https://www.genecards.org/), Jvenn (http://jvenn.toulouse.inra.fr/app/index.html)[19], STRING 11.0 (https://string-db.org/), and Chemical Book (https://www.chemicalbook.com/ProductIndex.aspx)[20]. The software used included Cytoscape version 3.7.2, R version 3.6.3, cluster Profiler, and SYBYL-X version 2.1.1.
Using the keywords "Radix Codonopsis", "Astragalus", "Coptis", "Poria", "Tangerine Peel", "Coix Seed", "Alismaceae", "Herba Portulacae", "Radix Sanguisorbae", "Radix Aucklandiae", "Radix Scutellariae" and "Radix Glycyrrhizae", we retrieved the active components of 12 TCM drugs in JPQCHSR in the TCMSP database. According to the pharmacokinetic principles, the active components were screened using the criteria including oral bioavailability (OB) ≥ 30% and drug-likeness (DL) ≥ 0.18. The relevant targets of the active components were extracted from the TCMSP database, and the targets of action were standardized and corrected using the human genes that have been validated in the UniProt database. Components without action targets and targets without standard names were deleted and active components and validated targets were collected, thus yielding the active components of JPQCHSR and the related targets of action in human beings.
Using the keyword “ulcerative colitis”, we searched the GeneCards database for UC-related targets.
Using the jvenn online analysis tool, we mapped the active component targets obtained in Section 1.2 with the disease targets in Section 1.3, and the intersection was the potential targets of JPQCHSR in the treatment of UC. Files containing TCM drugs, active components, targets, disease information and their attribute files were imported into the Cytoscape version 3.7.2 to construct a visual network of "TCM Drugs-Active Components-Targets-Diseases”. With the help of the "Network analyzer" plug-in in the software, the topological properties of network nodes were analyzed, and the key active components of JPQCHSR for treating UC were obtained according to the “degree” values.
The "Active components-Diseases" intersection target genes obtained in Section 1.4 were imported into the STRING 11.0 database (http://string-db.org), and the species were limited to "homo sapiens". Data with a confidence level higher than 0.900 were selected, and isolated nodes were removed. Then, a protein-protein interaction (PPI) network was constructed. According to the “degree” values, the core targets of JPQCHSR in the treatment of UC were screened.
Based on the cluster Profiler package in the R 3.6.3 software, we performed GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment analyses on the potential targets of active components. The results were filtered based on P values (P < 0.05). R 3.6.3 software was used to visualize the results, during which bubble charts and bar charts were drawn.
The Chemical Book database was searched to obtain the active components (mol. files), and the results were imported into the SYBYL-X 2.1.1 software for energy optimization. The PDB files of the crystal structure of the core target proteins were downloaded from the RSCB PDB database. After a series of optimization steps (including ligand extraction, hydrogenation, and water extraction) in the SYBYL-X 2.1.1 software, molecular docking was performed using the Surflex-Dock module in the software, and the binding activity was evaluated through function scoring.
After the TCM drugs in JPQCHSR were searched in the TCMSP database, 251 active components were obtained. Using the OB ≥ 30% and a DL index ≥ 0.18 as potential active compounds and excluding both non-target compounds and duplicate compounds, 181 active components corresponding to 302 targets were derived. The active components and targets of action of JPQCHSR are summarized in Table 1.
| No. | MOL ID | Compound | Source | No. | MOL ID | Compound | Source |
| 1 | MOL001006 | poriferasta-7,22E-dien-3beta-ol | Codonopsis | 92 | MOL004806 | euchrenone | Radix Glycyrrhizae |
| 2 | MOL002140 | Perlolyrine | Codonopsis | 93 | MOL004808 | glyasperin B | Radix Glycyrrhizae |
| 3 | MOL003036 | ZINC03978781 | Codonopsis | 94 | MOL004810 | glyasperin F | Radix Glycyrrhizae |
| 4 | MOL004355 | Spinasterol | Codonopsis | 95 | MOL004811 | Glyasperin C | Radix Glycyrrhizae |
| 5 | MOL005321 | Frutinone A | Codonopsis | 96 | MOL004814 | Isotrifoliol | Radix Glycyrrhizae |
| 6 | MOL006774 | stigmast-7-enol | Codonopsis | 97 | MOL004815 | (E)-1-(2,4-dihydroxyphenyl)-3-(2,2-dimethylchromen-6-yl)prop-2-en-1-one | Radix Glycyrrhizae |
| 7 | MOL007059 | 3-beta-Hydroxymethyllenetanshiquinone | Codonopsis | 98 | MOL004820 | kanzonols W | Radix Glycyrrhizae |
| 8 | MOL007514 | methyl icosa-11,14-dienoate | Codonopsis | 99 | MOL004824 | (2S)-6-(2,4-dihydroxyphenyl)-2-(2-hydroxypropan-2-yl)-4-methoxy-2,3-dihydrofuro[3,2-g]chromen-7-one | Radix Glycyrrhizae |
| 9 | MOL008393 | 7-(beta-Xylosyl)cephalomannine_qt | Codonopsis | 100 | MOL004827 | Semilicoisoflavone B | Radix Glycyrrhizae |
| 10 | MOL008397 | Daturilin | Codonopsis | 101 | MOL004828 | Glepidotin A | Radix Glycyrrhizae |
| 11 | MOL008400 | glycitein | Codonopsis | 102 | MOL004829 | Glepidotin B | Radix Glycyrrhizae |
| 12 | MOL008407 | (8S,9S,10R,13R,14S,17R)-17-[(E,2R,5S)-5-ethyl-6-methylhept-3-en-2-yl]-10,13-dimethyl-1,2,4,7,8,9,11,12,14,15,16,17-dodecahydrocyclopenta[a]phenanthren-3-one | Codonopsis | 103 | MOL004833 | Phaseolinisoflavan | Radix Glycyrrhizae |
| 13 | MOL008411 | 11-Hydroxyrankinidine | Codonopsis | 104 | MOL004835 | Glypallichalcone | Radix Glycyrrhizae |
| 14 | MOL000033 | (3S,8S,9S,10R,13R,14S,17R)-10,13-dimethyl-17-[(2R,5S)-5-propan-2-yloctan-2-yl]-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-3-ol | Astragalus | 105 | MOL004838 | 8-(6-hydroxy-2-benzofuranyl)-2,2-dimethyl-5-chromenol | Radix Glycyrrhizae |
| 15 | MOL000371 | 3,9-di-O-methylnissolin | Astragalus | 106 | MOL004841 | Licochalcone B | Radix Glycyrrhizae |
| 16 | MOL000378 | 7-O-methylisomucronulatol | Astragalus | 107 | MOL004848 | licochalcone G | Radix Glycyrrhizae |
| 17 | MOL000379 | 9,10-dimethoxypterocarpan-3-O-β-D-glucoside | Astragalus | 108 | MOL004849 | 3-(2,4-dihydroxyphenyl)-8-(1,1-dimethylprop-2-enyl)-7-hydroxy-5-methoxy-coumarin | Radix Glycyrrhizae |
| 18 | MOL000380 | (6aR,11aR)-9,10-dimethoxy-6a,11a-dihydro-6H-benzofurano[3,2-c]chromen-3-ol | Astragalus | 109 | MOL004855 | Licoricone | Radix Glycyrrhizae |
| 19 | MOL000387 | Bifendate | Astragalus | 110 | MOL004856 | Gancaonin A | Radix Glycyrrhizae |
| 20 | MOL000433 | FA | Astragalus | 111 | MOL004857 | Gancaonin B | Radix Glycyrrhizae |
| 21 | MOL000439 | isomucronulatol-7,2'-di-O-glucosiole | Astragalus | 112 | MOL004863 | 3-(3,4-dihydroxyphenyl)-5,7-dihydroxy-8-(3-methylbut-2-enyl)chromone | Radix Glycyrrhizae |
| 22 | MOL000442 | 1,7-Dihydroxy-3,9-dimethoxy pterocarpene | Astragalus | 113 | MOL004864 | 5,7-dihydroxy-3-(4-methoxyphenyl)-8-(3-methylbut-2-enyl)chromone | Radix Glycyrrhizae |
| 23 | MOL001454 | berberine | Rhizoma Coptis | 114 | MOL004866 | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-6-(3-methylbut-2-enyl)chromone | Radix Glycyrrhizae |
| 24 | MOL002894 | berberrubine | Rhizoma Coptis | 115 | MOL004879 | Glycyrin | Radix Glycyrrhizae |
| 25 | MOL002903 | (R)-Canadine | Rhizoma Coptis | 116 | MOL004882 | Licocoumarone | Radix Glycyrrhizae |
| 26 | MOL002904 | Berlambine | Rhizoma Coptis | 117 | MOL004883 | Licoisoflavone | Radix Glycyrrhizae |
| 27 | MOL002907 | Corchoroside A_qt | Rhizoma Coptis | 118 | MOL004884 | Licoisoflavone B | Radix Glycyrrhizae |
| 28 | MOL000622 | Magnograndiolide | Rhizoma Coptis | 119 | MOL004885 | licoisoflavanone | Radix Glycyrrhizae |
| 29 | MOL000785 | palmatine | Rhizoma Coptis | 120 | MOL004891 | shinpterocarpin | Radix Glycyrrhizae |
| 30 | MOL002668 | Worenine | Rhizoma Coptis | 121 | MOL004898 | (E)-3-[3,4-dihydroxy-5-(3-methylbut-2-enyl)phenyl]-1-(2,4-dihydroxyphenyl)prop-2-en-1-one | Radix Glycyrrhizae |
| 31 | MOL000273 | (2R)-2-[(3S,5R,10S,13R,14R,16R,17R)-3,16-dihydroxy-4,4,10,13,14-pentamethyl-2,3,5,6,12,15,16,17-octahydro-1H-cyclopenta[a]phenanthren-17-yl]-6-methylhept-5-enoic acid | Poria cocos | 122 | MOL004903 | liquiritin | Radix Glycyrrhizae |
| 32 | MOL000275 | trametenolic acid | Poria cocos | 123 | MOL004904 | licopyranocoumarin | Radix Glycyrrhizae |
| 33 | MOL000279 | Cerevisterol | Poria cocos | 124 | MOL004907 | Glyzaglabrin | Radix Glycyrrhizae |
| 34 | MOL000282 | ergosta-7,22E-dien-3beta-ol | Poria cocos | 125 | MOL004908 | Glabridin | Radix Glycyrrhizae |
| 35 | MOL000283 | Ergosterol peroxide | Poria cocos | 126 | MOL004910 | Glabranin | Radix Glycyrrhizae |
| 36 | MOL005815 | Citromitin | Tangerine Peel | 127 | MOL004911 | Glabrene | Radix Glycyrrhizae |
| 37 | MOL005828 | nobiletin | Tangerine Peel | 128 | MOL004912 | Glabrone | Radix Glycyrrhizae |
| 38 | MOL001323 | Sitosterol alpha1 | Coix Seed | 129 | MOL004913 | 1,3-dihydroxy-9-methoxy-6-benzofurano[3,2-c]chromenone | Radix Glycyrrhizae |
| 39 | MOL001494 | Mandenol | Coix Seed | 130 | MOL004914 | 1,3-dihydroxy-8,9-dimethoxy-6-benzofurano[3,2-c]chromenone | Radix Glycyrrhizae |
| 40 | MOL008121 | 2-Monoolein | Coix Seed | 131 | MOL004915 | Eurycarpin A | Radix Glycyrrhizae |
| 41 | MOL000953 | CLR | Coix Seed | 132 | MOL004924 | (-)-Medicocarpin | Radix Glycyrrhizae |
| 42 | MOL000831 | Alisol B monoacetate | Alisma | 133 | MOL004935 | Sigmoidin-B | Radix Glycyrrhizae |
| 43 | MOL000849 | 16β-methoxyalisol B monoacetate | Alisma | 134 | MOL004941 | (2R)-7-hydroxy-2-(4-hydroxyphenyl)chroman-4-one | Radix Glycyrrhizae |
| 44 | MOL000853 | alisol B | Alisma | 135 | MOL004945 | (2S)-7-hydroxy-2-(4-hydroxyphenyl)-8-(3-methylbut-2-enyl)chroman-4-one | Radix Glycyrrhizae |
| 45 | MOL000856 | alisol C monoacetate | Alisma | 136 | MOL004948 | Isoglycyrol | Radix Glycyrrhizae |
| 46 | MOL002464 | 1-Monolinolein | Alisma | 137 | MOL004949 | Isolicoflavonol | Radix Glycyrrhizae |
| 47 | MOL000862 | [(1S,3R)-1-[(2R)-3,3-dimethyloxiran-2-yl]-3-[(5R,8S,9S,10S,11S,14R)-11-hydroxy-4,4,8,10,14-pentamethyl-3-oxo-1,2,5,6,7,9,11,12,15,16-decahydrocyclopenta[a]phenanthren-17-yl]butyl] acetate | Alisma | 138 | MOL004957 | HMO | Radix Glycyrrhizae |
| 48 | MOL001439 | arachidonic acid | Purslane | 139 | MOL004959 | 1-Methoxyphaseollidin | Radix Glycyrrhizae |
| 49 | MOL003578 | Cycloartenol | Purslane | 140 | MOL004961 | Quercetin der. | Radix Glycyrrhizae |
| 50 | MOL002773 | beta-carotene | Purslane | 141 | MOL004966 | 3'-Hydroxy-4'-O-Methylglabridin | Radix Glycyrrhizae |
| 51 | MOL006657 | isobetanidin | Purslane | 142 | MOL000497 | licochalcone a | Radix Glycyrrhizae |
| 52 | MOL006662 | isobetanin_qt | Purslane | 143 | MOL004974 | 3'-Methoxyglabridin | Radix Glycyrrhizae |
| 53 | MOL005399 | alexandrin_qt | Radix Sanguisorbae | 144 | MOL004978 | 2-[(3R)-8,8-dimethyl-3,4-dihydro-2H-pyrano[6,5-f]chromen-3-yl]-5-methoxyphenol | Radix Glycyrrhizae |
| 54 | MOL005853 | methyl-2,3,6-tri-O-galloyl-β-D-glucopyranoside | Radix Sanguisorbae | 145 | MOL004980 | Inflacoumarin A | Radix Glycyrrhizae |
| 55 | MOL005858 | 3,7,8-Tri-O-methylellagic acid | Radix Sanguisorbae | 146 | MOL004985 | icos-5-enoic acid | Radix Glycyrrhizae |
| 56 | MOL005864 | methyl-6-O-galloyl-β-D-glucopyranoside | Radix Sanguisorbae | 147 | MOL004988 | Kanzonol F | Radix Glycyrrhizae |
| 57 | MOL005869 | daucostero_qt | Radix Sanguisorbae | 148 | MOL004989 | 6-prenylated eriodictyol | Radix Glycyrrhizae |
| 58 | MOL010813 | Benzo[a]carbazole | Radix Aucklandiae | 149 | MOL004990 | 7,2',4'-trihydroxy-5-methoxy-3-arylcoumarin | Radix Glycyrrhizae |
| 59 | MOL010828 | cynaropicrin | Radix Aucklandiae | 150 | MOL004991 | 7-Acetoxy-2-methylisoflavone | Radix Glycyrrhizae |
| 60 | MOL001689 | acacetin | Scutellaria | 151 | MOL004993 | 8-prenylated eriodictyol | Radix Glycyrrhizae |
| 61 | MOL000173 | wogonin | Scutellaria | 152 | MOL004996 | gadelaidic acid | Radix Glycyrrhizae |
| 62 | MOL000228 | (2R)-7-hydroxy-5-methoxy-2-phenylchroman-4-one | Scutellaria | 153 | MOL000500 | Vestitol | Radix Glycyrrhizae |
| 63 | MOL002714 | baicalein | Scutellaria | 154 | MOL005000 | Gancaonin G | Radix Glycyrrhizae |
| 64 | MOL002909 | 5,7,2,5-tetrahydroxy-8,6-dimethoxyflavone | Scutellaria | 155 | MOL005001 | Gancaonin H | Radix Glycyrrhizae |
| 65 | MOL002910 | Carthamidin | Scutellaria | 156 | MOL005003 | Licoagrocarpin | Radix Glycyrrhizae |
| 66 | MOL002913 | Dihydrobaicalin_qt | Scutellaria | 157 | MOL005007 | Glyasperins M | Radix Glycyrrhizae |
| 67 | MOL002914 | Eriodyctiol (flavanone) | Scutellaria | 158 | MOL005008 | Glycyrrhiza flavonol A | Radix Glycyrrhizae |
| 68 | MOL002915 | Salvigenin | Scutellaria | 159 | MOL005012 | Licoagroisoflavone | Radix Glycyrrhizae |
| 69 | MOL002917 | 5,2',6'-Trihydroxy-7,8-dimethoxyflavone | Scutellaria | 160 | MOL005016 | Odoratin | Radix Glycyrrhizae |
| 70 | MOL002925 | 5,7,2',6'-Tetrahydroxyflavone | Scutellaria | 161 | MOL005017 | Phaseol | Radix Glycyrrhizae |
| 71 | MOL002927 | Skullcapflavone II | Scutellaria | 162 | MOL005018 | Xambioona | Radix Glycyrrhizae |
| 72 | MOL002928 | oroxylin a | Scutellaria | 163 | MOL005020 | dehydroglyasperins C | Radix Glycyrrhizae |
| 73 | MOL002932 | Panicolin | Scutellaria | 164 | MOL002879 | Diop | Codonopsis Scutellaria |
| 74 | MOL002933 | 5,7,4'-Trihydroxy-8-methoxyflavone | Scutellaria | 165 | MOL000449 | Stigmasterol | Codonopsis Scutellaria Coix Seed Radix Aucklandiae |
| 75 | MOL002934 | NEOBAICALEIN | Scutellaria | 166 | MOL003896 | 7-Methoxy-2-methyl isoflavone | Codonopsis Radix Glycyrrhizae |
| 76 | MOL002937 | DIHYDROOROXYLIN | Scutellaria | 167 | MOL000006 | luteolin | Codonopsis Purslane |
| 77 | MOL000525 | Norwogonin | Scutellaria | 168 | MOL000211 | Mairin | Astragalus Radix SanguisorbaeRadix Aucklandiae Radix Glycyrrhizae |
| 78 | MOL000552 | 5,2'-Dihydroxy-6,7,8-trimethoxyflavone | Scutellaria | 169 | MOL000239 | Jaranol | Astragalus Radix Glycyrrhizae |
| 79 | MOL000073 | ent-Epicatechin | Scutellaria | 170 | MOL000296 | hederagenin | Astragalus Poria cocos |
| 80 | MOL001490 | bis[(2S)-2-ethylhexyl] benzene-1,2-dicarboxylate | Scutellaria | 171 | MOL000354 | isorhamnetin | Astragalus Radix Glycyrrhizae |
| 81 | MOL008206 | Moslosooflavone | Scutellaria | 172 | MOL000392 | formononetin | Astragalus Radix Glycyrrhizae |
| 82 | MOL010415 | 11,13-Eicosadienoic acid, methyl ester | Scutellaria | 173 | MOL000417 | Calycosin | Astragalus Radix Glycyrrhizae |
| 83 | MOL012245 | 5,7,4'-trihydroxy-6-methoxyflavanone | Scutellaria | 174 | MOL000422 | kaempferol | Astragalus Purslane Radix SanguisorbaeRadix Glycyrrhizae |
| 84 | MOL012246 | 5,7,4'-trihydroxy-8-methoxyflavanone | Scutellaria | 175 | MOL000098 | quercetin | Astragalus Rhizoma Coptis PurslaneRadix SanguisorbaeRadix Glycyrrhizae |
| 85 | MOL012266 | rivularin | Scutellaria | 176 | MOL002897 | epiberberine | Rhizoma CoptisScutellaria |
| 86 | MOL001484 | Inermine | Radix Glycyrrhizae | 177 | MOL001458 | coptisine | Rhizoma CoptisScutellaria |
| 87 | MOL001792 | DFV | Radix Glycyrrhizae | 178 | MOL000359 | sitosterol | Tangerine Peel Coix Seed Alisma Radix Aucklandiae Scutellaria Radix Glycyrrhizae |
| 88 | MOL002311 | Glycyrol | Radix Glycyrrhizae | 179 | MOL004328 | naringenin | Tangerine Peel Radix Glycyrrhizae |
| 89 | MOL002565 | Medicarpin | Radix Glycyrrhizae | 180 | MOL005100 | 5,7-dihydroxy-2-(3-hydroxy-4-methoxyphenyl)chroman-4-one | Tangerine Peel Purslane |
| 90 | MOL003656 | Lupiwighteone | Radix Glycyrrhizae | 181 | MOL000358 | beta-sitosterol | Purslane Radix SanguisorbaeScutellaria |
| 91 | MOL004805 | (2S)-2-[4-hydroxy-3-(3-methylbut-2-enyl)phenyl]-8,8-dimethyl-2,3-dihydropyrano[2,3-f]chromen-4-one | Radix Glycyrrhizae |
Using the key word “ulcerative colitis”, a total of 4622 UC-related targets were obtained in the GeneCards database.
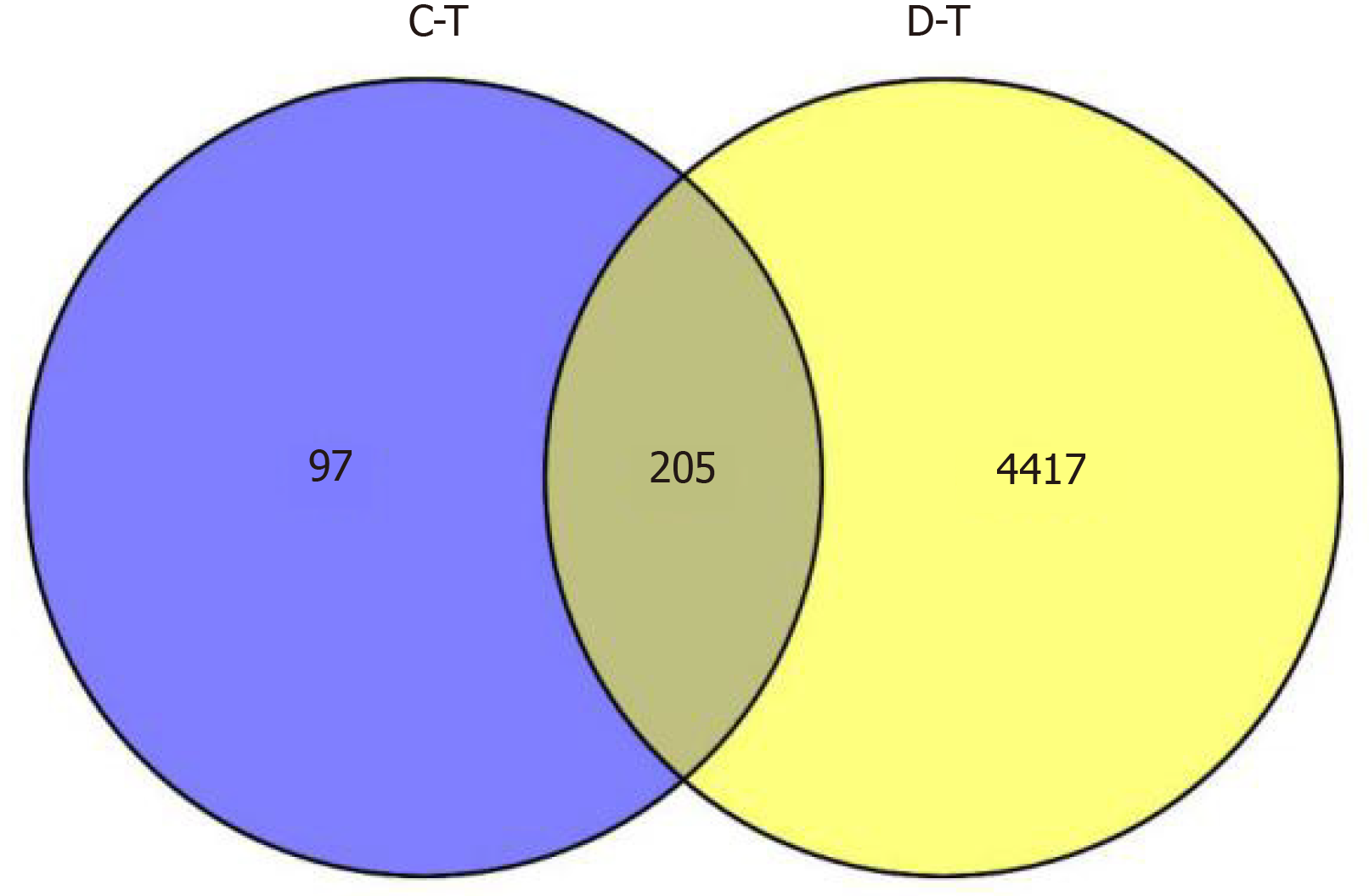
Using the jvenn online analysis tool, after the intersection of the active component targets and the disease targets, a total of 205 potential targets of JPQCHSR in the treatment of UC were obtained (Figure 1). Then, a visual network of "TCM Drugs - Active Components - Targets - Diseases” was established using the Cytoscape version 3.7.2 (Figure 2). The network diagram contains 383 nodes, which included 205 target gene nodes, 165 active component nodes, 12 TCM drug nodes, and 1 disease node. With the help of the "Network analyzer" plug-in in the software, we analyzed the topological properties of the network nodes. The average node “degree” value of the compounds in the network was calculated to be 12.72. Table 2 shows the topological information of the active components with a “degree” value of ≥ 13. The degree of a node is the number of edges connected to the node. A higher degree means a more important role that a node plays in the network. As shown in Table 2, 77 compounds including quercetin (degree = 121), luteolin (degree = 52), kaempferol (degree = 46), calycosin (degree = 38), naringenin (degree = 32), nobiletin (degree = 30), baicalein (degree = 29), arachidonic acid (degree = 29), and formononetin (degree = 27) had degree values higher than the mean degree value. Thus, these components were predicted preliminarily as the core compounds of JPQCHSR for the treatment of UC.
| Name | Degree | Name | Degree |
| MOL000098 | 121 | MOL004857 | 17 |
| MOL000006 | 52 | MOL004849 | 17 |
| MOL000422 | 46 | MOL004835 | 17 |
| MOL000173 | 38 | MOL004808 | 17 |
| MOL004328 | 32 | MOL003656 | 17 |
| MOL005828 | 30 | MOL012266 | 16 |
| MOL002714 | 29 | MOL004915 | 16 |
| MOL001439 | 29 | MOL004883 | 16 |
| MOL000392 | 27 | MOL004864 | 16 |
| MOL003896 | 25 | MOL004856 | 16 |
| MOL000497 | 24 | MOL004833 | 16 |
| MOL000378 | 24 | MOL004820 | 16 |
| MOL000354 | 24 | MOL004815 | 16 |
| MOL002773 | 22 | MOL002934 | 16 |
| MOL004959 | 21 | MOL005012 | 15 |
| MOL000358 | 21 | MOL004961 | 15 |
| MOL008206 | 20 | MOL004885 | 15 |
| MOL005003 | 20 | MOL004884 | 15 |
| MOL004978 | 20 | MOL004863 | 15 |
| MOL004974 | 20 | MOL004841 | 15 |
| MOL004891 | 20 | MOL004810 | 15 |
| MOL004828 | 20 | MOL002933 | 15 |
| MOL001689 | 20 | MOL000552 | 15 |
| MOL000500 | 20 | MOL005008 | 14 |
| MOL004991 | 19 | MOL004990 | 14 |
| MOL004966 | 19 | MOL004911 | 14 |
| MOL004811 | 19 | MOL004827 | 14 |
| MOL002565 | 19 | MOL002927 | 14 |
| MOL008400 | 18 | MOL002917 | 14 |
| MOL005007 | 18 | MOL000449 | 14 |
| MOL004912 | 18 | MOL005020 | 13 |
| MOL004824 | 18 | MOL005017 | 13 |
| MOL002928 | 18 | MOL004904 | 13 |
| MOL000417 | 18 | MOL004879 | 13 |
| MOL005016 | 17 | MOL004848 | 13 |
| MOL005000 | 17 | MOL004814 | 13 |
| MOL004957 | 17 | MOL000380 | 13 |
| MOL004908 | 17 | MOL000371 | 13 |
| MOL004907 | 17 |
| Name | Degree | Name | Degree | Name | Degree |
| STAT3 | 49 | CDKN1A | 20 | AR | 14 |
| AKT1 | 46 | PRKCA | 19 | HIF1A | 14 |
| TP53 | 46 | CASP3 | 19 | ALB | 14 |
| MAPK1 | 44 | TIMP1 | 19 | PTGS2 | 14 |
| MAPK3 | 43 | FN1 | 19 | PPARA | 14 |
| JUN | 42 | CASP8 | 19 | PPARG | 13 |
| TNF | 39 | STAT1 | 19 | GSK3B | 13 |
| RELA | 35 | PRKCD | 18 | IGF2 | 13 |
| HSP90AA1 | 32 | IL2 | 18 | CCNA2 | 13 |
| MAPK14 | 32 | BCL2 | 18 | TNFRSF1A | 13 |
| IL6 | 31 | TGFB1 | 18 | MMP2 | 13 |
| MAPK8 | 30 | IL4 | 18 | ERBB2 | 13 |
| VEGFA | 28 | IL1B | 18 | F2 | 12 |
| FOS | 28 | BCL2L1 | 17 | CXCL2 | 12 |
| EGFR | 25 | NFKBIA | 17 | RAF1 | 12 |
| RB1 | 24 | MMP9 | 17 | NOS3 | 12 |
| CTNNB1 | 24 | PRKCB | 16 | E2F1 | 12 |
| MYC | 24 | CDK1 | 16 | CHUK | 12 |
| CCND1 | 22 | IL10 | 16 | XIAP | 12 |
| ESR1 | 22 | CREB1 | 15 | IGFBP3 | 12 |
| RXRA | 21 | CCL2 | 15 | CCNB1 | 12 |
| CXCL8 | 21 | CDK2 | 15 | CDK4 | 12 |
| NR3C1 | 20 | IFNG | 15 | PCNA | 12 |
| EGF | 20 | MDM2 | 14 | MMP3 | 12 |
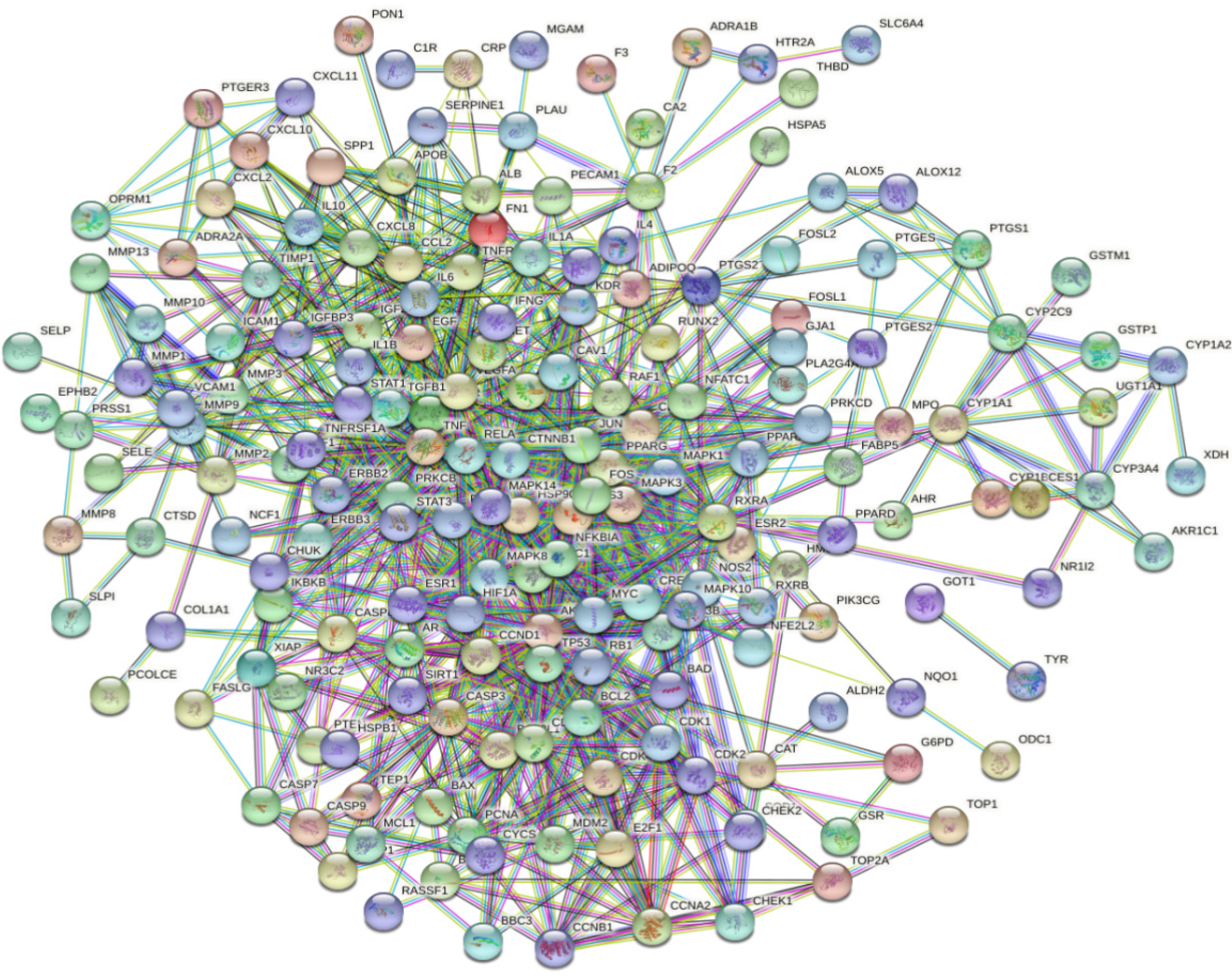
In order to understand the interactions among the UC-related target proteins after JPQCHSR treatment, the effective targets were imported into the STRING database for further analysis. The species default was "Homo sapiens", and the minimum interaction score threshold was set to 0.900. After the isolated nodes were removed, a PPI network graph was constructed (Figure 3). The network graph contained a total of 182 nodes and 1021 edges, with the average node degree value being 11.22. Table 3 shows the information of targets with degree values of ≥ 12. A total of 72 targets including the signal transducer and activator of transcription 3 (STAT3, degree = 49), protein kinase B (AKT1, degree = 46), tumor protein p53 (TP53, degree = 46), mitogen-activated protein kinase 1 (MAPK1, degree = 44), mitogen-activated protein kinase 3 (MAPK3, degree = 43), Jun N-terminal kinase (JNK, degree = 42), and tumor necrosis factor (TNF, degree = 39) might be the core targets of JPQCHSR in the treatment of UC.
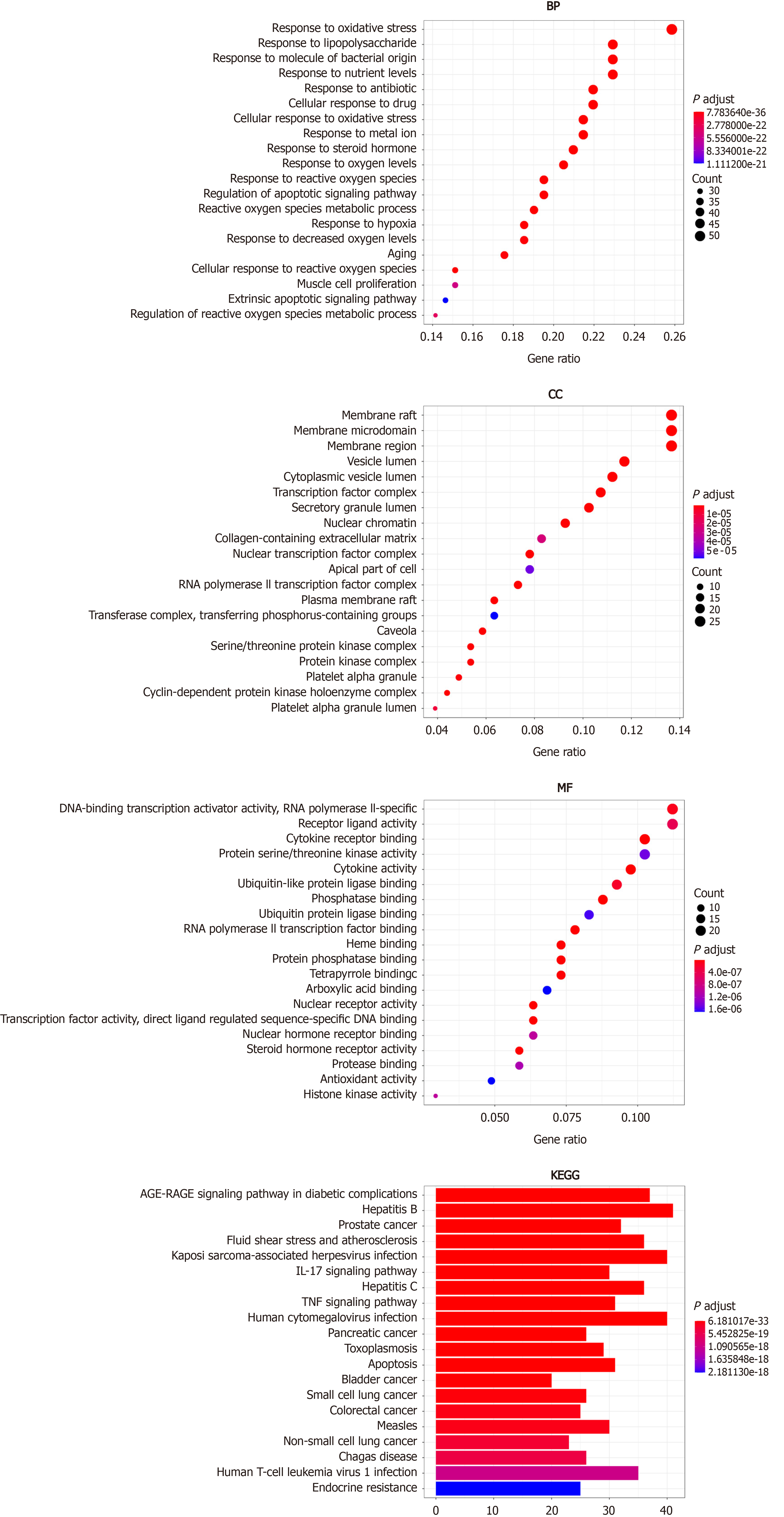
The R 3.6.3 software was used to perform GO and KEGG enrichment analyses on 205 intersection targets, during which a total of 2861 GO terms and 171 KEGG pathways were obtained. The GO enrichment analysis was based on three aspects: Biological process (BP), cellular component (CC), and molecular function (MF). The enriched GO terms included 2681 BPs, 93 CCs, and 171 MFs. With P value set at < 0.05 and according to the number of enriched genes, the top 20 terms were used to draw a bubble chart (Figure 4), in which the vertical axis represents the function annotation, the horizontal axis represents the gene ratio, the bubble size represents the number of enriched genes, and the bubble color represents P value. BPs involved response to oxidative stress, response to lipopolysaccharide, response to molecule of bacterial origin and response to antibiotics; CCs involved membrane raft, membrane microdomain, membrane region, vesicle lumen, and cytoplasmic vesicle lumen; and MFs involved transcription factor activity, preceptor ligand activity, cytokine receptor binding, protein serine/threonine kinase activity, and phosphatase binding.
The results of the KEGG pathway enrichment are displayed as a bar graph, in which the length of the bars represents the number of enriched genes and the color depth refers to P values. As shown in Figure 4, the results of KEGG metabolic pathway enrichment involved infection, apoptosis, nutrition, immunity, inflammation, and other pathways, which included AGE-RAGE signaling pathway, Kaposi sarcoma-associated herpesvirus infection, IL-17 signaling pathway, TNF signaling pathway, and human cytomegalovirus infection.
Molecular docking is a computer-aided virtual screening technology that predicts the binding mode and affinities between ligands and their receptors, in accordance with geometric matching, energy matching, and other principles of interaction. It is generally believed that the docking score > 4.0 indicates that the docking molecules have certain binding activity with the target, the docking score > 5.0 indicates good binding activity, and the docking score > 7.0 indicates strong binding activity[21].
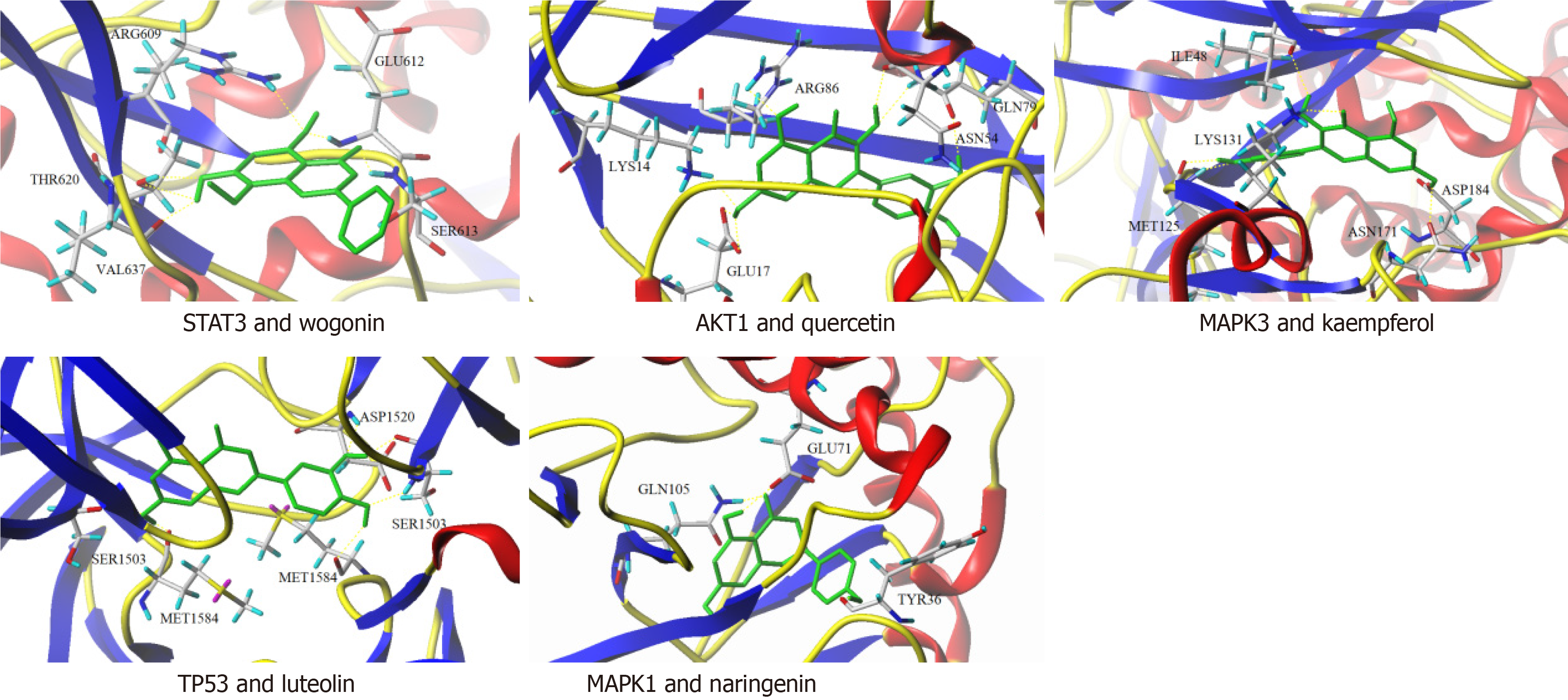
We selected compounds with the top ten “degree” values in Section 2.3 and target proteins with top five “degree” values in Section 2.4 for molecular docking. Except for baicalein, the remaining nine compounds had certain binding activity with AKT1, TP53, MAPK1, and MAPK3; among them, quercetin, wogonin, and naringenin had a docking score higher than 5.0 with AKT1, TP53, MAPK1, and MAPK3, suggesting good binding activity; nobiletin had a docking score higher than 7.0 with AKT1, MAPK1, and MAPK3, showing strong binding activity; all the docking scores of arachidonic acid with AKT1, TP53, MAPK1, and MAPK3 were higher than 7.0, indicating strong binding activity. As shown in the molecular docking pattern diagram (Figure 5), wogonin stably bound to the active site of STAT3 through hydrogen bonding interactions with amino acids including ARG609, GLU612, SER613, THR620, and VAL637 on the STAT3 target protein; through hydrogen bond interactions with LYS14, GLU17, ASN54, ARG86 and GLN79 on the AKT1 target protein, quercetin bound to the active site of AKT1 stably; luteolin stably bound to the active site of TP53 through SER1503, ASP1520, and MET1584 on the TP53 target protein; naringenin stably bound to the active site of MAPK1 through TYR36, GLU71, and GLN105 on the MAPK1 target protein; and kaempferol stably bound to the active site of MAPK3 through ILE48, MET125, LYS131, ASN171, and ASP184 on MAPK3 target protein.
The incidence of UC has been increasing over the past years. It is characterized by a long disease course and complex pathogenic mechanisms. Western medicine-based treatments rely mainly on hormones and biological agents[22-25]. Although these treatments may achieve short-term therapeutic effects, they may cause severe adverse reactions with long-term use. Based on the combination of the data mining method and network pharmacology approach[26-28], we performed comprehensive and systematic prediction for the effect of JPQCHSR in treating UC, which has provided some new insights into future basic research and clinical application of JPQCHSR.
Using network pharmacology approach, we obtained 181 active components and 302 corresponding targets, among which there were 205 UC-related targets. As shown in the visual network of "TCM Drugs-Active Components-Targets-Diseases”, the key components of JPQCHSR in treating UC included quercetin, luteolin, kaempferol, calycosin, naringenin, nobiletin, baicalein, arachidonic acid, and formononetin. Quercetin is a plant flavonoid that is found in many types of fruits and vegetables[29,30]. It has anti-inflammatory, anti-oxidant, and anti-cancer activities[31-34]. Studies have shown that quercetin can lower the expressions of MMP2, MMP9, TLR4, NF-κBp65, and cadherin E; by suppressing the migration and invasion of Caco-2 cells via inhibiting the Toll-like Receptor 4/NF-κB pathway, quercetin may be effective in treating colitis[35]. Luteolin is a natural antioxidant and has the functions of scavenging oxygen free radicals and protecting cells. Li et al[36] found that luteolin could significantly reduce the expressions of inflammatory factors such as iNOS, TNF-α and IL-6 and increase the activities of superoxide dismutase and catalase in colon tissues; it was expected that luteolin might suppress experimental colitis via the Nrf2 pathway. In addition, luteolin can also exert its anti-inflammatory, anti-apoptotic, and anti-autophagy effects by inhibiting JNK1/2, p38, PI3K/AKT, NF-κB, and STAT3 pathways and inducing ERK1/2, thereby playing a role in the treatment of colitis[37]. Kaempferol is a flavonoid compound with anti-apoptosis, anti-inflammatory, and other pharmacological activities. It can play an anti-osteoarthritis role by down-regulating the expression of miR-146a[38].
As shown in the PPI network, 72 targets including STAT3, AKT1, TP53, MAPK1, MAPK3, JUN, and TNF may be the core targets of JPQCHSR in treating UC. Signal Transducer and Activator of Transcription (STAT) pathway is essential for cell proliferation and differentiation[39] and is involved in the pathogenesis of colorectal cancer. Research has shown that STAT3 expression in T cells, macrophages, and epithelial cells in patients with colitis is directly related to the severity of inflammation[40]. TP53, a tumor suppressor gene, is closely related to the occurrence of colon cancer and may serve as a key target for UC prevention and treatment[41,42].
Our enrichment analyses showed that the effective targets were involved in many BPs including response to oxidative stress, response to lipopolysaccharide, molecular response to bacterial origin, and response to antibiotics; they also participated in membrane rafts, membrane microdomains, membrane region, vesicle lumen, cytoplasmic vesicle lumen, and other CCs; finally, they were involved in transcription factor activity, preceptor ligand activity, cytokine receptor binding, protein serine/ threonine kinase activity, phosphatase binding, and other MFs. The 205 effective targets exerted their effects in treating UC mainly through AGE-RAGE signaling pathway, Kaposi sarcoma-associated herpesvirus infection, IL-17 signaling pathway, TNF signaling pathway, and human cytomegalovirus infection, which also reflected the mechanisms of TCM drugs in treating diseases via multiple components, multiple targets, and multiple pathways. The IL-17 pathway is involved in autoimmune diseases, self-defense, and regulation of autoimmune balance[43-46]. As a strong proinflammatory cytokine, IL-17 can increase cell permeability and promote the production of other pro-inflammatory factors and chemokines, thus playing an important role in the pathogenesis of UC[47-52].
Molecular docking showed that the active compounds of JPQCHSR for treating UC had certain affinities with the core targets. The active compounds interacted with the amino acids of the target proteins through hydrogen bonding, thereby stably binding to the active sites of the target proteins.
In summary, using the network pharmacology and molecular docking technology, we predicted that the active components such as quercetin, luteolin, and kaempferol in JPQCHSR act on targets including STAT3, AKT1, TP53, MAPK1, MAPK3, JUN, and TNF via IL-17, TNF, and other signaling pathways to reduce the expressions of inflammatory factors and repair intestinal mucosal damage, thereby exerting their roles in the treatment UC.
Traditional Chinese medicine has played an important role in the treatment of ulcerative colitis (UC), but the specific mechanism of action has not been clarified, which needs further research.
To provide objective basis for the treatment of UC with traditional Chinese medicine.
To investigate the potential mechanism of Jianpi Qingchang Huashi Recipe (JPQCHSR) for the treatment of UC based on network pharmacology and molecular docking.
Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform was used to extract the active components and action targets of JPQCHSR.
Through database analysis, a total of 181 active components, 302 targets and 205 therapeutic targets were obtained for JPQCHSR. The key compounds include quercetin, luteolin, kaempferol, etc. The core targets involved STAT3, AKT1, TP53, MAPK1, MAPK3, JUN, TNF, etc. Total 2861 items were obtained by GO enrichment analysis, and 171 items were obtained by KEGG pathway enrichment analysis. The results of molecular docking showed that the key active components in JPQCHSR had certain affinity with the core target.
The treatment of UC with JPQCHSR is a complex process of multi-component, multi-target and multi-pathway regulation.
Traditional Chinese medicine has a huge potential mechanism in the treatment of UC.
Manuscript source: Unsolicited manuscript
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Nakaji K S-Editor: Gao CC L-Editor: Filipodia P-Editor: Li JH
| 1. | Gajendran M, Loganathan P, Jimenez G, Catinella AP, Ng N, Umapathy C, Ziade N, Hashash JG. A comprehensive review and update on ulcerative colitis. Dis Mon. 2019;65:100851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 343] [Article Influence: 57.2] [Reference Citation Analysis (0)] |
| 2. | Koning M, Ailabouni R, Gearry RB, Frampton CM, Barclay ML. Use and predictors of oral complementary and alternative medicine by patients with inflammatory bowel disease: a population-based, case-control study. Inflamm Bowel Dis. 2013;19:767-778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 3. | Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel JF. Ulcerative colitis. Lancet. 2017;389:1756-1770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2199] [Cited by in RCA: 2487] [Article Influence: 310.9] [Reference Citation Analysis (2)] |
| 4. | Porter RJ, Kalla R, Ho GT. Ulcerative colitis: Recent advances in the understanding of disease pathogenesis. F1000Res. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 146] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 5. | Weizman AV, Ahn E, Thanabalan R, Leung W, Croitoru K, Silverberg MS, Steinhart AH, Nguyen GC. Characterisation of complementary and alternative medicine use and its impact on medication adherence in inflammatory bowel disease. Aliment Pharmacol Ther. 2012;35:342-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 89] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 6. | Limketkai BN, Gordon M, Mutlu EA, De Silva PS, Lewis JD. Diet Therapy for Inflammatory Bowel Diseases: A Call to the Dining Table. Inflamm Bowel Dis. 2020;26:510-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 7. | Opheim R, Hoivik ML, Solberg IC, Moum B; IBSEN Study Group. Complementary and alternative medicine in patients with inflammatory bowel disease: the results of a population-based inception cohort study (IBSEN). J Crohns Colitis. 2012;6:345-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 8. | Dossett ML, Davis RB, Lembo AJ, Yeh GY. Complementary and alternative medicine use by US adults with gastrointestinal conditions: Results from the 2012 National Health Interview Survey. Am J Gastroenterol. 2014;109:1705-1711. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 9. | Zheng L, Zhang YL, Dai YC, Chen X, Chen DL, Dai YT, Tang ZP. Jianpi Qingchang decoction alleviates ulcerative colitis by inhibiting nuclear factor-κB activation. World J Gastroenterol. 2017;23:1180-1188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 42] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 10. | Chen YL, Zheng YY, Dai YC, Zhang YL, Tang ZP. Systems pharmacology approach reveals protective mechanisms of Jian-Pi Qing-Chang decoction on ulcerative colitis. World J Gastroenterol. 2019;25:2603-2622. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 16] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 11. | Dai YC, Zheng L, Zhang YL, Chen X, Chen DL, Wang LJ, Tang ZP. Jianpi Qingchang decoction regulates intestinal motility of dextran sulfate sodium-induced colitis through reducing autophagy of interstitial cells of Cajal. World J Gastroenterol. 2017;23:4724-4734. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 31] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 12. | Wu M, Li P, An Y, Ren J, Yan D, Cui J, Li D, Li M, Wang M, Zhong G. Phloretin ameliorates dextran sulfate sodium-induced ulcerative colitis in mice by regulating the gut microbiota. Pharmacol Res. 2019;150:104489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 190] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 13. | Zhang R, Zhu X, Bai H, Ning K. Network Pharmacology Databases for Traditional Chinese Medicine: Review and Assessment. Front Pharmacol. 2019;10:123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 352] [Cited by in RCA: 771] [Article Influence: 128.5] [Reference Citation Analysis (0)] |
| 14. | Saikia S, Bordoloi M. Molecular Docking: Challenges, Advances and its Use in Drug Discovery Perspective. Curr Drug Targets. 2019;20:501-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 282] [Article Influence: 56.4] [Reference Citation Analysis (0)] |
| 15. | Kataria R, Khatkar A. Molecular Docking of Natural Phenolic Compounds for the Screening of Urease Inhibitors. Curr Pharm Biotechnol. 2019;20:410-421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 16. | Liu CX, Lui R, Fan HR, Xiao XF, Chen XP, Xu HY, Lin YP. Network Pharmacology Bridges Traditional Application and Modern Development of Traditional Chinese Medicine. Chin Herb Med (English). 2015;7:3-17. |
| 17. | Ferreira LG, Dos Santos RN, Oliva G, Andricopulo AD. Molecular docking and structure-based drug design strategies. Molecules. 2015;20:13384-13421. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1277] [Cited by in RCA: 1177] [Article Influence: 117.7] [Reference Citation Analysis (0)] |
| 18. | Ru J, Li P, Wang J, Zhou W, Li B, Huang C, Guo Z, Tao W, Yang Y, Xu X, Li Y, Wang Y, Yang L. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1623] [Cited by in RCA: 3148] [Article Influence: 286.2] [Reference Citation Analysis (0)] |
| 19. | Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. jvenn: an interactive Venn diagram viewer. BMC Bioinformatics. 2014;15:293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 822] [Cited by in RCA: 1544] [Article Influence: 140.4] [Reference Citation Analysis (0)] |
| 20. | Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11591] [Cited by in RCA: 22257] [Article Influence: 1712.1] [Reference Citation Analysis (0)] |
| 21. | Lohning AE, Levonis SM, Williams-Noonan B, Schweiker SS. A Practical Guide to Molecular Docking and Homology Modelling for Medicinal Chemists. Curr Top Med Chem. 2017;17:2023-2040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 95] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 22. | Li S, Zhang B. Traditional Chinese medicine network pharmacology: theory, methodology and application. Chin J Nat Med. 2013;11:110-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 627] [Article Influence: 57.0] [Reference Citation Analysis (0)] |
| 23. | Wu CW, Lu L, Liang SW, Chen C, Wang SM. [Application of drug-target prediction technology in network pharmacology of traditional Chinese medicine]. Zhongguo Zhong Yao Za Zhi. 2016;41:377-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 24. | Yang M, Chen JL, Xu LW, Ji G. Navigating traditional chinese medicine network pharmacology and computational tools. Evid Based Complement Alternat Med. 2013;2013:731969. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 75] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 25. | Huo MQ, Peng S, Ren Y, Shu Z, Zhang YL, Qiao YJ. [Discovery and application of traditional Chinese medicine efficacy markers based on systematic traditional Chinese medicine]. Zhongguo Zhong Yao Za Zhi. 2020;45:3245-3250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 26. | Hua Z, Zhai FT, Tian J, Gao CF, Xu P, Zhang F, Liu SJ, Dong K, Du XF, Zhang Z, Yang G. Effectiveness and safety of oral Chinese patent medicines as adjuvant treatment for unstable angina pectoris on the national essential drugs list of China: a protocol for a systematic review and network meta-analysis. BMJ Open. 2019;9:e026136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 27. | Zhou J, Wang Q, Xiang Z, Tong Q, Pan J, Wan L, Chen J. Network Pharmacology Analysis of Traditional Chinese Medicine Formula Xiao Ke Yin Shui Treating Type 2 Diabetes Mellitus. Evid Based Complement Alternat Med. 2019;2019:4202563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 28. | Hao da C, Xiao PG. Network pharmacology: a Rosetta Stone for traditional Chinese medicine. Drug Dev Res. 2014;75:299-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 202] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 29. | Zhou C, Liu L, Zhuang J, Wei J, Zhang T, Gao C, Liu C, Li H, Si H, Sun C. A Systems Biology-Based Approach to Uncovering Molecular Mechanisms Underlying Effects of Traditional Chinese Medicine Qingdai in Chronic Myelogenous Leukemia, Involving Integration of Network Pharmacology and Molecular Docking Technology. Med Sci Monit. 2018;24:4305-4316. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 30. | Jarrell JT, Gao L, Cohen DS, Huang X. Network Medicine for Alzheimer's Disease and Traditional Chinese Medicine. Molecules. 2018;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 31. | Luo TT, Lu Y, Yan SK, Xiao X, Rong XL, Guo J. Network Pharmacology in Research of Chinese Medicine Formula: Methodology, Application and Prospective. Chin J Integr Med. 2020;26:72-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 442] [Article Influence: 73.7] [Reference Citation Analysis (0)] |
| 32. | Yuan H, Ma Q, Cui H, Liu G, Zhao X, Li W, Piao G. How Can Synergism of Traditional Medicines Benefit from Network Pharmacology? Molecules. 2017;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 181] [Cited by in RCA: 304] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 33. | Granato M, Rizzello C, Gilardini Montani MS, Cuomo L, Vitillo M, Santarelli R, Gonnella R, D'Orazi G, Faggioni A, Cirone M. Quercetin induces apoptosis and autophagy in primary effusion lymphoma cells by inhibiting PI3K/AKT/mTOR and STAT3 signaling pathways. J Nutr Biochem. 2017;41:124-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 178] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 34. | Tang SM, Deng XT, Zhou J, Li QP, Ge XX, Miao L. Pharmacological basis and new insights of quercetin action in respect to its anti-cancer effects. Biomed Pharmacother. 2020;121:109604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 380] [Article Influence: 63.3] [Reference Citation Analysis (0)] |
| 35. | Han M, Song Y, Zhang X. Quercetin Suppresses the Migration and Invasion in Human Colon Cancer Caco-2 Cells Through Regulating Toll-like Receptor 4/Nuclear Factor-kappa B Pathway. Pharmacogn Mag. 2016;12:S237-S244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 36. | Li Y, Shen L, Luo H. Luteolin ameliorates dextran sulfate sodium-induced colitis in mice possibly through activation of the Nrf2 signaling pathway. Int Immunopharmacol. 2016;40:24-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 83] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 37. | Vukelić I, Detel D, Batičić L, Potočnjak I, Domitrović R. Luteolin ameliorates experimental colitis in mice through ERK-mediated suppression of inflammation, apoptosis and autophagy. Food Chem Toxicol. 2020;145:111680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 47] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 38. | Jiang R, Hao P, Yu G, Liu C, Yu C, Huang Y, Wang Y. Kaempferol protects chondrogenic ATDC5 cells against inflammatory injury triggered by lipopolysaccharide through down-regulating miR-146a. Int Immunopharmacol. 2019;69:373-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 39. | Oda K, Hamanishi J, Matsuo K, Hasegawa K. Genomics to immunotherapy of ovarian clear cell carcinoma: Unique opportunities for management. Gynecol Oncol. 2018;151:381-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 103] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 40. | Kasembeli MM, Bharadwaj U, Robinson P, Tweardy DJ. Contribution of STAT3 to Inflammatory and Fibrotic Diseases and Prospects for its Targeting for Treatment. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 141] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 41. | Lu X, Yu Y, Tan S. p53 expression in patients with ulcerative colitis - associated with dysplasia and carcinoma: a systematic meta-analysis. BMC Gastroenterol. 2017;17:111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 42. | Elmashad NM, Ziada DH, Hasby EA, Mohamed AEM. Immunohistochemical expression of proinflammatory enzyme COX-2 and p53 in ulcerative colitis and its associated dysplasia and colorectal carcinoma. J Microsc Ultrastruct. 2016;4:195-202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 43. | Singh S, Fumery M, Sandborn WJ, Murad MH. Systematic review with network meta-analysis: first- and second-line pharmacotherapy for moderate-severe ulcerative colitis. Aliment Pharmacol Ther. 2018;47:162-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 189] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 44. | Yang H, Li Y, Shen S, Gan D, Han C, Wu J, Wang Z. Network Pharmacology-Based Investigation into the Mechanisms of Quyushengxin Formula for the Treatment of Ulcerative Colitis. Evid Based Complement Alternat Med. 2019;2019:7870424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 45. | Tao W, Xu X, Wang X, Li B, Wang Y, Li Y, Yang L. Network pharmacology-based prediction of the active ingredients and potential targets of Chinese herbal Radix Curcumae formula for application to cardiovascular disease. J Ethnopharmacol. 2013;145:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 368] [Cited by in RCA: 453] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 46. | Wang ZY, Liu JG, Li H, Yang HM. Pharmacological Effects of Active Components of Chinese Herbal Medicine in the Treatment of Alzheimer's Disease: A Review. Am J Chin Med. 2016;44:1525-1541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 122] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 47. | Pan Y, Kong LD. High fructose diet-induced metabolic syndrome: Pathophysiological mechanism and treatment by traditional Chinese medicine. Pharmacol Res. 2018;130:438-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 48. | Xin P, Kuang HX, Li XL, Wang Y, Zhang BM, Bu H, Wang ZB, Meng YH, Wang YH, Wang QH. [Proteomics and its application to determine mechanism of action of traditional Chinese medicine]. Zhongguo Zhong Yao Za Zhi. 2018;43:904-912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 49. | Yoo M, Shin J, Kim H, Kim J, Kang J, Tan AC. Exploring the molecular mechanisms of Traditional Chinese Medicine components using gene expression signatures and connectivity map. Comput Methods Programs Biomed. 2019;174:33-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 50. | Su M, Gong XJ, Zhou X. [Research progress in mechanism of traditional Chinese medicine active ingredients against cervical cancer]. Zhongguo Zhong Yao Za Zhi. 2019;44:675-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 51. | Li L, Liu H, Shi W, Yang J, Xu D, Huang H, Wu L. Insights into the Action Mechanisms of Traditional Chinese Medicine in Osteoarthritis. Evid Based Complement Alternat Med. 2017;2017:5190986. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 52. | Huo X, Lu F, Qiao L, Li G, Zhang Y. A Component Formula of Chinese Medicine for Hypercholesterolemia Based on Virtual Screening and Biology Network. Evid Based Complement Alternat Med. 2018;2018:1854972. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |