Published online Aug 16, 2021. doi: 10.12998/wjcc.v9.i23.6846
Peer-review started: April 14, 2021
First decision: May 11, 2021
Revised: May 24, 2021
Accepted: June 4, 2021
Article in press: June 4, 2021
Published online: August 16, 2021
Processing time: 113 Days and 11.3 Hours
A palatal radicular groove is an unusual developmental deformity of the tooth, which may serve as a channel linking the periodontal and periapical inflammation, and yet no literature could be obtained analyzing microbiota within the palatal radicular grooves.
Four patients diagnosed with palatal radicular groove and concomitant periodontal-endodontic deformity in permanent maxillary lateral incisors were enrolled in this work. Twelve bacterial samples from 4 patients were collected from different parts of the palatal radicular groove during intentional replantation surgery. Illumina sequencing was performed to analyze the taxonomical composition and microbiome structure inside the palatal grooves, and 1162 operational taxonomic units were obtained. The phyla of Firmicutes and Proteobacteria predominated in most of the samples. An unknown genus from the Bacillaceae family, Lactococcus, and Porphyromonas were the most abundant genera identified. There was no difference in the microbiota richness and diversity in three sections of the groove.
The unique ecological niches inside the palatal grooves harbored bacterial communities that shared some component features of both the endodontic and periodontal infections. The existence of palatal groove may play an interaction bridge between the root apex and tooth cervix and thus impair the outcome of traditional therapeutic methods such as root canal treatment and periodontal management.
Core Tip: Microbial communities dwelling in the palatal grooves are as complex as those related to endodontic and periodontal infections. The existence of palatal groove may bridge interactions between the root apex and dental cervix and thus impair the outcome of traditional therapeutic methods such as root canal treatment and periodontal management.
- Citation: Tan XL, Chen X, Fu YJ, Ye L, Zhang L, Huang DM. Diverse microbiota in palatal radicular groove analyzed by Illumina sequencing: Four case reports. World J Clin Cases 2021; 9(23): 6846-6857
- URL: https://www.wjgnet.com/2307-8960/full/v9/i23/6846.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i23.6846
A palatal radicular groove is an unusual developmental deformity of the tooth, with the groove usually starting near the cingulum of the incisor, running apically, and terminating at various lengths along the root[1]. The existence of palatal grooves is rare, with a prevalence ranging from 2.8%-8.5%[2]. According to previous studies, palatal grooves can be classified into three types based on the depths of grooves generated via cone-beam computed tomographic scanning[3,4]. Type I represents the shallowest grooves, while types II and III of palatal grooves can extrude and affect the relevant shape of the root canal, thus increasing the difficulty of root canal therapy as well as providing extra ecological niches for potential oral pathogens. When encountering beneficial dysbiosis environment in the local area, bacterial inhabitants in these niches tend to invade alongside the groove[5], which may damage the periodontal tissues, generate localized and deep periodontal pocket extending along the palatal groove and consequently cause root surface and root canal infections including pulp necrosis and/or apical periodontitis, and finally present the concomitant periodontal-endodontic deformity.
Due to the close relationship between the microbiota existing on the root surface and the local inflammation around the palatal groove, a comprehensive understanding of the etiology and pathogenesis of the bacterial community composition inside the palatal grooves may be helpful in developing effective prevention and successful treatment strategies for palatal grooves concomitant to periodontal-endodontic deformity[6,7]. Studies on apex microbiome have shown that the apical region of the root canal system drives a more diverse and obligated anaerobe community than the coronal region due to the complex anatomic structures on the apical end and the sharp reduction of oxygen and nutrient gradients from fresh to protein-rich regions[6,8]. It has also been reported that the microbial community presenting in combined endodontic–periodontal lesions is complex and more diverse than previously thought[9]. It is also the case with palatal radicular grooves in terms of structural complexity such as the depths of the groove and periodontal pocket are highly variable in different clinical cases[3,5]. Thus, it is reasonable to extrapolate that the microbial community may be subject to various changes of the palatal grooves. However, the exact etiology of palatal radicular grooves and the difference of microbiota com
Recently, emerging molecular genetic techniques, such as high-throughput sequencing technologies, gradually became the optimal methods to comprehensively characterize microbiota from human ecological niches. This will aid in better understanding of the role of prokaryotes in the pathogenesis of infectious diseases in the oral cavity[9,12].
In this case report, we uncovered the highly polymicrobial communities in the palatal radicular groove samples associated with periodontal-endodontic lesions utilizing the Illumina MiSeq sequencing technology to analyze the microbial compositions inside the palatal grooves to initially provide evidence for unveiling the potential etiology and pathogenesis of these polymicrobial communities as well as reference for optional therapeutic strategy for this kind of disease.
All patients reported to our department with chief complaints of pus discharge in their maxillary incisor regions.
All patients found pus discharge in their maxillary incisor regions 1-4 wk ago.
There was no history of trauma, and all the patients were systemically healthy.
The personal and family history was noncontributory.
Patient information, such as age, sex, probing depth, etc. were accurately recorded in Table 1.
| Patient ID | Age in yr | Sex | Tooth number1 | Groove type | Location ofthe groove | Probing depth of the localized periodontal pocket in mm | Groove’s stop, examined during intentional replantation |
| 1 | 28 | Male | 7 | II | Mesial | 8 | Junction of the medium and apical thirds of the root |
| 2 | 33 | Male | 7 | I | Distal | 10 | Junction of the medium and apical thirds of the root |
| 3 | 36 | Male | 7 | II | Distal | 12 | Root apex |
| 4 | 44 | Female | 7 | I | Distal | 12 | Approaching the root apex (about 3 mm above the apex) |
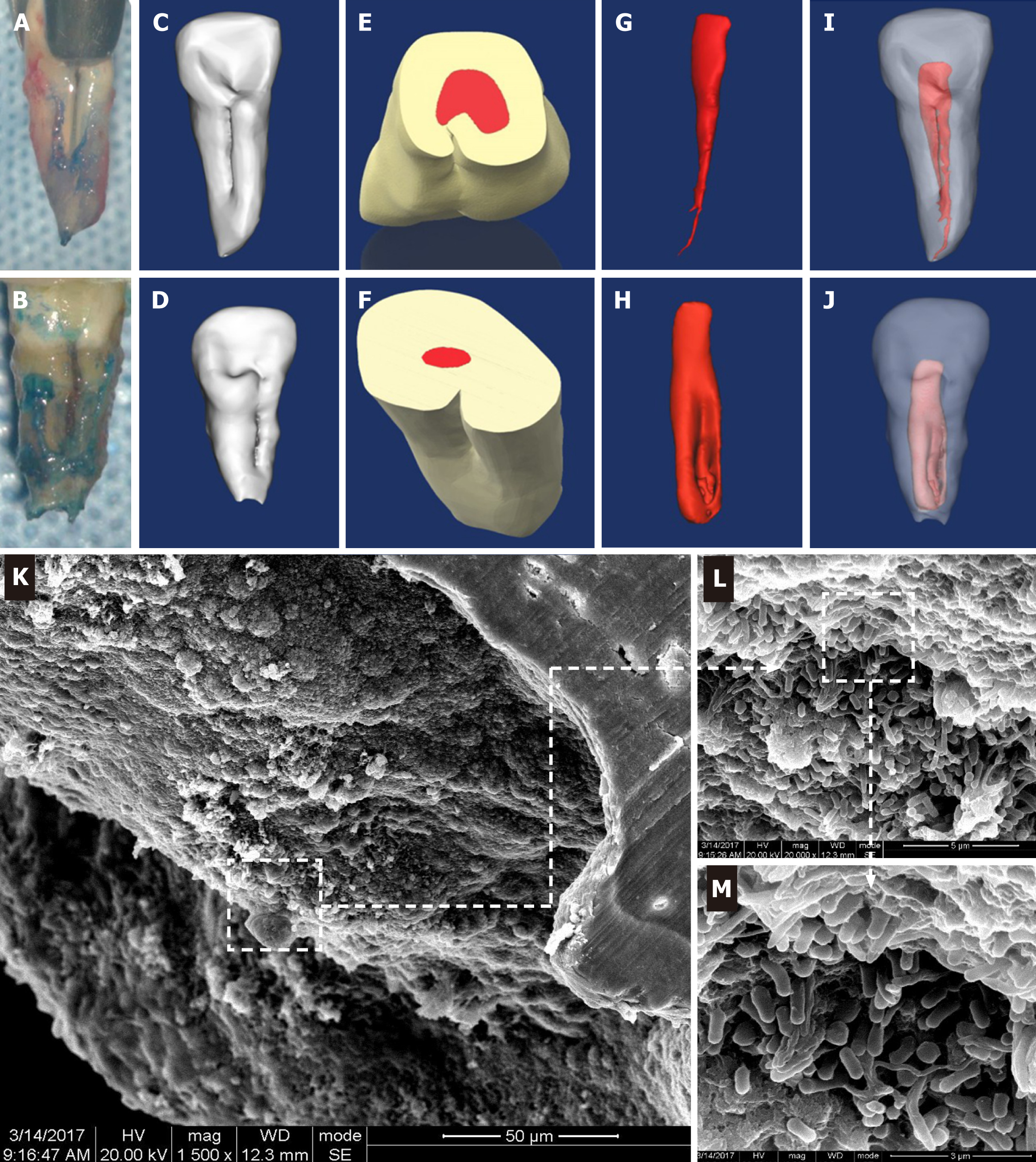
Tooth extraction and sample collection: Intentional replantation was applied to treat these cases after root canal treatment. The tooth and the operating field were carefully decontaminated and disinfected with 1% povidone-iodine followed by 0.1% chlorhexidine solution. After the tooth was minimally invasively extracted from the alveolar bone, the palatal groove was observed directly (Figure 1A and 1B). We trisected the palatal groove region into three equal segments namely the apical 1/3 (GJ), medium 1/3 (GZ), and cervical 1/3 (GG). Twelve bacteria samples were collected from the three regions of the 4 teeth under strict aseptic conditions. Briefly, the root surface was gently washed with sterile saline to remove possible contamination during extraction, then sterile microsurgery curettes were used to scrape debris from the surface of the groove. The obtained debris was then transferred into cryotubes containing TE buffer (10 mmol/L Tris-HCl, 0.1 mmol/L EDTA, pH 7.6) and immediately frozen at −20 °C before further extraction and sequencing procedures.
DNA extraction and sequencing: Genomic DNA of the collected samples was extracted with the Power Soil DNA Kit (MoBio Laboratories, Carlsbad, CA, United States) according to the manufacturer’s instructions. Concentration of the generated DNA was examined by a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, United States). For high-throughput sequencing, the V4 variable regions of the 16S rRNA gene were amplified using primers[13] according to the Illumina 16S Metagenomic Sequencing Library Preparation instructions. The Illumina MiSeq was performed by Personalbio Biomedical Technology Co. Ltd. (Shanghai, China) as described elsewhere[14].
Scanning electron microscope examination: The teeth apexes were then cut during the intentional replantation procedure, immersed into 2.5% glutaraldehyde solution, and stored overnight at 4 °C, followed by gradient dehydration using alcohol at consecutive concentrations of 30%, 50%, 80%, 85%, 90%, 95%, and 100%. Then, the samples were rinsed in phosphate-buffer solution twice, followed by dehydration in a critical point device (Denton Vacuum DCP-1; Denton Vacuum, Moorestown, NJ) and gold sputter coating (Denton Vacuum Desk II; Denton Vacuum). The surface of the root tip was then examined under a scanning electron microscope (SEM) operated at 15 kV (Jeol JSM–5600 LV, Akishima, Tokyo, Japan).
Three-dimensional reconstruction for each case was conducted after cone-beam computed tomographic scanning using Mimics 17.0 software (Materialise Company, Belgium) using a 0.13 mm interval thickness with voxel sizes of 60 mm × 60 mm × 60 mm as described elsewhere[3].
Three-dimensional reconstruction of the infected teeth based on cone-beam computed tomographic scanning revealed that the 4 cases enrolled in this study could be classified into either type I or type II. Both type I (Figure 1C) and type II (Figure 1D) had a palatal groove that started from the cingulum and extended to the apex along the root surface, which was in agreement with the intraoperative awareness during intentional replantation (Figure 1A and 1B), but the groove depth of type II (Figure 1E) was deeper than that of type I (Figure 1F), and the type II groove extruded into the root canal to form a “C” shape. Simultaneously, the shape of the pulp cavity (Figure 1G and 1H) and its relationship with teeth (Figure 1I and 1J) were correspondingly changed.
SEM results suggested that there were highly polymicrobial communities dwelling in the apexes of the infected teeth. Analyses of the SEM photo captures revealed a rugged radicular surface and bacterial biofilms that mainly consisted of kinds of long bacillus, short bacillus, and coccus surrounded and covered the apical foramen and the external radicular surface (Figure 1K-M).
A total of 1162 operational taxonomic units (OTUs) were obtained via the MiSeq, and these OTUs could be taxonomically assigned into 24 bacterial phyla, 51 classes, 91 orders, 185 families, and 339 genera. The numbers of OTUs and subsequently assigned taxa were not statistically different in the three parts of the root (P > 0.05) (Table 2). Of the OTUs, 67% were ubiquitously distributed in the full length of the palatal groove. There were 38 OTUs that could only be located on the apical portion of the root, followed by 33 OTUs exclusive to the cervical segment, and only 4 OTUs uniquely distributed in the middle third of the root.
| Sample ID | Phylum | Class | Order | Family | Genus | Species |
| GJ1 | 6861 | 686 | 679 | 598 | 391 | 92 |
| GZ1 | 513 | 513 | 501 | 452 | 327 | 75 |
| GG1 | 477 | 477 | 465 | 422 | 308 | 56 |
| GJ2 | 622 | 622 | 610 | 531 | 362 | 84 |
| GZ2 | 543 | 543 | 534 | 460 | 313 | 74 |
| GG2 | 583 | 583 | 567 | 500 | 340 | 75 |
| GJ3 | 573 | 573 | 560 | 479 | 287 | 70 |
| GZ3 | 428 | 428 | 419 | 374 | 244 | 52 |
| GG3 | 635 | 635 | 620 | 548 | 348 | 79 |
| GJ4 | 480 | 480 | 469 | 420 | 281 | 60 |
| GZ4 | 444 | 444 | 434 | 377 | 240 | 48 |
| GG4 | 402 | 402 | 391 | 367 | 258 | 41 |
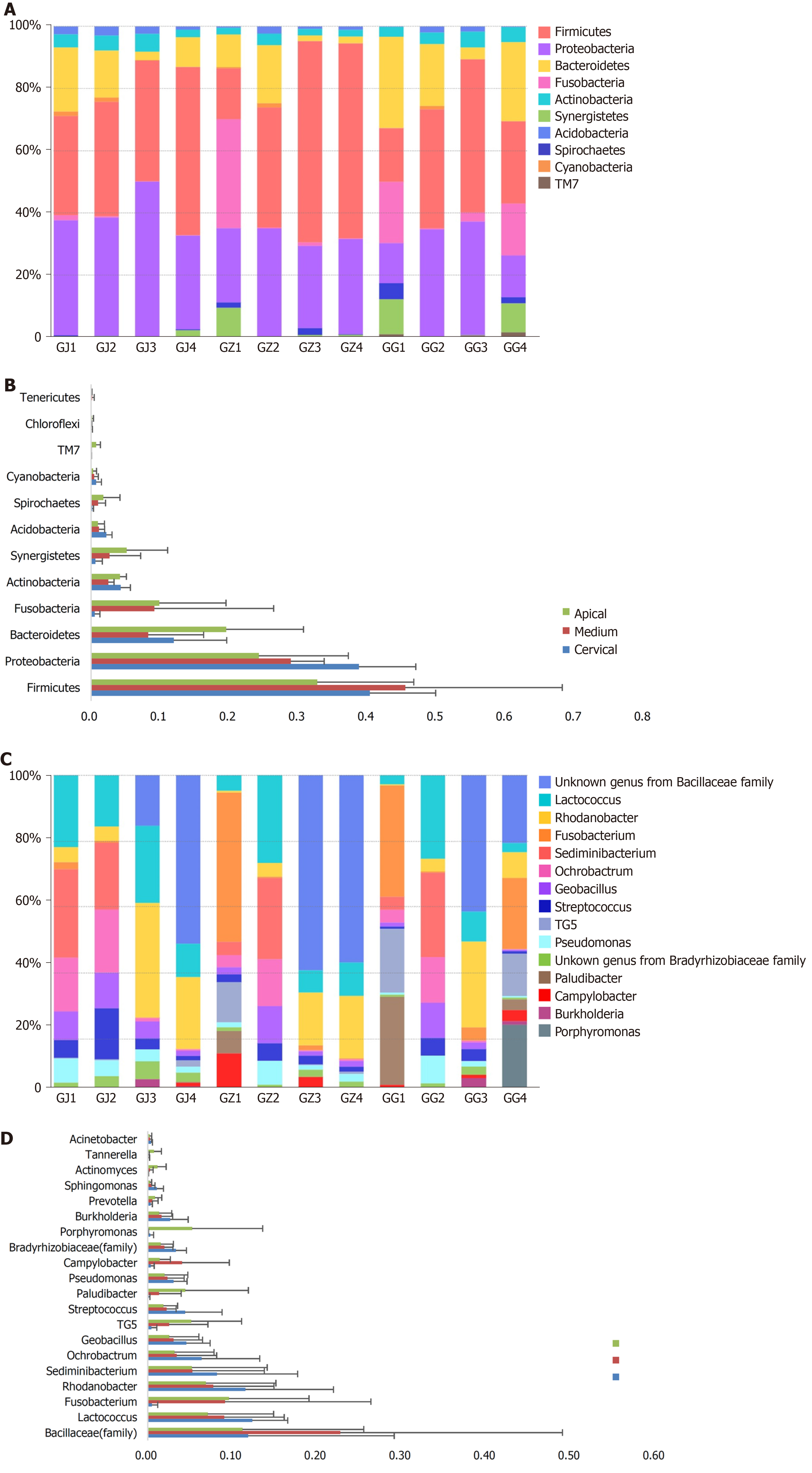
Regarding the phyla division, Firmicutes (average 40.7% ± 15.2%) and Proteobacteria (average 32.2% ± 9.0%) predominated in most of the samples, followed by Bacteroidetes (average 12.2% ± 8.8%) and Fusobacteria (average 5.6 ± 10.8%), while there were also some exceptions that one of the middle 1/3 samples (GZ1) was predominated by Fusobacteria and Proteobacteria, and one of the cervical samples (GG1) harbored mostly the Bacteroidetes and Fusobacteria (Figure 2A). Viewing spatially, cervical portion of the roots harbored 40% of the Proteobacteria detected, much more than the other two parts (Figure 2B). However, Firmicutes was more frequently detected in the middle third samples (GZ). Some phyla including Synergistetes, Spirochaetes and TM7, though less abundant, were mostly exhibited at the apical end (Figure 2B).
Among the identified 339 genera, an unknown genus from the Bacillaceae family (15.2%), genera of Lactococcus (9.5%), Porphyromonas (8.7%), Rhodanobacter (6.4%), and Sediminibacterium (6.2%) were the only five genera that exhibited an abundance above 5%. Lactococcus was the most abundant genus in the apical samples (GJ, average 12.4 ± 3.6%), while the genus from the Bacillaceae family was the most abundant in both middle (GZ) and cervical (GG) samples (average 22.8 ± 22.7% and 11.2 ± 12.4%, respectively) (Figure 2C and 2D). Three genera, namely Peptococcus, Acholeplasma, and Brooklawnia, were found exclusively in the apical samples (GJ) with an incredibly low abundance (< 0.1%). Some genera that are frequently associated with endodontic infections were found below the top 30 abundance: Prevotella ranked number 30 (0.7%), Sphingomonas ranked number 31 (0.6%), Actinomyces ranked number 32 (0.5%), Tannerella ranked number 42 (0.3%), Acinetobacter ranked number 43 (0.3%), Capnocytophaga ranked number 57 (0.1%), and Enterococcus ranked number 181 (below 0.1%).
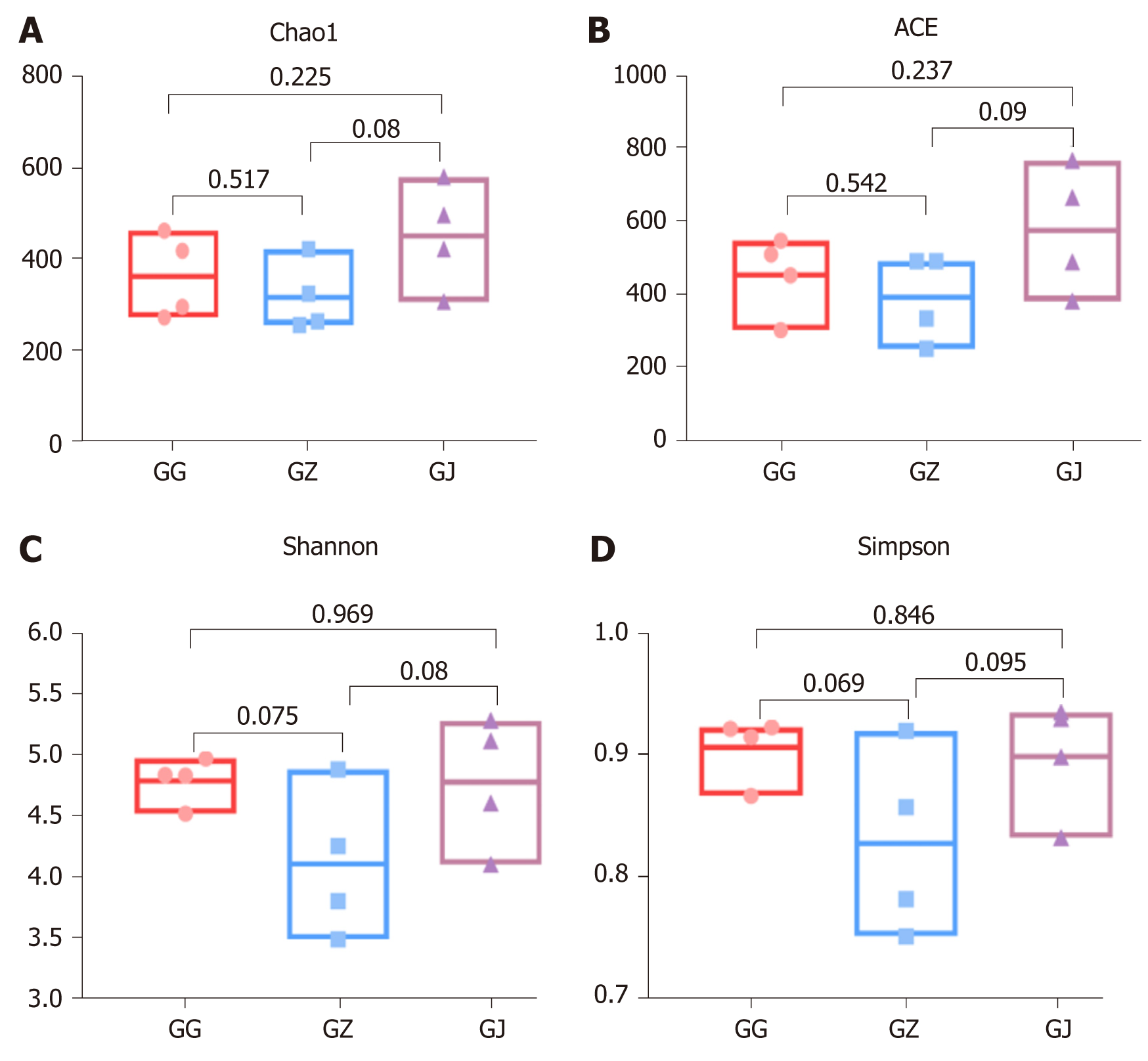
All 690 taxa (final taxon to genus level) were used to perform alpha diversity analyses. Chao1 and angiotensin converting enzyme richness estimates were not significantly different (P > 0.05) among the three segments (Figure 3A and 3B). Likewise, comparison of the Shannon and Simpson indexes indicated that there were no significant differences (P > 0.05) in bacterial community diversity in the cervical, middle, and apical parts of the palatal groove (Figure 3C and 3D).
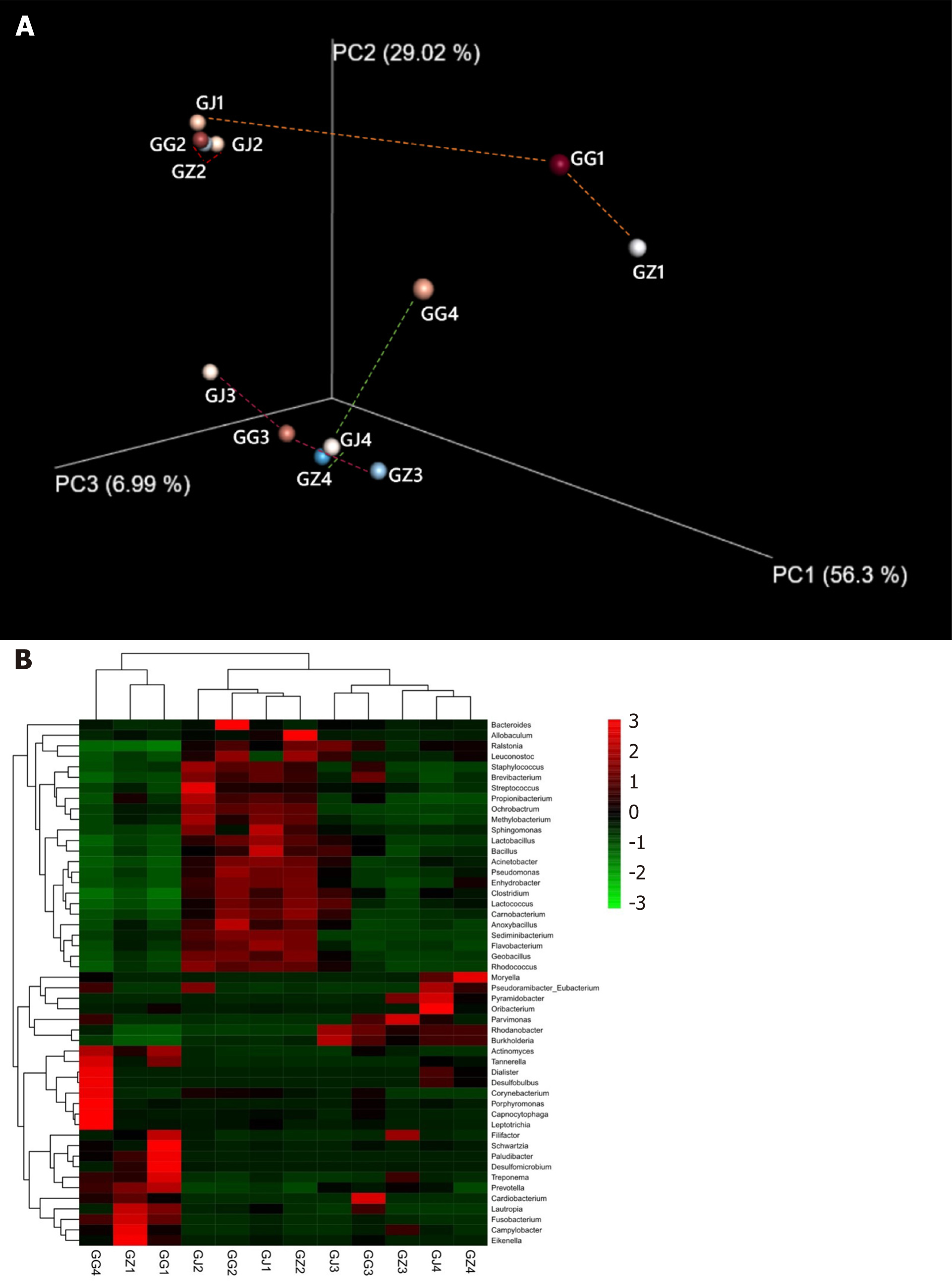
Bacterial community structures in the collected samples were compared using UniFrac based on the phylogenetic relationships of representative reads from different samples, and the weighted UniFrac distances were utilized to conduct a principal coordinate analysis. The results suggested that samples originating from the same root had similar UniFrac distance (0.176 ± 0.110) to the average distance calculated from all samples (0.227 ± 0.090; P > 0.05), meaning that microbial communities distributed in a ubiquitous pattern in all the tested samples regardless of whether the samples were collected from the same tooth or not. However, the data contained a few strong outliers (samples GG4, GG1, and GZ1), which exhibited significantly longer distances to the other samples. Visualization of the UniFrac analysis on phylogenetic distances amongst the different samples showed that patients 2 and 3 tooth samples clustered together, while the apical part of patient 1 (GJ1) and the cervical portion of patient 4 (GG4) were way too far from their counterparts derived from the same root (Figure 4A). Similar results could also be obtained from the gradient heatmap describing abundances of the top 50 abundant genera in different samples (Figure 4B). Samples GJ2, GG2, GJ1, and GZ2 were clustered together as their microbial communities were all rich in genera such as Streptococcus, Anoxybacillus, Sphingomonas, and Lactococcus. Adjacent to the aforementioned four samples, GJ3, GG3, GZ3, GJ4, and GZ4 also grouped together, exhibiting higher contents of Rhodanobacter and Burkholderia. On the other hand, the three outliners, GG4, GZ1, and GG1, were found to be dominated by periodontal infection-related genera including Porphyromonas, Tannerella, Fusobacterium, Treponema, and Prevotella.
Four patients were diagnosed with palatal radicular grooves in permanent maxillary lateral incisors.
Four patients were treated with nonsurgical root canal therapy in combination with intentional replantation in the Department of Conservative Dentistry and Endodontics of the West China Hospital of Stomatology at Sichuan University.
All patients could achieve a soundness prognosis at 1 year follow-up with excellent periradicular healing.
In this study for the first time, we reported the microbial composition and structure inside the niches of palatal grooves. Our results suggested that the ecological niches inside the palatal grooves harbored a distinct and highly polymicrobial community. Palatal radicular grooves are always found to be involved in periodontal-endodontic deformity. Like several other human endogenous infections including endodontic infections and periodontitis[15-17], palatal grooves accommodating a set of highly organized bacterial communities can serve as a favorable channel for the invasion and progression of periodontal-endodontic lesions. But unfortunately, traditional therapeutic measures such as root canal treatment and periodontal management lack effectiveness in treating this kind of disease.
Recently, cases were reported to handle this condition with innovative strategies including root canal treatment followed by intentional replantation[3,18] and enamel matrix augmented periodontal regeneration[19], but the long-term follow-ups are necessary to illustrate the effectiveness of these cases. In this case report, under the intentional replantation procedure, the clinician could clean a vast amount of bacterial plaque, granulation tissues, and caries that were often noted on the surface of the grooves. Thus, intentional replantation is likely to be a sufficient treatment for clearing the apical- and periodontal-infected tissues. However, a long-time follow-up is still needed concerning the possible complications such as alternative absorption under intentional replantation. In this context, the exploration and identification of community profiles involved in palatal radicular grooves may represent an important step towards a better understanding of the pathogenesis of the disease as well as the establishment of more effective therapeutic protocols[20].
With the aid of high-throughput sequencing techniques, researchers are able to characterize the apical periodontitis associated microbiota in a more accurate and effective way, and majority of the results suggested that the most frequently detected bacterial populations during apical inflammations were classified within the phyla of Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Fusobacteria, where Proteo
Intriguingly, in a study conducted by Vengerfeldt et al[20] Proteobacteria was the most abundant phyla in the root sample of an apical abscess patient, and the high proportion of Proteobacteria detected in our case might be part of the reason why all the patients enrolled in this experiment suffered from pus discharge in their maxillary incisor regions. Fusobacterium, which occupied 93% of the bacterial population within the phylum of Fusobacteria detected in our current research, has regularly been associated with periodontal disease and has also been related to polymicrobial infections due to bacterial synergism[17,22]. More specifically, high abundance of Fusobacterium was detected in the cervical portion of patient 1 and 4 and the middle portion of patient 1, while Porphyromonas, another periodontal pathogenic genus and the third abundant genus detected in all the samples, was also enriched in the cervical part of patient 4. The relatively high dominance of genera related to periodontal infection in the region of palatal groove might indicate its microbiological resemblance with the subgingival niches, implying the community shift between these niches, and the potential related polymicrobial synergy might also add difficulties to the control and treatment of the palatal groove deformity associated periodontal-endodontic infections.
The top two most abundant genera were unknown members of the Bacillaceae family and the genus Lactococcus. Bacillus pumilus from the Bacillaceae family has been isolated from endodontic and periodontal lesions, which shows elastin and collagen dissolution activities, and is deemed as a potential virulence pathogen contributing to apical and periodontal tissue damage during inflammation[23]. Another well-known Bacillus species Bacillus subtilis is widely used as the standard strain in the construction of endodontic infection models[24,25]. Lactcoccus lactis has been isolated from endodontic biofilms associated with apical periodontitis and cellulitis and exhibited multidrug resistance[26]. This evidence implies that members isolated from the family of Bacillaceae and the genus of Lactococcus in this case might possess some endodontic and periodontal pathogenic potential as well and are worthy of further investigation.
While most of the genera obtained via MiSeq were endodontic associated bacteria, most of these taxa were at low abundances. Taxa from the genera Rhodanobacter and Sediminibacterium, however, turned out to be the third and fifth most abundant genus in our analysis, respectively. Rhodanobacter and Sediminibacterium have always been isolated from the soil and sediment microbiota[27,28], and we were the first to detect these two genera in endodontic-periodontal infection. Considering the relatively high dominance of these genera in our samples, it is urgent to elaborate the involvements and pathogenesis of Rhodanobacter and Sediminibacterium in endodontic inflammations. Some genera frequently associated with endodontic infections, such as Enterococcus[6], was found in low abundance (0.02%) in the full length of the palatal grooves, which may suggest that the apical bacteria composition has changed in the palatal radicular groove context and indicated potential apical-cervical communication and substance shift by means of the palatal grooves.
It is well known that the more apical the root is, the more complex the networks of canals are, and this phenomenon would result in a sharp decrease of fresh nutrients and oxygen in the apical area of root canals compared to their coronal counterparts[8,12], and thus more obligate anaerobes and fastidious bacteria can be detected in the apical region[29]. However, though SEM examination revealed highly complex biofilms forming on the apical end of the root, we have not found significant differences of bacterial distribution among the apical, middle, and cervical samples in our current research. This might be attributable to the limit of our sample capacity. Meanwhile, another possible explanation could be that the existence of palatal groove serves as a bypath for the apex and periodontal tissue, and this in turn reduces the nutrition and oxygen gradient between the apical and cervical segments, which end up with a similar distribution of microbial communities through the palatal grooves.
Within the limitation of this study, we demonstrated some features of the microbiota in palatal radicular groove with concomitant periodontal-endodontic deformity. The unique ecological niches inside the palatal grooves harbored bacterial communities that shared the composition of both the endodontic and periodontal infection related microbiota and indicated the potential communication between the apical and cervical parts of the groove, which may impair the outcome of traditional therapeutic methods such as root canal treatment and periodontal management. Some taxa such as Firmicutes, Proteobacteria, Fusobacterium, and Rhodanobacter were highlighted for their relatively high abundance, and their accurate functions in the initiation and progression of palatal groove associated endodontic-periodontal infections still needs further exploration.
Manuscript source: Unsolicited manuscript
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ferreira GSA, Liakina V S-Editor: Fan JR L-Editor: Filipodia P-Editor: Zhang YL
| 1. | Lara VS, Consolaro A, Bruce RS. Macroscopic and microscopic analysis of the palato-gingival groove. J Endod. 2000;26:345-350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 29] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 2. | Cho YD, Lee JE, Chung Y, Lee WC, Seol YJ, Lee YM, Rhyu IC, Ku Y. Collaborative Management of Combined Periodontal-endodontic Lesions with a Palatogingival Groove: A Case Series. J Endod. 2017;43:332-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 3. | Tan X, Zhang L, Zhou W, Li Y, Ning J, Chen X, Song D, Zhou X, Huang D. Palatal Radicular Groove Morphology of the Maxillary Incisors: A Case Series Report. J Endod. 2017;43:827-833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 4. | Castelo-Baz P, Ramos-Barbosa I, Martín-Biedma B, Dablanca-Blanco AB, Varela-Patiño P, Blanco-Carrión J. Combined Endodontic-Periodontal Treatment of a Palatogingival Groove. J Endod. 2015;41:1918-1922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 5. | Marsh PD, Zaura E. Dental biofilm: ecological interactions in health and disease. J Clin Periodontol. 2017;44 Suppl 18:S12-S22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 290] [Article Influence: 41.4] [Reference Citation Analysis (0)] |
| 6. | Ozok AR, Persoon IF, Huse SM, Keijser BJ, Wesselink PR, Crielaard W, Zaura E. Ecology of the microbiome of the infected root canal system: a comparison between apical and coronal root segments. Int Endod J. 2012;45:530-541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 80] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 7. | Takahama A, Rocas IN, Faustino ISP, Alves FRF, Azevedo RS, Gomes CC, Araujo-Filho WR, Siqueira JF. Association between bacteria occurring in the apical canal system and expression of bone-resorbing mediators and matrix metalloproteinases in apical periodontitis. Int Endod J. 2018;51:738-746. [RCA] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 8. | Pham LC, van Spanning RJ, Röling WF, Prosperi AC, Terefework Z, Ten Cate JM, Crielaard W, Zaura E. Effects of probiotic Lactobacillus salivarius W24 on the compositional stability of oral microbial communities. Arch Oral Biol. 2009;54:132-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 55] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 9. | Sánchez-Sanhueza G, Bello-Toledo H, González-Rocha G, Gonçalves AT, Valenzuela V, Gallardo-Escárate C. Metagenomic study of bacterial microbiota in persistent endodontic infections using Next-generation sequencing. Int Endod J. 2018;51:1336-1348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 10. | Simon JH, Dogan H, Ceresa LM, Silver GK. The radicular groove: its potential clinical significance. J Endod. 2000;26:295-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 26] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 11. | Kishan KV, Hegde V, Ponnappa KC, Girish TN, Ponappa MC. Management of palato radicular groove in a maxillary lateral incisor. J Nat Sci Biol Med. 2014;5:178-181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 12. | Manoil D, Al-Manei K, Belibasakis GN. A Systematic Review of the Root Canal Microbiota Associated with Apical Periodontitis: Lessons from Next-Generation Sequencing. Proteomics Clin Appl. 2020;e1900060. [RCA] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 13. | Chen H, Hu HY, Chen QQ, Shi ML, Jin RC. Successful start-up of the anammox process: Influence of the seeding strategy on performance and granule properties. Bioresour Technol. 2016;211:594-602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 84] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 14. | Ji L, Zhang Q, Fu X, Zheng L, Dong J, Wang J, Guo S. Feedback of airborne bacterial consortia to haze pollution with different PM2.5 levels in typical mountainous terrain of Jinan, China. Sci Total Environ. 2019;695:133912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 15. | Montagner F, Jacinto RC, Signoretti FG, Sanches PF, Gomes BP. Clustering behavior in microbial communities from acute endodontic infections. J Endod. 2012;38:158-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 16. | Gomes BP, Berber VB, Kokaras AS, Chen T, Paster BJ. Microbiomes of Endodontic-Periodontal Lesions before and after Chemomechanical Preparation. J Endod. 2015;41:1975-1984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 139] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 17. | Mombelli A. Microbial colonization of the periodontal pocket and its significance for periodontal therapy. Periodontol 2000. 2018;76:85-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 224] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 18. | Yan H, Xu N, Wang H, Yu Q. Intentional Replantation with a 2-segment Restoration Method to Treat Severe Palatogingival Grooves in the Maxillary Lateral Incisor: A Report of 3 Cases. J Endod. 2019;45:1543-1549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 19. | Corbella S, Alberti A, Zotti B, Francetti L. Periodontal Regenerative Treatment of Intrabony Defects Associated with Palatal Grooves: A Report of Two Cases. Case Rep Dent. 2019;2019:8093192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 20. | Vengerfeldt V, Špilka K, Saag M, Preem JK, Oopkaup K, Truu J, Mändar R. Highly diverse microbiota in dental root canals in cases of apical periodontitis (data of illumina sequencing). J Endod. 2014;40:1778-1783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 21. | Qian W, Ma T, Ye M, Li Z, Liu Y, Hao P. Microbiota in the apical root canal system of tooth with apical periodontitis. BMC Genomics. 2019;20:189. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 22. | Yun KH, Lee HS, Nam OH, Moon CY, Lee JH, Choi SC. Analysis of bacterial community profiles of endodontically infected primary teeth using pyrosequencing. Int J Paediatr Dent. 2017;27:56-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 23. | Johnson BT, Shaw LN, Nelson DC, Mayo JA. Extracellular proteolytic activities expressed by Bacillus pumilus isolated from endodontic and periodontal lesions. J Med Microbiol. 2008;57:643-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 10] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 24. | Böhm AL, Koch M, Rosiwal S, Burkovski A, Karl M, Grobecker-Karl T. Electrochemical Disinfection of Experimentally Infected Teeth by Boron-Doped Diamond Electrode Treatment. J Clin Med. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 25. | Subha N, Prabhakar V, Koshy M, Abinaya K, Prabu M, Thangavelu L. Efficacy of peracetic acid in rapid disinfection of Resilon and gutta-percha cones compared with sodium hypochlorite, chlorhexidine, and povidone-iodine. J Endod. 2013;39:1261-1264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 26. | Kaboré WAD, Dembélé R, Bagré TS, Konaté A, Boisramé S, Chevalier V, Konsem T, Traoré AS, Barro N. Characterization and Antimicrobial Susceptibility of Lactococcus lactis Isolated from Endodontic Infections in Ouagadougou, Burkina Faso. Dent J (Basel). 2018;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 27. | Dahal RH, Chaudhary DK, Kim J. Rhodanobacter hydrolyticus sp. nov., a novel DNA- and tyrosine-hydrolysing gammaproteobacterium isolated from forest soil. Int J Syst Evol Microbiol. 2018;68:2580-2586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 28. | Song Y, Jia J, Liu D, Choi L, Wang G, Li M. Sediminibacterium roseum sp. nov., isolated from sewage sediment. Int J Syst Evol Microbiol. 2017;67:4674-4679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 29. | Kreft JU. Biofilms promote altruism. Microbiology (Reading). 2004;150:2751-2760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 205] [Article Influence: 9.8] [Reference Citation Analysis (0)] |