Published online Jul 26, 2021. doi: 10.12998/wjcc.v9.i21.6056
Peer-review started: March 14, 2021
First decision: March 27, 2021
Revised: April 6, 2021
Accepted: May 15, 2021
Article in press: May 15, 2021
Published online: July 26, 2021
Processing time: 128 Days and 17.9 Hours
Familial hemophagocytic lymphohistiocytosis type 2 (FHL2) is a rare genetic disorder presenting with fever, hepatosplenomegaly, and pancytopenia secondary to perforin-1 (PRF1) mutation. FLH2 has been described in Chinese but usually presents after 1 year old. We describe a female Chinese neonate with FHL2 secondary to compound heterozygous PRF1 mutation with symptom onset before 1 mo old. We review Chinese FHL2 patients in the literature for compa
A 15-d-old female neonate was referred to our hospital for persistent fever and thrombocytopenia with diffuse petechiae. She was born to a G5P3 mother at 39 wk and 4 d via cesarean section secondary to breech presentation. No resuscitation was required at birth. She was described to be very sleepy with poor appetite since birth. She developed a fever up to 39.5°C at 7 d of life. Leukocytosis, anemia, and thrombocytopenia were detected at a local medical facility
A literature review identified 75 Chinese FHL2 patients, with only five presenting in the first year of life. Missense and frameshift mutations are the most common PRF1 mutations in Chinese, with 24.8% having c.1349C>T followed by 11.6% having c.65delC. The c.658G>C mutation has only been reported once in the literature and our case suggests it can be pathogenic, at least in the presence of another pathogenic mutation such as c.1066C>T.
Core Tip: We report the case of a newborn infant with familial hemophagocytic lymphohistiocytosis who had clinical manifestations by the age of 7 d. The genetic test revealed a compound heterozygous mutation of c.658G>C (p.Gly220Arg) and c.1066C>T (p.Arg356Trp). The two mutations carried by the index cases were not commonly seen variants in Chinese PRF1 mutations. The clinical manifestation of our case strongly suggests that c.658G>C (p.Gly220Arg) is also a pathogenic variant.
- Citation: Bi SH, Jiang LL, Dai LY, Wang LL, Liu GH, Teng RJ. Familial hemophagocytic lymphohistiocytosis type 2 in a female Chinese neonate: A case report and review of the literature. World J Clin Cases 2021; 9(21): 6056-6066
- URL: https://www.wjgnet.com/2307-8960/full/v9/i21/6056.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i21.6056
Familial hemophagocytic lymphohistiocytosis (FHL), a type of hemophagocytic lymphohistiocytosis (HLH), is a rare disorder of the immune system presenting with excessive inflammatory syndrome caused by activated T cells and histiocytosis, which affects almost all ages with 70%-80% presenting in the first year of life[1]. Its clinical manifestations include fever, pancytopenia, hepatosplenomegaly, hypertriglyceridemia, hypofibrinogenemia, hepatitis, neurological symptoms, etc[2]. FHL can be divided into five subtypes (FHL1 to 5) according to the causative genetic mutation. The pathogenic genes of FHL1 remain unknown, while PRF1, UNC13D, STX11, and STXBP2 are specific genes for type 2 to 5, respectively[3]. Nearly 58% of all reported FHL patients have PRF1 gene mutations, with more than 100 FHL2 cases with identified PRF1 variants reported so far[4]. Here we report a case of FHL2 initially presenting with recurrent fever and jaundice by 1 wk of life secondary to a compound heterozygous PRF1 mutation. PRF1 gene mutations in the Chinese population are also reviewed.
A 15-day-old female neonate was referred to our hospital for persistent fever starting from the age of 7 d and thrombocytopenia with diffuse petechiae.
The patient was born to a G5P2 mother at 39 wk and 4 d via cesarean section secondary to breech presentation. No resuscitation was required at birth. She was described to be very sleepy with poor appetite since birth. She developed a fever up to 39.5°C at 7 d of life. Leukocytosis, anemia, and thrombocytopenia were detected at a local medical facility.
No resuscitation was required at birth.
The parents were healthy and not blood-related. The elder sister was 13 years old in good health. Another brother died at the age of 3 mo, without known etiology but manifested with recurrent fever and jaundice. The mother also experienced two spontaneous abortions. One aunt had chronic thrombocytopenia and anemia without an established diagnosis.
Physical examination upon admission revealed hypotonia with poor response to stimuli. Extensive bruises and petechia, pallor, and a soft but flat anterior fontanelle were noticed. She had mild shallow tachypnea without retraction or rales. Her heart rate was 150 per minute without a murmur. Her abdomen was soft but distended with palpable margins of the liver while the tip of the spleen reached 3.5 cm below the costal margin.
Abdominal ultrasound showed gallbladder edema, and moderate amount of abdominal and pelvic effusion. Hemogram showed pancytopenia with a hemoglobin level of 77 g/L, platelet count of 11 × 109/L, and white blood cell count of 2.7 × 109/L with 10.4% neutrophils and 78.5% lymphocytes. Serum ferritin level was 16322.60 ng/mL. Bone marrow examination showed hemophagocytic histiocytes; NK cell killing activity was decreased, and soluble interleukin-2 receptor (sCD25) was 12702 pg/mL. We also detected an elevated D-dimer, abnormal coagulation profile, alanine aminotransferase 1041.0 IU/L, aspartate aminotransferase 2390.0 IU/L, and negative Epstein-Barr virus detection (DNA < 400 copies/mL).
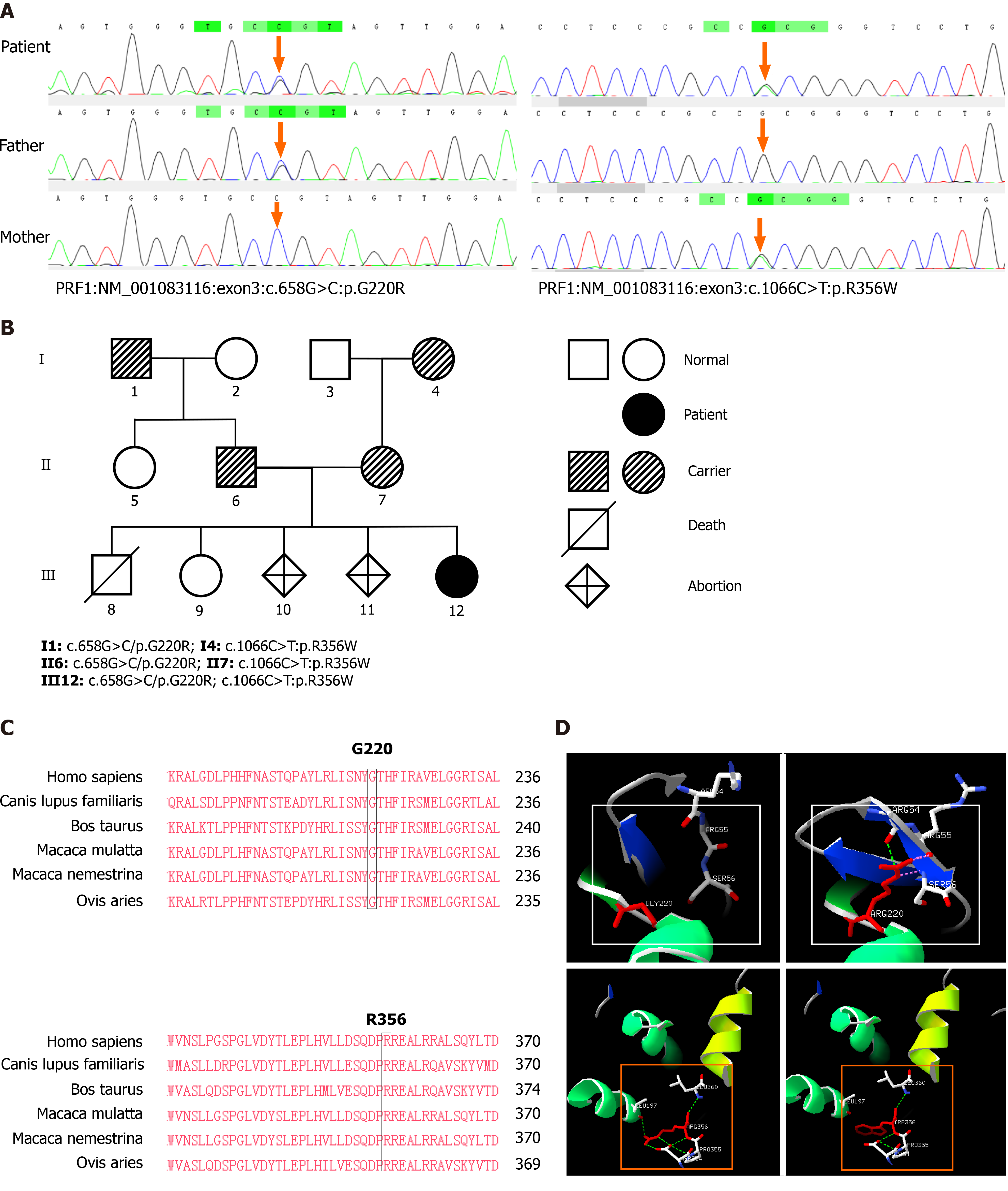
A genetic study showed compound heterozygous PRF1 mutation involving c.658G>C (p.Gly220Arg) and c.1066C>T (p.Arg356Trp). The diagnosis of FHL2 was thus established. Trio-clinical exome sequencing was performed on the extended family members for the purpose of genetic counselling after approval from the hospital ethical committee. Written consent was obtained from the parents for reporting this case. Peripheral blood was collected into an EDTA tube, and genomic DNA was extracted after gene fragmentation, amplification and purification, and library construction; the targeted sequence was captured by the Illumina MiSeq high-throughput sequencing platform, covering approximately 4200 known HLH genes. The reference sequence was the hg19 genome, the reliable mutation spectrum was obtained after the invalid mutations were filtered through biological information analysis, and the suspected pathogenic mutations selected after the hazard prediction analysis of the mutation sites. Sanger verification was carried out. The amplified products were sequenced by ABI 3500 analyzer.
Polyphen_2[5], SIFT[6], and Mutation Taster[7] online analysis platform were used to predict the biological hazardous variants, search public SNP databases for variant sites, check the incidence frequency of variant sites in the normal population, and curate the output determined according to the "ACMG Genetic Variation Classification Standards and Guidelines "(ACMG) Rating[8] for the variation. Mega 7.0 software was used to conduct conservative analysis of the variant sites. Swiss-model online analysis platform (https://swissmodel.expasy.org/) was used to search for the three-dimensional PRF1 protein structure. Protein structure was obtained from the PDB protein structure database (http://www.rcsb.org/) and DeepView software was used for visual display and analysis to obtain the result after optimization.
Results obtained from Sanger sequencing confirmed the father and paternal grandfather were heterozygous for c.658G>C mutation while the mother and maternal grandmother were heterozygous for c.1066C>T mutation (Figure 1A and B). The distribution frequency of c.658G>C in the ExAC database was 0.00005, and the frequency of c.1066C>T was zero. Both mutation sites had been reported in ClinVar and HGMD databases. According to the "ACMG Genetic Variation Classification Standards and Guidelines"[9], c.658G>C is rated as possible pathogenic while c.1066C>T is rated as a pathogenic variant. Judging from our index case with her very early onset clinical presentation, the c.658G>C may be classified as a pathogenic variant, at least in the presence of another pathogenic variant.
The homology analysis indicates that Gly220 and Arg356 are highly conserved between different species (Figure 1C), and the structure simulation indicates that c.658G>C (p.Gly220Arg) mutation makes Arg220 collide with Arg55 and Ser56, which affects the second level of Arg55. The c.1066C>T mutation breaks the hydrogen bond between Trp356 and Leu197, Asp354, Pro355, and the steric hindrance of the benzene ring structure in Trp can change the local spatial conformation and affect protein function (Figure 1D).
FHL2.
Our patient received only supportive treatment, including vitamin K1 injection, transfusions, anti-infective therapy, and hepatoprotective medications such as glycyrrhizin and glutathione. Unfortunately, hematopoietic stem cell transplantation (HSCT) was refused by the family due to financial difficulty.
Our patient passed away at the age of 3 mo at home with fever and jaundice.
Wanfang database, CNKI full-text database, and Pubmed database were used for our literature search. Keywords of "PRF1", "Perforin 1", "Familial Hemophagocytic Lymphohistiocytosis type 2", "Hemophagocytic Lymphohistiocytosis type 2" were used as the input and search for "Lymphohistiocytosis" and "Hemophagocytic Lymphohistiocytosis" to obtain the reported PRF1 gene mutation in the Chinese population.
We identified a total of 75 Chinese FHL2 patients with complete clinical phenotype and genotype reported between 2000 and 2020 (Table 1). The age of onset ranged between 45 days and 56 years, with only six (8%) presenting before the age of 1 year. There were 43 males and 32 females (1.34:1). Overall, 56 PRF1 gene mutations were identified. The hotspot mutation sites were c.1349C>T (24.8%), c.65delC (11.6%), and c.503G>A (9.3%); the main type of mutations was missense mutations (72%), followed by frameshift mutations (24%)[3,10-23].
| Case | Age of onset | Gender | Variant | Amino acid change(s) | Type of mutation | Ref. |
| 1 | 6 yr | M | c.C1066T | p.R356W | Missense mutation | [10] |
| 2 | 19 yr | M | c.C1349T | p.T450M | Missense mutation | |
| 3 | 27 yr | M | c.65delC | p.P22RfsX29 | Frameshift mutation | |
| c.G503A | p.S168N | Missense mutation | ||||
| 4 | 11 yr | M | c.T172C | p.S58P | Missense mutation | |
| c.1083_1094del | p.361_365del | Frameshift mutation | ||||
| 5 | 9 yr | F | c.G218A | p.C73Y | Missense mutation | |
| c.G394A | p.G132R | Missense mutation | ||||
| 6 | 17 yr | F | c.A380G | p.N127S | Missense mutation | |
| c.853_855delAAG | p.K285del | Frameshift mutation | ||||
| 7 | 20 yr | F | c.G503A | p.S168N | Missense mutation | |
| c.C1349T | p.T450M | Missense mutation | ||||
| 8 | 1 yr | F | c.C673T | p.R225W | Missense mutation | |
| c.T1535G | p.L512R | Missense mutation | ||||
| 9 | 2 mo | M | c.853_855delAAG | p.K285del | Frameshift mutation | |
| c.C1349T | p.T450M | Missense mutation | ||||
| 10 | 6 yr | F | c.853_855delAAG | p.K285del | Frameshift mutation | |
| c.C1349T | p.T450M | Missense mutation | ||||
| 11 | 3 yr | M | c.G984A | p.W328X | Nonsense mutation | |
| c.C1349T | p.T450M | Missense mutation | ||||
| 12 | 2 yr | F | c.1090_1091delCT | p.T364fsX93 | Frameshift mutation | |
| c.C1349T | p.T450M | Missense mutation | ||||
| 13 | 1 yr | F | c.C10T | p.R4C | Missense mutation | |
| 14 | 10 yr 9 mo | M | c.65delC | p.P22RfsX29 | Frameshift mutation | |
| 15 | 14 yr | M | c.G503A | p.S168N | Missense mutation | |
| 16 | 13 yr | M | c.G503A | p.S168N | Missense mutation | |
| 17 | 3 yr | M | c.G503A | p.S168N | Missense mutation | |
| 18 | 24 yr | M | c.C1005A | p.S335R | Missense mutation | |
| 19 | 26 yr | M | c.1419delC | p.T474QfsX5 | Frameshift mutation | |
| 20 | 1 yr 6 mo | F | c.1083_1094del | p.361_365del | Frameshift mutation | [11] |
| c.634T>C | p.Y212H | Missense mutation | ||||
| 21 | 4 yr 11 mo | F | c.1349C>T | p.T450M | Missense mutation | |
| c.853_855delAAG | p.K285del | Frameshift mutation | ||||
| 22 | 9 yr | F | c.1349C>T | p.T450M | Missense mutation | |
| c.1306G>T | p.D436Y | Missense mutation | ||||
| 23 | 1 yr 9 mo | M | c.65delC | p.P22RfsX29 | Frameshift mutation | |
| c.148G>A | p.V50M | Missense mutation | ||||
| 24 | 7 yr | M | c.C10T | p.R4C | Missense mutation | [12] |
| p.R33H | p.R33H | Missense mutation | ||||
| 25 | 6 yr | M | p.V50L | p.V50L | Missense mutation | |
| 26 | 4 yr | M | C1465A>T | p.R489W | Missense mutation | |
| 27 | 25 yr | M | c.65delC | p.P22RfsX29 | Frameshift mutation | [13] |
| c.916G>A | p.G306S | Missense mutation | ||||
| 28 | 13 yr | F | c.503G>A | p.S168N | Missense mutation | [14] |
| c.1177T>C | p.C393R | Missense mutation | ||||
| 29 | 8 yr | M | c.1349C>T | p.T450M | Missense mutation | [3] |
| c.445G>A | p.G149S | Missense mutation | ||||
| 30 | 48 yr | M | c.916G>A | p.G306S | Missense mutation | [15] |
| c.822C>T | p.A274= | Synomynous mutation | ||||
| c.900C>T | p.H300= | Synomynous mutation | ||||
| 31 | 2 yr 2 mo | F | c.10 C>T | p.R4C | Missense mutation | [16] |
| 32 | 8 yr 5 mo | c.1349CT | p.T450M | Missense mutation | [17] | |
| c.1273dupT | p.W425fsX457 | Frameshift mutation | ||||
| 33 | 5 yr 2 mo | M | c.305C>T | p.C102F | Missense mutation | [18] |
| 34 | 14 yr 5 mo | F | c.503G>A | p.S168N | Missense mutation | |
| 35 | 5 yr 2 mo | F | c.503G>A | p.S168N | Missense mutation | |
| c.1349C>T | p.T450M | Missense mutation | ||||
| 36 | 45 D | F | c.65delC | p.P22Rfs*29 | Frameshift mutation | [19] |
| c.65delC | p.P22Rfs*29 | Frameshift mutation | ||||
| 37 | 13 yr 8 mo | F | c.1349C>T | p.T450M | Missense mutation | [20] |
| c.1450G>A | c.1450G>A | Missense mutation | ||||
| 38 | 2 mo | M | c.1349C>T | p.T450M | Missense mutation | |
| c.853_855delAAG | p.K285del | Frameshift mutation | ||||
| 39 | 4 yr | F | c.133G>A | c.133G>A | Missense mutation | |
| c.116C>A | c.116C>A | Missense mutation | ||||
| 40 | 4 yr 2 mo | F | c.1349C>T | p.T450M | Missense mutation | |
| c.65delC | c.65delC | Frameshift mutation | ||||
| 41 | 3 yr | F | c.1349G>A | c.1349G>A | Missense mutation | |
| c.218C>T | c.218C>T | Missense mutation | ||||
| 42 | 5 mo | F | c.673C>T | c.673C>T | Missense mutation | |
| c.1535T>G | c.1535T>G | Missense mutation | ||||
| 43 | 4 yr 11 mo | F | c.1349C>T | p.T450M | Missense mutation | |
| c.853_855delAAG | p.K285del | Frameshift mutation | ||||
| 44 | 10 mo | F | c.1349C>T | p.T450M | Missense mutation | |
| c.1090_1091delCT | p.T364fsX93 | Frameshift mutation | ||||
| 45 | 11 yr 3 mo | M | c.1349C>T | p.T450M | Missense mutation | |
| c.503G>A | p.S168N | Missense mutation | ||||
| 46 | 6 yr | F | c.133G>A | c.133G>A | Missense mutation | |
| c.394G>A | c.394G>A | Missense mutation | ||||
| 47 | 8 mo | M | c.562C>G | p.P188A | Missense mutation | [21] |
| 48 | 1 yr | M | c.98G>A | p.R33H | Missense mutation | |
| 49 | 1 yr | M | c.65delC | p.P22RfsX29 | Frameshift mutation | |
| c.1349C>T | p.T450M | Missense mutation | ||||
| 50 | 1 yr | M | c.65delC | p.P22RfsX29 | Frameshift mutation | |
| c.1349C>T | p.T450M | Missense mutation | ||||
| 51 | 2 yr | M | c.1349C>T | p.T450M | Missense mutation | |
| c.1103T>A | p.L368Q | Missense mutation | ||||
| 52 | 2 yr | M | c.1349C>T | p.T450M | Missense mutation | |
| c.1491T>A | p.C497X | Nonsense mutation | ||||
| 53 | 3 yr | M | c.1349C>T | p.T450M | Missense mutation | |
| 54 | 3 yr | M | c.634T>C | p.Y212H | Missense mutation | |
| c.65delC | p.P22RfsX29 | Frameshift mutation | ||||
| 55 | 3 yr | F | c.673C>T | p.R225W | Missense mutation | |
| c.1304C>T | p.T435M | Missense mutation | ||||
| 56 | 4 yr | M | c.742G>A | p.G248R | Missense mutation | |
| 57 | 4 yr | M | c.65delC | p.P22RfsX29 | Frameshift mutation | |
| c.148G>A | p.V50M | Missense mutation | ||||
| 58 | 5 yr | M | c.65delC | p.P22RfsX29 | Frameshift mutation | |
| c.1349C>T | p.T450M | Missense mutation | ||||
| 59 | 6 yr | M | c.394G>A | p.G132R | Missense mutation | |
| 60 | 6 yr | F | c.445G>A | p.G149S | Missense mutation | |
| c.1349C>T | p.T450M | Missense mutation | ||||
| 61 | 9 yr | M | c.1349C>T | p.T450M | Missense mutation | |
| 62 | 10 yr 9 mo | F | c.503G>A | p.S168N | Missense mutation | |
| c.215G>A | p.T72N | Missense mutation | ||||
| 63 | 19 yr | M | c.1066C>T | p.R356W | Missense mutation | |
| 64 | 56 yr | M | c.65delC | p.P22RfsX29 | Frameshift mutation | |
| c.503G>A | p.S168N | Missense mutation | ||||
| 65 | 18 yr | F | c.1168C>T | p.R390X | Nonsense mutation | [22] |
| c.1349C>T | p.T450M | Missense mutation | ||||
| 66 | 19 yr | M | c.1349C>T | p.T450M | Missense mutation | |
| 67 | 18 yr | M | c.172T>C | p.S58P | Missense mutation | |
| c.1083_1094del | p.361_365del | Frameshift mutation | ||||
| 68 | 54 yr | F | c.65delC | p.P22RfsX29 | Frameshift mutation | |
| c.674G>A | p.R225Q | Missense mutation | ||||
| 69 | 27 yr | M | c.503G>A | p.S168N | Missense mutation | |
| c.65delC | p.P22RfsX29 | Frameshift mutation | ||||
| 70 | 18 yr | F | c.1090_109ldel | p.T364fsX93 | Frameshift mutation | |
| c.1349C>T | p.T450M | Missense mutation | ||||
| 71 | 18 yr | M | c.65delC | p.P22RfsX29 | Frameshift mutation | |
| c.1349C>T | p.T450M | Missense mutation | ||||
| 72 | 18 yr | F | c.380A>G | p.N127S | Missense mutation | |
| c.853_855delAAG | p.K285del | Frameshift mutation | ||||
| 73 | 18 yr | F | c.46C>T | p.P16S | Missense mutation | |
| c.1066C>T | p.R356W | Missense mutation | ||||
| 74 | 15 yr | M | c.1349C>T | p.T450M | Missense mutation | [23] |
| c.282C>A | p.N94K | Missense mutation | ||||
| 75 | 12 yr | M | c.1349C>T | p.T450M | Missense mutation | |
| c.282C>A | p.T450M | Missense mutation |
FHL is a kind of HLH with an autosomal or X-linked recessive genetic mutation that is commonly present in infants or young children. About 90% of the children are younger than 2 years old, and the incidence is about 0.12 per 100,000[24]. Our literature review showed that only 8% (6/75) of Chinese FHL2 cases, excluding this reported case, were younger than 1 year old, which is in sharp contrast to the literature. However, we cannot exclude the possibility of reporting bias. The prognosis is known to be worse with a younger age of presentation. The main clinical features are persistent fever, hepatosplenomegaly, pancytopenia, increased ferritin level, and decreased NK cell activity[25], which can be detected in 20%-73% of the patients at initial clinical presentation. Patients with FHL may also present with neurological symptoms, including seizures, facial paralysis, gait instability, or even coma. It is worth noting that FHL is more susceptible to neurologic involvement than the non-familial HLH[11,19].
Our index case had a very early onset, by the age of 7 d, and was the youngest one in the reported Chinese cases. Although there was no proof that the deceased eldest brother was also a case of FHL2, his clinical presentation was highly suggestive of another case of FHL2. The pancytopenia, splenomegaly, increased serum ferritin, and decreased NK cell activity detected by the age of 15 d in our reported case were typical for FHL. Although there was no clinically evident neurologic involvement, the diagnosis of FHL could be established by fulfilling the criteria of HLH-2004 guidelines[25].
Five subtypes of FHL have been identified. Except for FHL1, which has no precise gene location being identified, each subtype is caused by its respective gene mutation such as PRF1, UNC13D, STX11, and STXBP2[11]. Mutations involving PRF1 and UNC13D are more pathogenic than STX11 and STXBP2, as shown in studies involving Italian, Turkish, and Korean cohorts[20]. PRF1 is a gene located in the 10q21-22 region containing three exons. The encoded human perforin protein precursor is mainly expressed in cytotoxic T lymphocytes (CTL) and NK cells, which plays an important role in immune regulation. Patients who carry PRF1 gene defects are vulnerable to infections, autoimmune diseases, and malignant tumors[26,27].
The PRF1 gene mutations reported in Chinese are different from those of other ethnic groups. We identified that c.1349C>T is the most common PRF1 mutation in Chinese FHL2 patients, accounting for 24.8% of all reported cases, followed by the c.65delC mutation, which accounts for 11.6%. The distribution of variants is in sharp contrast to that reported in Turkish patients with the most prevalent one (approximately 74%) being the c.1122G>A (p.W374X), a variant not reported in Chinese FHL2 patients. About one-third of Japanese FHL2 patients have c.1090-1091delCT variant that only occurs in 2% of Chinese FHL2 cases. The hotspot mutation in Chinese FHL2 is in the protein kinase C conserved region 2. This domain is the key region for PRF1 protein to initiate membrane perforation and to maintain cytotoxic activity. The conformational change of the mutated PRF1 reduces the cytotoxic activity of the protein and causes disease[28]. Our case is the second one with c.658G>C (p.Gly220Arg), which has not been confirmed as a pathogenic variant at this moment. However, our patient's rapid progressive manifestation strongly suggests the c.658G>C (p.Gly220Arg) is a true pathogenic variant, at least in the presence of another known pathogenic variant.
FHL2 is a rapidly progressing immune disorder with a high mortality rate if not treated. Most untreated patients will die of severe infection or have multiple organ dysfunction syndrome. FHL2 treatment is by supportive measures and chemotherapy until allogeneic HSCT can be available[25]. A recent comprehensive study of the HLH-94/2004 treatment regimens showed an overall response rate of 72.7% (complete response rate of 55.5%) and a 3-year overall survival rate of 74.7%, with an overall incidence of side effects at 18.2%. Although chemotherapy can temporarily alleviate the symptoms, it cannot eliminate the genetic basis of immune deficiency. HSCT is generally considered the curative treatment for FHL. Studies have shown that patients who fail the transplantation can still have long-term survival[24]. Our patient received only supportive treatment, including vitamin K1 injection, transfusions, anti-infective therapy, and hepatoprotective medications such as glycyrrhizin and glutathione. Unfortunately, HSCT was refused by the family due to financial difficulty. Our patient passed away at the age of 3 mo at home with fever and jaundice.
In summary, we report the case of a newborn infant with FHL2 who had clinical manifestations by the age of 7 d. The genetic test revealed a compound heterozygous mutation of c.658G>C (p.Gly220Arg) and c.1066C>T (p.Arg356Trp). The two mutations carried by the index cases were not commonly seen in Chinese PRF1 mutations. The clinical manifestation of our case strongly suggests that c.658G>C (p.Gly220Arg) is also a pathogenic variant.
We would like to express our thanks to Maggie Teng (Washington University in St. Louis) and Michelle Teng (University of Wisconsin - Madison) for their help in editing the manuscript.
Manuscript source: Unsolicited manuscript
Specialty type: Genetics and heredity
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: El Chazli Y, Garg A S-Editor: Gong ZM L-Editor: Wang TQ P-Editor: Xing YX
| 1. | Gholam C, Grigoriadou S, Gilmour KC, Gaspar HB. Familial haemophagocytic lymphohistiocytosis: advances in the genetic basis, diagnosis and management. Clin Exp Immunol. 2011;163:271-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 117] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 2. | Osińska I, Popko K, Demkow U. Perforin: an important player in immune response. Cent Eur J Immunol. 2014;39:109-115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 124] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 3. | Sun S, Guo X, Zhu Y, Yang X, Li Q, Gao J. [Analysis of clinical phenotype and genetic mutations of a pedigree of familial hemophagocytic lymphohistiocytosis]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2014;31:570-573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 4. | Sheth J, Patel A, Shah R, Bhavsar R, Trivedi S, Sheth F. Rare cause of Hemophagocytic Lymphohistiocytosis due to mutation in PRF1 and SH2D1A genes in two children - a case report with a review. BMC Pediatr. 2019;19:73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 5. | Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248-249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10971] [Cited by in RCA: 10419] [Article Influence: 694.6] [Reference Citation Analysis (0)] |
| 6. | Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2340] [Cited by in RCA: 2191] [Article Influence: 168.5] [Reference Citation Analysis (0)] |
| 7. | Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2345] [Cited by in RCA: 2868] [Article Influence: 260.7] [Reference Citation Analysis (0)] |
| 8. | Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Lyon E, Ward BE; Molecular Subcommittee of the ACMG Laboratory Quality Assurance Committee. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med. 2008;10:294-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 560] [Cited by in RCA: 636] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 9. | Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405-424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19696] [Cited by in RCA: 22240] [Article Influence: 2224.0] [Reference Citation Analysis (0)] |
| 10. | Zhang J, Sun Y, Shi X, Zhang R, Wang Y, Xiao J, Cao J, Gao Z, Wang J, Wu L, Wei W, Wang Z. Genotype characteristics and immunological indicator evaluation of 311 hemophagocytic lymphohistiocytosis cases in China. Orphanet J Rare Dis. 2020;15:112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 11. | Feng WX, Yang XY, Li JW, Gong S, Wu Y, Zhang WH, Han TL, Zhuo XW, Ding CH, Fang F. Neurologic Manifestations as Initial Clinical Presentation of Familial Hemophagocytic Lymphohistiocytosis Type2 Due to PRF1 Mutation in Chinese Pediatric Patients. Front Genet. 2020;11:126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 12. | Zhou XH, Luo JM, Bin Q, Huang XH. [Expression of porforin and granzyme B in familial hemophagocytic lymphohistiocytosis]. Zhonghua Xue Ye Xue Za Zhi. 2016;37:227-232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 13. | Gao L, Dang X, Huang L, Zhu L, Fang M, Zhang J, Xu X, Li T, Zhao L, Wei J, Zhou J. Search for the potential "second-hit" mechanism underlying the onset of familial hemophagocytic lymphohistiocytosis type 2 by whole-exome sequencing analysis. Transl Res. 2016;170:26-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 14. | Liu HX, Tong CR, Wang H, Zhu J, Wang F, Cai P, Teng W, Yang JF, Zhang YL, Lu DP. [An analysis of etiological and genetic factors of a patient with familial hemophagocytic lymphohistiocytosis]. Zhonghua Nei Ke Za Zhi. 2011;50:132-135. [PubMed] |
| 15. | Wang YN, Wang Z, Wang XL. [A case report of adult onset of primary hemophagocytic syndrome with literature review]. Zhonghua Xue Ye Xue Za Zhi. 2012;33:291-293. [PubMed] |
| 16. | Chen KL, Li H, Li JX, Xiong H. Gene mutation associated with hemophagocytic lymphohistiocytosis in children. Linchuang Erke Zazhi. 2017;35:616-619. [DOI] [Full Text] |
| 17. | Ding Q, Guo X, Li Q. [Analysis of PRF1gene variant in a child with late-onset familial hemophagocytic lymphohistiocytosis type 2 and severe central nervous system disease]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2019;36:592-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 18. | Lu G, Xie ZD, Shen KL, Wu RH, Jin YK, Yang S, Liu CY. [Clinical analysis and follow-up study of Epstein-Barr virus associated-hemophagocytic lymphohistiocytosis in childhood]. Zhonghua Er Ke Za Zhi. 2010;48:121-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 19. | Ji Q, Wang G, Xu W. Familial Hemophagocytic Lymphohistiocytosis Type 2 in a Chinese Infant with PRF1 Homozygous Mutation: a Case Report. Clin Lab. 2020;66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 20. | Zhang L, Li Z, Liu W, Ma H, Wang T, Zhang R. Genetic characterization of pediatric primary hemophagocytic lymphohistiocytosis in China: a single-center study. Ann Hematol. 2019;98:2303-2310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 21. | Chen X, Wang F, Zhang Y, Teng W, Wang M, Nie D, Zhou X, Wang D, Zhao H, Zhu P, Liu H. Genetic variant spectrum in 265 Chinese patients with hemophagocytic lymphohistiocytosis: Molecular analyses of PRF1, UNC13D, STX11, STXBP2, SH2D1A, and XIAP. Clin Genet. 2018;94:200-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 22. | Jin Z, Wang Y, Wang J, Zhang J, Wu L, Gao Z, Lai W, Wang Z. Primary hemophagocytic lymphohistiocytosis in adults: the utility of family surveys in a single-center study from China. Orphanet J Rare Dis. 2018;13:17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 23. | Liu C, Li M, Wu X, Yao X, Zhao L. Type 2 familial hemophagocytic lymphohistiocytosis in half brothers: A case report. Medicine (Baltimore). 2018;97:e11577. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 24. | Ma H, Zhang R, Zhang L, Wei A, Zhao X, Yang Y, Liu W, Li Z, Qin M, Wang T. Treatment of pediatric primary hemophagocytic lymphohistiocytosis with the HLH-94/2004 regimens and hematopoietic stem cell transplantation in China. Ann Hematol. 2020;99:2255-2263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (1)] |
| 25. | Henter JI, Horne A, Aricó M, Egeler RM, Filipovich AH, Imashuku S, Ladisch S, McClain K, Webb D, Winiarski J, Janka G. HLH-2004: Diagnostic and therapeutic guidelines for hemophagocytic lymphohistiocytosis. Pediatr Blood Cancer. 2007;48:124-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3075] [Cited by in RCA: 3579] [Article Influence: 198.8] [Reference Citation Analysis (1)] |
| 26. | Stepp SE, Dufourcq-Lagelouse R, Le Deist F, Bhawan S, Certain S, Mathew PA, Henter JI, Bennett M, Fischer A, de Saint Basile G, Kumar V. Perforin gene defects in familial hemophagocytic lymphohistiocytosis. Science. 1999;286:1957-1959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 861] [Cited by in RCA: 856] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 27. | Mian A, Kumari K, Kaushal S, Fazal F, Kodan P, Batra A, Kumar P, Baitha U, Jorwal P, Soneja M, Sharma MC, Biswas A. Fatal familial hemophagocytic lymphohistiocytosis with perforin gene (PRF1) mutation and EBV-associated T-cell lymphoproliferative disorder of the thyroid. Autops Case Rep. 2019;9:e2019101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 28. | An O, Gursoy A, Gurgey A, Keskin O. Structural and functional analysis of perforin mutations in association with clinical data of familial hemophagocytic lymphohistiocytosis type 2 (FHL2) patients. Protein Sci. 2013;22:823-839. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |