Published online Jul 6, 2021. doi: 10.12998/wjcc.v9.i19.4890
Peer-review started: January 23, 2021
First decision: February 28, 2021
Revised: March 14, 2021
Accepted: May 26, 2021
Article in press: May 26, 2021
Published online: July 6, 2021
Processing time: 151 Days and 18.3 Hours
Hepatocellular carcinoma (HCC) is the most common type of cancer among primary malignant tumors of the liver and is a consequential cause of cancer-related deaths worldwide. In recent years, uncovering the molecular mechanisms involved in the development and behavior of this tumor has led to the identification of multiple potential treatment targets. Despite the vast amount of data on this topic, HCC remains a challenging tumor to treat due to its aggressive behavior and complex molecular profile. Therefore, the number of studies aiming to elucidate the mechanisms involved in both carcinogenesis and tumor progression in HCC continues to increase. In this context, the close association of HCC with viral hepatitis has led to numerous studies focusing on the direct or indirect involvement of viruses in the mechanisms contributing to tumor development and behavior. In line with these efforts, this review was undertaken to highlight the current understanding of the molecular mechanisms by which hepatitis B virus (HBV) and hepatitis C virus (HCV) participate in oncogenesis and tumor progression in HCC and summarize new findings. Cumulative evidence indicates that HBV DNA integration promotes genomic instability, resulting in the overexpression of genes related to cancer development, metastasis, and angiogenesis or inactivation of tumor suppressor genes. In addition, genetic variations in HBV itself, especially preS2 deletions, may play a role in malignant transformation. Epigenetic dysregulation caused by both viruses might also contribute to tumor formation and metastasis by modifying the methylation of DNA and histones or altering the expression of microRNAs. Similarly, viral proteins of both HBV and HCV can affect pathways that are important anticancer targets. The effects of these two viruses on the Hippo-Yap-Taz pathway in HCC development and behavior need to be investigated. Additional, comprehensive studies are also needed to determine these viruses' interaction with integrins, farnesoid X, and the apelin system in malignant transformation and tumor progression. Although the relationship of persistent inflammation caused by HBV and HCV hepatitis with carcinogenesis is well defined, further studies are warranted to decipher the relationship among inflammasomes and viruses in carcinogenesis and elucidate the role of virus-microbiota interactions in HCC development and progression.
Core Tip: Hepatocellular carcinoma remains an aggressive tumor, despite extensive studies on its ontogeny and prognosis. Although the occurrence of this tumor in both hepatitis B virus (HBV) and hepatitis C virus (HCV) backgrounds indicates the effect of persistent inflammation in malignant transformation, the viral effects are not limited to the impaired microenvironment. Recent studies have revealed complex mechanisms that reflect concerted and cumulative effects of chronic inflammation-related alter
- Citation: Elpek GO. Molecular pathways in viral hepatitis-associated liver carcinogenesis: An update. World J Clin Cases 2021; 9(19): 4890-4917
- URL: https://www.wjgnet.com/2307-8960/full/v9/i19/4890.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i19.4890
Primary liver cancer is a major global health problem. Its incidence and mortality are increasing, in contrast to the decreasing incidence of many organ cancers worldwide. Indeed, hepatocellular carcinoma (HCC), which accounts for 90% of liver malignancies, is the 5th and 9th most common cancer in men and women, respectively, and is the 3rd leading cause of cancer-related deaths[1,2]. Despite exten
Although risk factors associated with the development of HCC are numerous, the damage caused by hepatitis B virus (HBV) and hepatitis C virus (HCV) infections plays a large role in the development and progression of HCC[1]. In this context, the detection of HBV and HCV in 50% and 30% of patients with HCC, respectively, and the higher prevalence of HCC in endemic regions of the world are strong indicators supporting the relationship between HCC and viral infection[3-6]. In addition, among patients with HBV infection worldwide, higher rates of HCC development and mortality in patients with a coinfection of HBV (24%) or HCV (1%) compared to those with HVB monoinfection also support the contribution of viruses to oncogenesis and tumor behavior[7-10]. Therefore, defining the mechanisms by which viruses participate in molecular events in HCC development and progression is paramount for identifying new treatment targets.
This review aims to provide an overview of basic and clinical studies dealing with the molecular regulation and cell biology of HBV and HCV that affect carcinogenesis and tumor behavior in HCC. Additionally, it addresses the current understanding of existing mechanisms to facilitate the identification of new therapeutic targets and strategies to combat this disease.
This virus is a member of the hepadnaviridae family with cellular tropism for hepatocytes[11-13]. HBV has a compact, partially double-stranded relaxed circular DNA genome (rcDNA) containing four overlapping open reading frames[14]. Viral protein products include three surface proteins [also known as large/pre-S1 (L-HBsAg), medium/pre-S2 (M-HBsAg) and small/major (S-HBsAg)], a core antigen (HBcAg), secreted "e" antigen (HBeAg), viral polymerase (reverse transcriptase, DNA polymerase, and RNaseH activity) and protein X (HBx), which play an important role in HBV pathogenesis and viral transcription[14].
Determination of viral genotypes in HBV has also been found to play an important role in assessing HCC risk. Among the ten subgenotypes of HBV (A to J), HCC risk from highest to lowest is ranked as genotypes C, B, F, D, and A[6,15]. It has been suggested that core promoter mutations (T1762/A1764) convey a higher risk for developing HCC, particularly in young and noncirrhotic subjects infected with HBV genotype B or C[16]. Finally, there is also evidence that in genotype C infections, the development of HCC can be predicted by mutations/deletions in the preS region[17]. Comprehensive information regarding viral replication and other features of HBV is available in many recent studies.
HCV, part of the Flaviviridae family, is a single-stranded RNA virus[18]. HCV infects hepatocytes because they contain both essential entry receptors and suitable cellular host factors (miR-122) for the virus[19,20]. As a function of HBV, three structural proteins (core, E1, and E2), as well as seven nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B), are involved[18]. Recently, two mini-core proteins containing the C-terminal portion of the p21 core nucleocapsid (but lacking the N-terminus) and translated from an alternative open reading frame at amino acids 70 and 91 have been described[21,22]. Their mutations were found to be correlated with an increased risk of HCC, insulin resistance, and failure of interferon therapies. Following viral replication and protein translation, the core protein is collected on a lipid droplet in the ER, allowing the nucleocapsid to form by collecting the encapsulated HCV to release the viral particles associated with very low-density lipoproteins from the cytoplasm[23-25]. These data are all evidence for the dependence of HCV on lipoproteins for survival.
There are 6 main genotypes (1 to 6) of HCV. However, findings regarding the HCV genotype and the risk of HCC development differ[6,10]. In the United States, HCV genotype 3 infections have been observed as conveying a higher risk for developing HCC than genotype 1 infections. Another study from Asia reported an increased risk of genotype 6 infections[26,27]. Although geographical distribution appears to affect these data, further studies are needed to examine the relationship between HCV genotypes and HCC risk.
The integration of HBV DNA into host chromosomes is not an essential step in the HBV life cycle. However, this phenomenon promotes the stimulation of carcinogenesis by causing instability of the host chromosomes and increasing the expression of genes related to cancer development, metastasis, and angiogenesis, or by inactivating tumor suppressor genes. Since the 1980s, when the integration of HBV DNA into the host genome was first reported, the genomic integration regions on the chromosomes have long been thought to be random. In recent years, whole-genome sequencing techniques of tumor tissues have revealed repetitive integration spots. In tumor tissues from patients with HBV-HCV, HBV DNA integration was observed in 80% of cases, and the frequency of integration was significantly higher in HCC than in nontumor areas[28,29]. Moreover, the integration sites in tumors tend to be in the vicinity of important promotor regions, such as CpG islands and telomeres, suggesting that preferential targeting of these regions is related to gene regulation[29-31]. Target genes affected by HBV genome integration include the following: tumor suppressor protein 53; telomerase reverse transcriptase; catenin beta 1 (CTNNB1); retinoic acid receptor beta; catenin delta 2; Axin1; AT-rich interactive domain-containing protein 1A (ARID1A); ARID1B; ARID2; myeloid/Lymphoid or mixed-lineage leukemia 3 (MLL3); MLL4; cyclin E1; cyclin A2; protein tyrosine phosphatase receptor type D; and unc-5 netrin receptor D[32-36]. Neuregulin 3, aryl-hydrocarbon receptor repressor, SUMO-specific peptidase 5, rho-associated coiled-coil containing protein kinase 1, fibronectin, angiopoietin 1 calcium signaling-related genes, platelet-derived growth factor, apoptosis-related genes, and ribosomal protein 60S-like genes are also affected by HBV[37-41].
On the other hand, the integration of HBV DNA into host chromosomes often results in fragmentation, rearrangement, and degradation of the viral genome, leading to structural alterations. The most frequent change occurs with a high frequency of integration in the HBx coding sequence, leading to the formation of chimeric transcripts containing both viral and host sequences and the expression of C-terminally truncated HBx[30,42,43]. This form of HBx has been shown to contribute to expanding cell lines with cancer stem cell-like properties, induce Wnt-5a, and participate in metastasis and cell proliferation[44-47]. It also stimulates oxidative DNA damage and promotes MMP-10 expression[48-50]. Additionally, it can also counteract the growth-suppressive and apoptotic effects of full-length HBx[51,52]. Hybrid transcripts formed by integration with host DNA (HBx-LINE1, HBx-S protein-coding region breaks, and truncated preS/S proteins) may also have an oncogenic effect[34,42,53].
Epigenetic changes include all chromatin changes, which can be reversible according to physiological conditions that include three interactive types, DNA methylation, histone modification, and RNA-related silencing without any changes in the DNA sequence.
DNA methylation: In addition to epigenetic modifications of HBV DNA itself, HBV may also play a role in the methylation of genes that affect the activation of pathways involved in carcinogenesis development. In HBx-transgenic mice, the contribution of hypomethylation of GpG islands to the downregulation of E and N-cadherins involved in epithelial-mesenchymal transition (EMT) was demonstrated[54]. This hypomethylation also affects Smad6 and Kcp, which are involved in Smad-dependent TGF-β and Wnt signaling pathways[54]. Moreover, genes with important roles in HCC development such as p16 (INK4A), GSTP1, CDH1 (E-cadherin), RASSF1A, and p21 (WAF1/CIP1), can be affected by HBV methylation[55]. In cell cultures, decreased transcriptional activity of CD82, which is both a tumor and metastasis suppressor, by HBV-mediated methylation also emphasizes the importance of viral-induced hypomethylation in carcinogenesis[56]. Ankyrin repeat-containing, SH3 domain-containing, and proline-rich-region-containing protein (ASPP) family members are newly identified apoptosis regulatory proteins. Recently, downregulation of the ASPP1 and ASPP2 genes was correlated with HBV-related DNA methylation in HCC-HBV[31]. In addition, HBx, which increases the recruitment of DNA methyltransferase 1 (DNMT1) and DNMT3a to ASPP2, influences the host genome in carcinogenesis. Furthermore, after the suggestion that HBx plays a role by increasing the recruitment of DNMT1 and DNMT3a to the promoter region of ASPP2 during methylation, many studies have shown that the effect of HBx on host promoter methylation is associated with oncogenesis and tumor behavior[31]. The HBx protein contributes to tumor aggressiveness by methylating the CpG1 island that contains the P53 repressor and metastasis-associated protein 1 (MTA1) promoters by recruiting DNMT3a and DNMT3b. Similarly, it participates in oncogenesis by methylating the SOCS-1 tumor suppressor[57,58]. HBx can modulate oncogenesis and tumor behavior by modulating tumor suppressors, such as RASSF1A, protocadherin-10, insulin-like growth factor-binding protein 3, and E-cadherin[59-62].
Histone modifications: For gene regulation, acetylation is important for the posttranslational modifications of histones and involves the transfer of acetyl groups from acetyl-CoA to lysine residues on their N-terminal tail. This phenomenon is regulated by histone acetyltransferases and histone deacetylases (HDACs) and allows the binding of trans-acting factors to promote gene activation. In particular, increased expression of HDACs can contribute to oncogenesis due to their negative effects on suppressor genes[63,64]. In an elegant study, Liu et al[65] demonstrated that HBx binds to methyl-CpG binding domain protein 2 (MBD2) and the transcriptional coactivator CREB1 binding protein (CBP)/p300, leading to activation of insulin-like growth factor 2 (IGF-2) in HCC. They also demonstrated that HBx promotes hypomethylation and acetylation of histone H4 in the IGF-2 promoter and facilitates the formation of the MBD2-HBx-CBP/p300 complex, leading to transcriptional activation.
MicroRNAs in HBV: MicroRNAs (miRs) are small noncoding RNA molecules involved in gene silencing, and their modulation in the liver is associated with hepatocarcinogenesis and tumor behavior, similar to many other cancers. In HBV-HCC, it has been noted that while the expression of miR-224, miR-545, miR-374a, and the miR-17-92 polycistron is increased[66-68], the expression levels of miR-145, miR-199b, let-7a, and miR-152 are decreased[67,69-71], supporting the distinct roles of miRs in these processes. Recently, upregulation of miR21, which is highly expressed in HBV-HCC, was found to be induced by HBx, leading to increased cell proliferation by inhibiting programmed cell death protein-4 and PTEN[72,73]. Li et al[74] suggested that miR21 upregulation is dependent on stimulation of IL-6, facilitating STAT3 activation. In the liver, impaired lipid metabolism results in cellular damage and lipid accumulation involved in oncogenesis. In HCC, the levels of miR205, which regulates the expression of acyl-CoA synthetase long-chain family member 1 (an important lipid metabolism enzyme), decrease, leading to excessive lipid synthesis and accumulation[75,76], and the suppressive effect of HBx on miR205 has also been demonstrated. The results of a recent comprehensive study on the alterations of miRs in the liver suggest that different miRs are involved during the different steps of hepatocarcinogenesis[77]. In HBV-HCC, the expression of miR-150, miR-342-3p, miR-663, miR-20b, miR-92a-3p, miR-376c-3p, and miR-92b was found to be altered. However, in HBV, alterations in miR-98, miR-375, miR-335, miR-199a-5p, and miR-22 have been described. There is evidence that some of these miRs may be potential biomarkers for detecting HCC developing in HBV backgrounds, while others may represent therapeutic targets[69]. However, further studies are warranted to elucidate the biological significance of alterations in miRs during oncogenesis.
HBV DNA polymerase is predisposed to errors, which can result in mutations in all four genes in the viral genome[30]. As described above, HBsAg proteins are in the form of three envelope proteins, L-HBsAg, M-HBsAg, and S-HBsAg. While L-HBsAg is involved in viral binding to the hepatocyte surface, the function of M-HBsAg is not fully understood. Nevertheless, increasingly more findings indicate that M-HBsAg deficiency does not affect viral replication or maturation. Among these three proteins, the most frequently observed S-HBsAg protein has its primary hydrophilic region (MHR) located between amino acids 99 and 169, containing the major antigenic determinant (determinant "a") between amino acids 127-147[30,31,77]. It has been shown that mutations in this determinant can lead to escape from vaccine-induced immunity[78]. In addition, various MHR mutations may lead to the reactivation of viral infection after an immunosuppressive therapy or may reduce the antibody development and antigenicity of HBsAg[79,80]. On the other hand, mutations in the preS sequence have been detected in the serum and tumor tissues of HBV-HCC patients[81,82]. PreS deletions may also disrupt the interaction of L-HBsAg with S-HBsAg[83]. This may lead to retention of the L-HBsAg protein in the ER. Indeed, ground glass hepatocyte formation in HBV hepatitis is caused by the accumulation of the L-HBsAg protein in the ER due to PreS mutation[84-86]. However, the accumulation of L-HBsAg proteins is not limited to ground glass formation but also leads to the induction of ER stress and subsequent oxidative DNA damage followed by malignant transformation with genomic instability[81,82]. Demonstrations of the association among preS or preS2 mutations with increased L-HBsAg expression and borderline dysplastic lesions and HCC development in experimental studies support the involvement of PreS deletions in carcinogenesis[81,87]. Another new mutant formed by the change in the 4th amino acid of the PreS1 protein (from tryptophan to proline or arginine) was found to be associated with an increased risk of HCC[88]. In addition, PreS mutants have also been shown to induce anchorage-independent cellular growth, inflammatory cytokines, the DNA repair gene OGG-1, and the expression of ER chaperones[84,89].
Among the naturally occurring nucleotide mutations involving all four HBV genes, the double nucleotide mutations A1762T and G1764A, which reside in the basal core promoter (BCP), greatly reduce the levels of precore protein mRNA and consequently HBeAg expression[79,90]. This mutant is often found in HBeAg-negative patients with chronic hepatitis[90]. This double mutation transforms a nuclear receptor binding site in BCP into an HNF1 transcription factor binding site and increases HBV replication[91,92]. Mutation of BCP usually occurs in the form of G1896A, which abolishes HBeAg expression by converting codon 28 of the precore sequence into the TAG termination codon[91,93]. This mutation differs across HBV genotypes due to the localization of G1896 in an epsilon (e) structure (stem-loop structure) required for the initiation of pgRNA encapsulation[94]. G1896A mutation is uncommon in HBV genotype A and may lead to destabilization of the (e) structure. However, in other types of HBV, it can increase viral replication by stabilizing the (e) structure[95]. Previous studies have shown that these mutations are associated with high HCC risk, while other BCP mutations (such as T1753C and C1766T) are associated with carcinogenesis[96-98]. In addition, Yan et al[99] showed that these mutations were associated with upregulation of S phase kinase-associated protein 2 and induction of p53 degradation.
HBx and gene expression: HBx does not bind DNA directly but can interact with multiple transcription factors to alter the expression of host and viral genes. This protein, which plays a crucial role in the prevalence of HCC, is a 154 amino acid peptide chain containing a C-terminal transactivation domain consisting of an N-terminal negative regulatory domain with a molecular mass of 17 kDa. HBx, which is formed from HBx gene transcription initiated by integrating viral DNA into the host genome, can be found in different subcellular regions[44]. Therefore, HBx acts differently depending on its subcellular localization within hepatocytes[100]. HBx in the nucleus performs viral replication both by increasing HBV gene expression and preventing HBV gene methylation[101-103]. Within the cytoplasm, HBx affects signaling pathways and transcription factors. On the other hand, it regulates protein degradation, cellular transcription, apoptosis, and cell proliferation by colocalization in the mitochondrial cytoplasm and nucleus[104]. Therefore, it is not surprising that HBx interacts with many factors and pathways directly and indirectly involved in carcinogenesis and tumor progression.
One of HBx’s mechanisms of action is its ability to bind to transcription factors involved in oncogenesis and the progression of HCC. These factors include nuclear factor kappa-Β (NF-κB), RNA polymerase binding protein, transcription factor II B, transcription factor II H, CBP/p300, activating transcription factor 2, and activating protein-2[105]. Furthermore, the effect of HBx on several transcription factors is not limited to being a binding partner and may further affect their function through distinct mechanisms. For instance, HBx not only binds to the cAMP response element binding protein (CREB) but also facilitates the participation of CREB in viral DNA by interfering with protein phosphatase 1 activity[105,106]. Similarly, HBx interferes with SIRT1, allowing the release of β-Catenin, which enables activation of the expression of cancer-promoting genes, such as cyclin-D1 and c-myc[107].
HBx also regulates gene expression through epigenetic modifications. Restriction of the expression of the secreted frizzled-associated proteins SFRP1 and SFRP5, members of the extracellular glycoprotein family, occurs through epigenetic silencing mediated by HBx[44,108,109]. In this process, HBx facilitates the binding of DNMT1 and DNMT3 to the promoters of these genes, leading to their hypermethylation[109]. Suppression of SFRP1 and SFRP5 allows transactivation of Wnt target genes, including c-myc and cyclin D1, consequently promoting EMT and malignant transformation[109]. The decrease in the expression of both SFRP1 and SFRP5 in HBV-HCC is also in line with these findings[108-110]. SUZ12 and ZNF198 are two transcription repressive complexes held together by binding to the long noncoding RNA HOTAIR. SUZ12 constitutes a subunit of polycomb repressor complex 2 (PRC2), and ZNF198 stabilizes the transcription suppressor complex (LSD1, Co-REST, and HDAC1). The effect of HBx on these two transcription suppressors leads to reduced PRC2 and LSD1/Co-REST/HDAC1 complexes of target genes, such as epithelial cell adhesion molecule (EpCAM), which play an important role in tumor development and progression[111,112].
HBx and DNA repair: Cells inhibit cell cycle progression by inactivating cyclin-dependent kinases (CDKs) in response to DNA damage. The absence of such a response can cause genetic instability, mutagenesis, and tumor growth. Accumulating evidence indicates that HBx facilitates the accumulation of DNA damage by interfering with cell cycle checkpoints and DNA repair[31]. HBx also affects DNA repair by competing with XPB/D to bind and sequester p53[113]. An important protein in the base excision repair pathway is human thymine DNA glycosylase (TDG), structurally similar to HBx. Van de Klundert et al[114] suggested that HBx causes the accumulation of DNA damage and cellular transformation by altering or preventing the function of TDG in the DNA repair process. There is also evidence that HBx stimulates CDK2 and CDC2, which play a role in the transition of the cell to S and M phases, forcing the cell to enter the proliferation cycle.
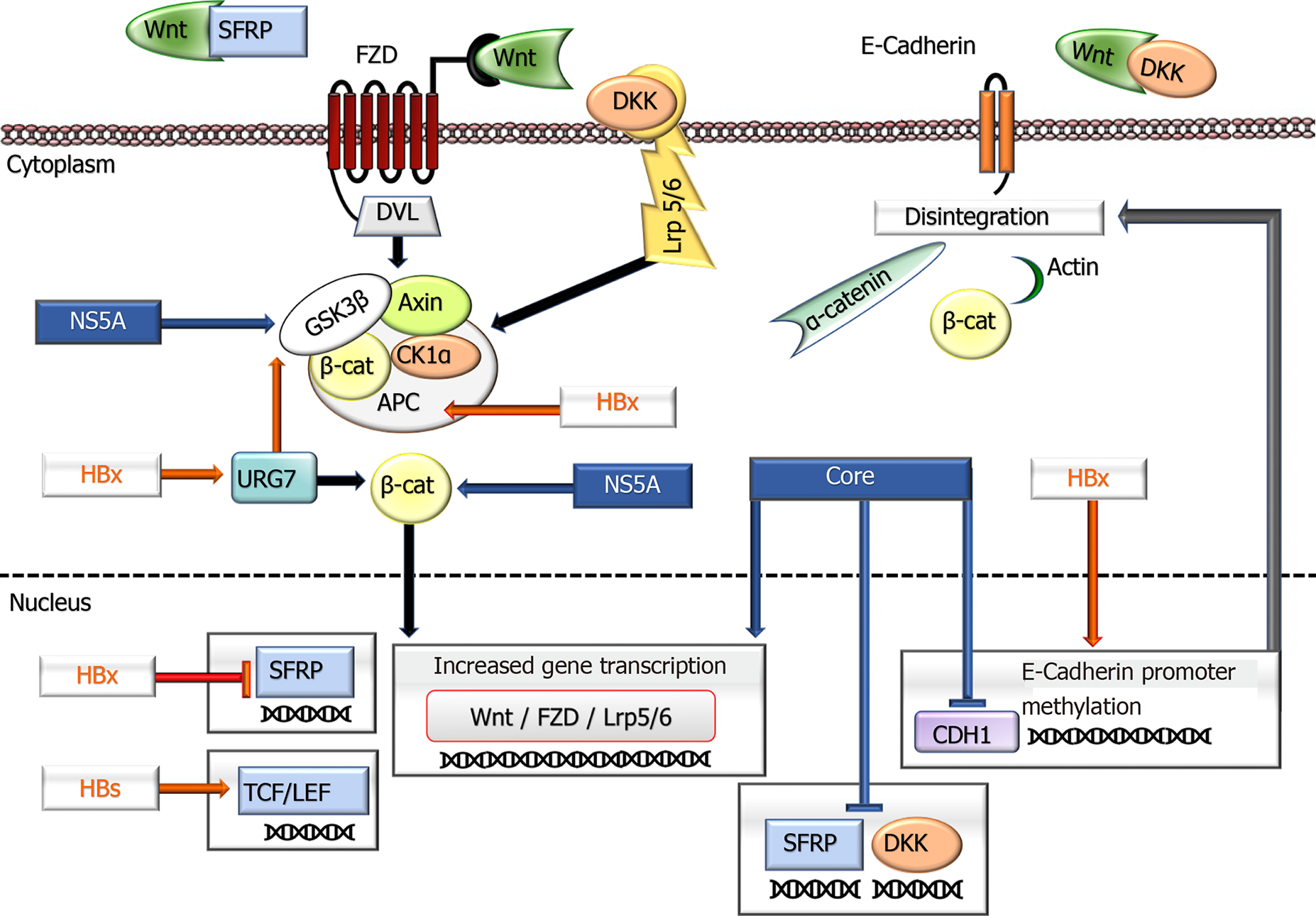
Wnt/β-Catenin pathway: It has been shown that CTNNB1, the gene encoding beta-catenin, is mutated in 40% of HBV-HCC cases[115]. Similarly, activation of Wnt/β-Catenin was noted with loss of function due to hypermethylation in the tumor suppressor APC gene, which is an important component of this pathway[116,117]. Detection of different expression levels of components in the canonical Wnt/β-Catenin pathway in HBV-related HCC (HBV-HCC) has presented findings supporting the role of HBV in oncogenesis. In such cases, higher expression of the regulators of Wnt/β-Catenin (Frizzled 2, Frizzled 7, Segment polarity protein disheveled homolog DVL-3, secreted frizzled-associated protein 4, Wnt-1 inducible signaling pathway protein 1 and fos-associated antigen 1) in tumor tissues compared to nontumoral mucosa and decreases in transducin-like enhancer protein, naked cuticle 1 (NKD1) and NKD2 gene expression indicate notable evidence for the involvement of this pathway in HBV-associated carcinogenesis[109,115-119].
In this context, the HBx protein activates the Wnt pathway by inhibiting E-cadherin in different ways (promoter hypermethylation, changes in SNAIL gene expression, and Src signal activation) and represses SFRP1 and SFRP5[109,115-118]. On the other hand, HBx can disrupt the degradation of β-Catenin through its ability to bind with APC. Again, inhibiting the GSK3b degradation complex and stimulating URG7 causes abnormal activation of the Wnt signaling pathway. As a result, the expression of c-myc, CTFG, and WISP2 increases[120,121]. HBx may also indirectly cause an increase in EpCAM from target genes of this pathway on miRs. It has been suggested that it may contribute to hepatocarcinogenesis, especially with the increase in c-myc onco
HBsAg affects the Wnt pathway by increasing the expression of lymphoid enhancing factor, a transcription factor in this pathway. Differences in the expression of this factor in nontumor areas and tumors (cytoplasmic vs nuclear) indicate the role of HBsAg in the Wnt pathway in carcinogenesis[122]. Viral influences on this pathway are summarized in Figure 1.
NF-κB pathway: NF-κB consists of 5 proteins [NF-κB1 (p105/p50), NF-κB2 (p100/p52), Rel A (p65), cRel and RelB], all of which contain a nuclear localization signal Rel homology region, while some (p65, cRel and RelB) have a transactivation region. More comprehensive information on this factor was presented in a recent review by Shokri et al[123], with viral contributions to the NF-κB pathway summa
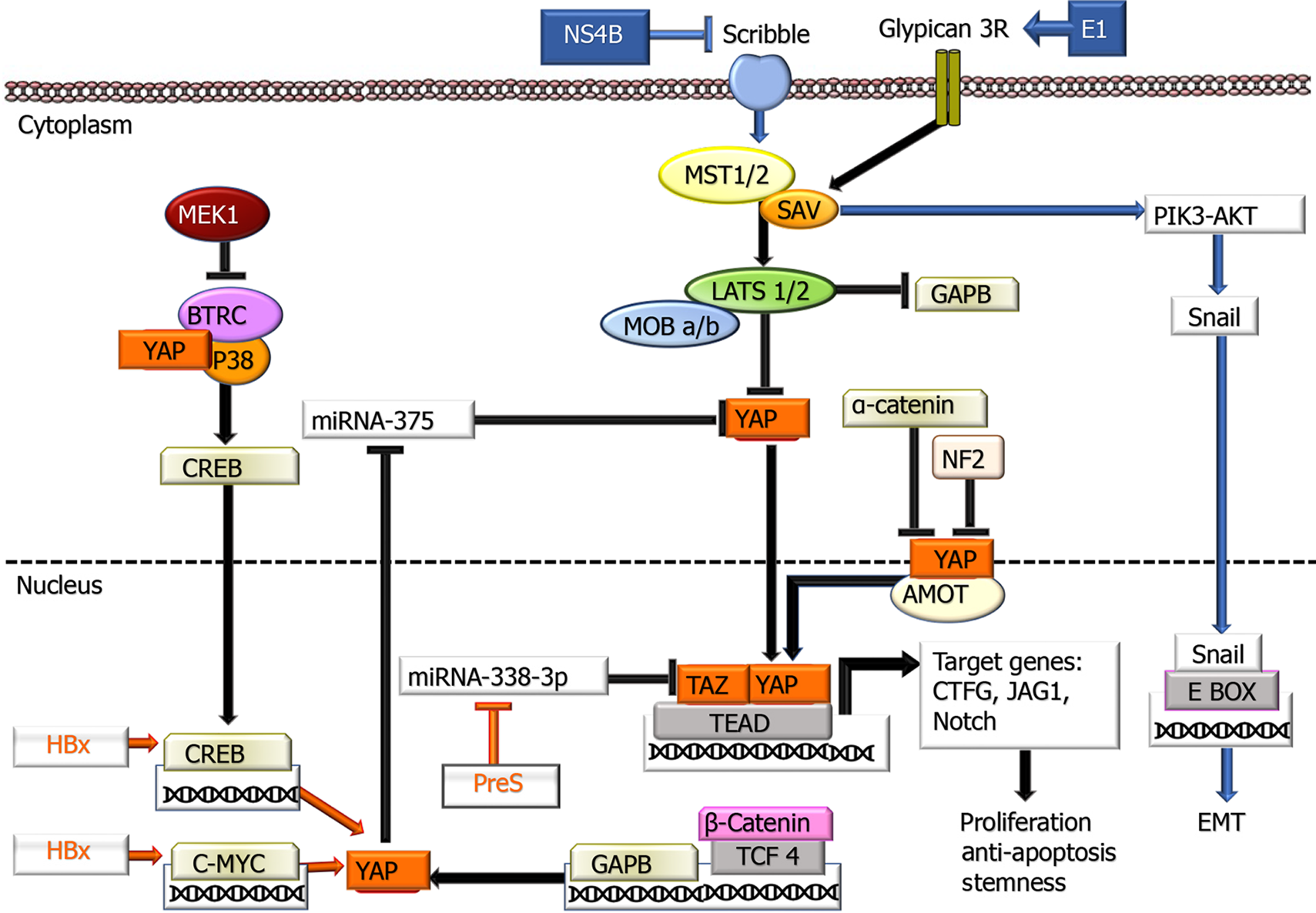
Hippo-YAP/TAZ pathway: In the liver, there is evidence that Hippo-YAP/TAZ inactivation plays a role in fibrosis, hepatocarcinogenesis, and tumor behavior. More detailed information on this factor is presented in a recent review by Moon et al[131], with viral contributions to the Hippo-YAP/TAZ pathway summarized in Figure 3. The primary region of the Hippo pathway, which is considered a critical tumor suppressor, consists of a protein complex. If the Hippo pathway is inactive or suppressed, YAP and TAZ are translocated to the nucleus and regulate the activation of transcription factors that play a role in cell proliferation, survival, miR processing, metastasis development, and maintaining stem cell continuity. Some studies have revealed that HBx expression in HCC tissues correlates with both the miR level and immunohistochemical nuclear expression of YAP[132,133] and its relationship with the cyclic adenosine monophosphate response element binding protein CREB pathway has been revealed[132]. In subsequent studies, there were also data demonstrating that HBx exerts its effect on p53 and ubiquitination of histone H2B, which plays a role in transcription control, through upregulation of FOX-1A and male-specific lethal 2 (MSL2), a ubiquitin E3 Ligase[134]. HBx also increases in YAP 1 by using NEDD with the E3 Ligase HDM2 to protect against ubiquitination and degradation[135]. In HBV-HCC cases, it has been shown that the c-terminal truncated middle surface protein of HBV, which is integrated into 30% of the host genome, increases TAZ activation by suppressing miR338-3p, promoting carcinogenesis[136]. Although these data on preS2, whose role in carcinogenesis has not been fully elucidated, are valuable, further studies are needed to address the effects of HBV on this pathway.
Angiogenesis pathways: In HCC, antitumor cell therapies, such as chemotherapy, are unsuccessful due to tumor cell heterogeneity. This has led to the application of new treatment options targeting relatively stable vascular cells in recent years. Currently, agents targeting angiogenesis are being used in the treatment of HCC. Vascular endothelial growth factor (VEGF) appears to be an important angiogenic factor in HBV-associated hepatocarcinogenesis. Together with VEGF in experimentally induced HBV-HCC mice, the increase in its expression supports this view[137]. In tumor tissues from patients with HBV-HCC, VEGF was also found to be upregulated, together with COX-2, and its expression correlates with microvessel density, which is a marker of angiogenesis[138]. In HBV-HCC, the primary inducer of VEGF expression is HBx. This protein increases VEGF expression in multiple ways in transfected cells and other cell cultures by stabilizing HIF-α or inducing mTOR and IKKβ[139]. In addition to VEGF, it also participates in angiogenesis by promoting increased expression and secretion of angiopoietin-2 (Ang-2) in liver tissue through activation of MAPK[139,140]. HBx may also contribute to HCC angiogenesis through the upregulation of MMPs that promote angiogenesis by disrupting the basement membrane and other extracellular matrix components and allowing endothelial cells to migrate into the surrounding tissue. The correlation between HBx expression and the regulation of MMP-2, MMP-9, MMP-14, and MMP protein levels and activities has been demon
The Pre-S protein has also been reported to enhance VEGF expression, thereby increasing the angiogenic environment produced by the virus[139].
Integrins: The integrin family (ITGs) consists of cell-surface glycoprotein receptors composed of 14a and 8b subunits found in at least 20 species. The combination of these subunits leads to the emergence of more than 100 integrin heterodimers, resulting in a diversity of receptor-ligand combinations that can produce different effects specific to cell adhesion, which play important roles in the invasion and metastatic properties of tumor cells[143,144]. ITGs play two important roles in cancer development and progression: they mediate uncontrolled cell growth and differentiation by transmitting signals from the ECM into the cell and facilitating the invasive properties of tumor cells by changing the content of the surrounding matrix. However, the roles of ITGs in HCC, as in other cancers, are not easy to determine due to the complexity of their structures and functions and require further investigation. Despite the complexity of the functions of ITG, while ITGβ can act as a receptor for collagen, fibronectin, and laminin, ITGβ4 acts solely on laminin by binding with ITGα6[143]. Laminin-induced a6b4 may play a role in tumor formation and progression by affecting multiple signaling pathways, including p53, Ras, RhoA, and epidermal growth factor receptor family, to activate signaling pathways involved in tumorigenesis and metastasis, including PI3K, AKT, and MAPK signaling[144]. In HCC, there is an increase in the expression of some subunits of ITGs (α1, α2, α3, α6, and β1), especially at the tumor margin. Among these, decreased expression of ITG5β1, a negative regulator of fibronectin signal transmission, in poorly differentiated tumors with high metastatic capacity is a finding that supports integrins having differential roles in HCC. The results of several studies have shown that integrin α6β4 plays a role in HCC independent of the differences among hepatitis virus types[143]. More recently, Kim et al[144] observed that the levels of ITGa6, a laminin receptor, were higher in patients and animals with HBV-HCC than in other groups, including HCV-HCC, and this was found to be associated with early migration and invasion of tumor cells. In particular, HBx has been found to be involved in the expression of this glycoprotein, and its suppression reduces the metastatic properties of tumor cells. It has also been indicated that the key signaling molecule between ITGa6 and HBx is the p53 tumor suppressor. Therefore, the use of ITGa6 as a therapeutic target has been suggested[144]. However, the relationships of integrins with other HBV proteins during carcinogenesis in HCC remain to be elucidated.
The farnesoid X receptor: The farnesoid X receptor (FXR), a member of the nuclear receptor family, primarily stimulates bile excretion by suppressing the import and synthesis of bile acids into hepatocytes. However, the protective effect of FXR in hepatocytes is not limited to preventing the toxicity of bile acids but also involves inhibition of lipogenesis and stimulation of insulin sensitivity[145]. Moreover, it enhances EMT effects induced by TGF-β. After being experimentally determined to be an initiator of liver regeneration, FXR has also been found to be involved in HCC carcinogenesis related to chronic liver injury[146]. FXR expression is significantly lower in the livers of patients with HCC. In Huh7 cell lines, Niu et al[147] demon
Unlike HBV, HCV, which does not integrate into the host genome, may play an important role in carcinogenesis and tumor behavior by creating epigenetic disorders[148,149].
DNA methylation: Since HCV viral proteins can be found in the nucleus, they can generate nuclear signals in the host DNA[150,151]. The finding that the core protein significantly increases the expression of DNMT-1 and histone deacetylase HDAC1 suggests that it is capable of epigenetically silencing tumor suppressor gene expression that prevents tumor growth and progression[152]. This protein causes EMT induction and disruption of Wnt/β-Catenin signaling by suppressing SFRP[152,153]. The effects of HCV on changes in the methylation of genes, including APC, p73, p14, and OGMT, suggest that it may play a role in epigenetic silencing in malignant transformation[154,155].
Histone modifications: Detection of viral-mediated acetylation of lysine 27 position histone 3 (H3K27Ac) in HCV-infected liver tissue suggests an effect of HCV on the disease through histone deacetylation[156,157]. H3K27Ac separates active from inactive/poised enhancer elements, promoting the transcription of associated genes, and is considered an activation mark during developmental states[158]. Moreover, the correlation between HCV-induced genome-wide H3K27Ac changes and the expression of genes at risk of developing cancer supports this virus's involvement in carcinogenesis through histone modification[156]. Another histone in HCV-mediated acetylation in carcinogenesis is histone H3 Lysine 9 (H3K9Ac), which shows parallel findings with H3K27Ac[159]. The fact that these epigenetic changes create a memory, or footprint, has been attributed to their detection persisting even after viral therapy[159,160]. Indeed, the presence of this epigenetic footprint is associated with aggressive tumor behavior in HCV-HCC cases, as well as increased cancer-risk gene expression after treatment in both experimental studies and patients with HCV hepatitis[156,159,161]. The persistence of the HCV-specific epigenetic footprint of 65 oncogenes has been detected both in the fibrotic liver of patients with cured HCV and in the liver of HCV-cured animals[156]. In addition, a fibrosis-related footprint of 1693 cancer-risk genes was observed in a study group including patients with fibrotic liver of HCV, and dysregulation of subgroups of these fibrosis-related epigenetic footprint genes have been defined as prognostic epigenetic signatures and may be useful in determining cancer risk[157]. An attempt was made to identify new treatments that could remove these footprints with various epidrugs[162].
miRs in HCV: Similar to HBV, HCV alters miR expression in a way that affects liver carcinogenesis[163-165]. miR-19a, miR-135-5p, miR-146a-5p, and miR-122 are also impaired by HCV infection and are associated with HCC development[165-167].
Wnt/β-Catenin pathway: In HCV-HCC, the frequency of CTNNB1 mutations is approximately 26%, and this rate is significantly higher than that in either HBV-HCC (12%) or nonviral HCC (21%)[168-170]. Parallel to this finding, experimental studies have shown that HCV infection leads to CTNNB1 mutation. It has been suggested that the NS3 protein of HCV may contribute to oncogenic transformation by disrupting DNA repair mechanisms, leading to CTTNB1 mutation[171]. It should be noted that these data are very striking for a virus that does not exhibit DNA integration, and further studies are warranted to determine causal relationships. However, the effect of HCV is not limited to the CTNNB1 mutation, and HCV-associated proteins also participate in the activation of Wnt/β-Catenin signaling[160]. The core protein is one of them. In particular, by regulating the hepatocyte transcription factor in the nucleus, the core protein can activate this pathway[172]. The core protein also induces the FZD receptor and low-density lipoprotein receptor-related protein 5/6 (LRP5/6), supporting their activities by inhibiting antagonists, such as FZD-related protein 2 and Dickkopf 1 (DDK1)[173-176]. In the early phase of HCV infection, DDK1 inhibition plays a role in epigenetic silencing of the promoters by enabling the recruitment of DNA methyltransferase 1 and histone deacetylase[175]. In addition, it contributes to the inhibition of β-Catenin sequestration by hypermethylation of the gene promoter of E-Cadherin (CDH1)[174].
On the other hand, NS5A, one of the proteins involved in the HCV replication complex, is known to stabilize GSK3b both by direct binding and indirect inhibition through the PIK3-AKT pathway, leading to activation of Wnt/β-Catenin signaling[172]. There is also evidence that HCV, similar to HBV, activates this pathway through its effects on miRs. The effect of HCV on the Wnt/β-Catenin pathway is summarized in Figure 1.
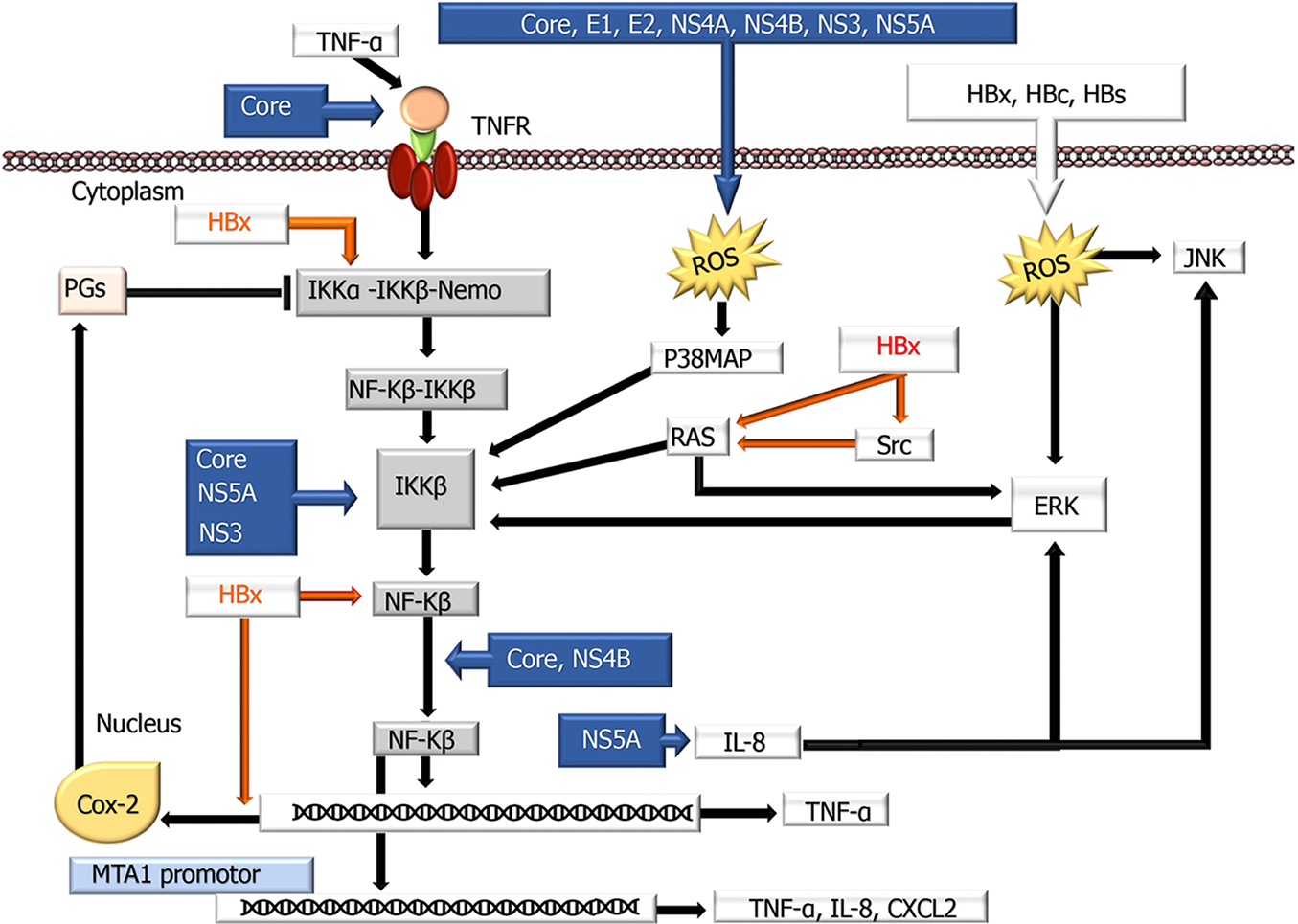
NF-κB pathway: In the liver, the role of NF-κB in inflammation, fibrosis, carcinogenesis, and tumor progression has been demonstrated in HCV-HCC. HCV may have different effects (both activator and inhibitor) on this pathway[123]. In previous studies, it has been reported that NF-κB is overexpressed in patients with HCV[177]. For instance, core proteins can induce the TNF receptor (TNFR), which plays a key role in the activation of this pathway, by direct induction or by mimicking proinflammatory cytokines, such as TNF-α[178,179]. This protein can also stimulate Th-17 cells that produce cytokines that stimulate CD161, IL-17A, IL-17F, IL-21, and IL-22 on TNFR, enabling NF-κB activation[179-182]. There is evidence that E1 and E2 proteins also increase TNF-α secretion[183]. In contrast, recent experimental studies have observed that HCV can downregulate this pathway by activating several genes that suppress NF-κB. The NS3 and NS5B proteins suppress NF-κB activation by TNF-α[184]. Considering the important role of inflammation and oxidative stress in liver diseases, it is not surprising that core E1, E2, NS3, NS4A, NS4B, and NS5A induce oxidative stress to increase NF-κB, and these signals, in turn, induce ROS production through mitochondrial effects[124,185]. Paracha et al[186] demonstrated that core, NS3, and NS5A proteins are effective in promoting Ca++ release from mitochondria. In addition, ROS induced by viral proteins activate phosphorylation of extracellular signal-regulated kinase (ERK), p38 mitogen-activated protein kinase (p38 MAPK), and c-Jun N-terminal kinase (JNK), which then promote NF-κB activation. Subsequently, activated NF-κB induces the expression of many genes, such as cyclooxygenase-2 (COX-2) and IL-8, by nuclear translocation. Chen et al[187] observed that the NS5A protein activates the NF-κB pathway and increases COX-2 expression by increasing ERK and JNK phosphorylation by inducing IL-8 production. On the other hand, COX-2 inhibits NF-κB activity by inhibiting IKK by bypassing the production of prostaglandins J2, A2, and A1. The role of HCV in this pathway is summarized in Figure 2.
Hippo-YAP/TAZ pathway: Similar to HBV, studies related to the Hippo-YAP-1 pathway are few, and most of them are experimental in HCV-HCC. The contribution of HCV to this pathway is presented in Figure 3. The HCV NS5B protein has been shown to play a role in EMT by inactivating the Hippo signaling pathway, upregulating Snail activity, and enabling PIK3/AKT activation[188]. In a few studies, there is evidence that the HCV E2 protein can mimic CD81, the major binding ligand for glypican 3. In this way, interestingly, it reduces Hippo activation and decreases YAP, attenuating proliferation[189]. Considering that CD81 is the entry route of HCV, it is concluded that due to the loss of CD81 in early carcinogenesis, this feature enables tumor cells to resist HCV during early HCC development, contributing to the clonal expansion of neoplastic cells compared to nontumor cells. Therefore, examining the possible effects of both factors on the Hippo pathway in carcinogenesis will enable us to determine whether this pathway represents a target for HCC therapy.
Angiogenesis pathways: Although various components of HCV have been reported to influence hepatocarcinogenesis, among them, the core protein has been reported to have the strongest potential associations with angiogenesis. It enhances the expression of angiogenic factors, including VEGF and Ang-2, by activating various growth factor signaling pathways, such as MAPK, PIK3, and JNK[190-193]. Some studies have reported that the core protein increases VEGF through androgen receptor-mediated transcription by activating STAT3[194]. On the other hand, despite the same correlation of androgen and VEGF expression in other studies, no increase in STAT3 was observed[195]. It has been suggested that this may be due to experimental methodological differences, such as the use of different genotypes of the core protein. Similarly, inconsistent results were obtained regarding the interaction of the core protein with the PIK3 and MAPK pathways during the induction of VEGF expression[196-198]. Recently, Shao et al[195] demonstrated that the HCV core protein's proangiogenic activity is dose-dependent and that the mechanism involves an increase in VEGF expression through activator protein-1 activation. The core protein can also induce angiogenesis by increasing the expression of endoglin (CD105), which plays an important role in TGF-β signaling[199].
After the apelin system (APJ) was shown to have a role in HCV-HCC during carcinogenesis progression in recent years, the relationship of this system with angiogenesis was revealed[200]. Although more research is needed to better understand apelin's role in HCC, Cabiati et al[201] recently demonstrated that higher expression of this angiogenic agent in patients with HCV-HCC is consistent with pathological staging and emphasized that APJ could be considered important for developing therapeutic targets that identify new pathways for cancer treatment.
In the study of gene expression for secreted proteins in HCV-infected hepatoma cells by applying microarray analysis, it was found that HCV exerts a proangiogenic effect by inducing the EGFR-ERK signaling pathway[202].
FXR: Although there are several studies on the role of FXR in the pathogenesis of HCV hepatitis, further studies are needed to understand its role in HCV-HCC.
At the onset of inflammation, hepatocyte differentiation and proliferation after exposure to acute injury or microbe patterns or danger-related molecular models (DAMPs) are directed toward liver repair[203]. However, persistent inflammation caused by cellular damage and loss triggers the immune response, leading to the development of HCC. Therefore, inflammation plays a very important role in hepatocarcinogenesis, similar to other types of cancer. The autoamplification cycle created by proinflammatory pathways induced by the release of DAMPs in necroinflammation, defined as cell death caused by the immune response, promotes hepatocarcinogenesis by disrupting genetic stability during DNA degradation[125]. In addition, during hepatocarcinogenesis, disruption of the immune system and release of cytokines that cause immune suppression (IL-10, IL-13, and TGF-β) promote malignant trans
Viral inflammation plays a significant role in the development of both HBV-HCC and HCV-HCC. In this section, the effects of viral inflammation on carcinogenesis will be discussed separately under three main headings, each of which has a significant role in HCC development.
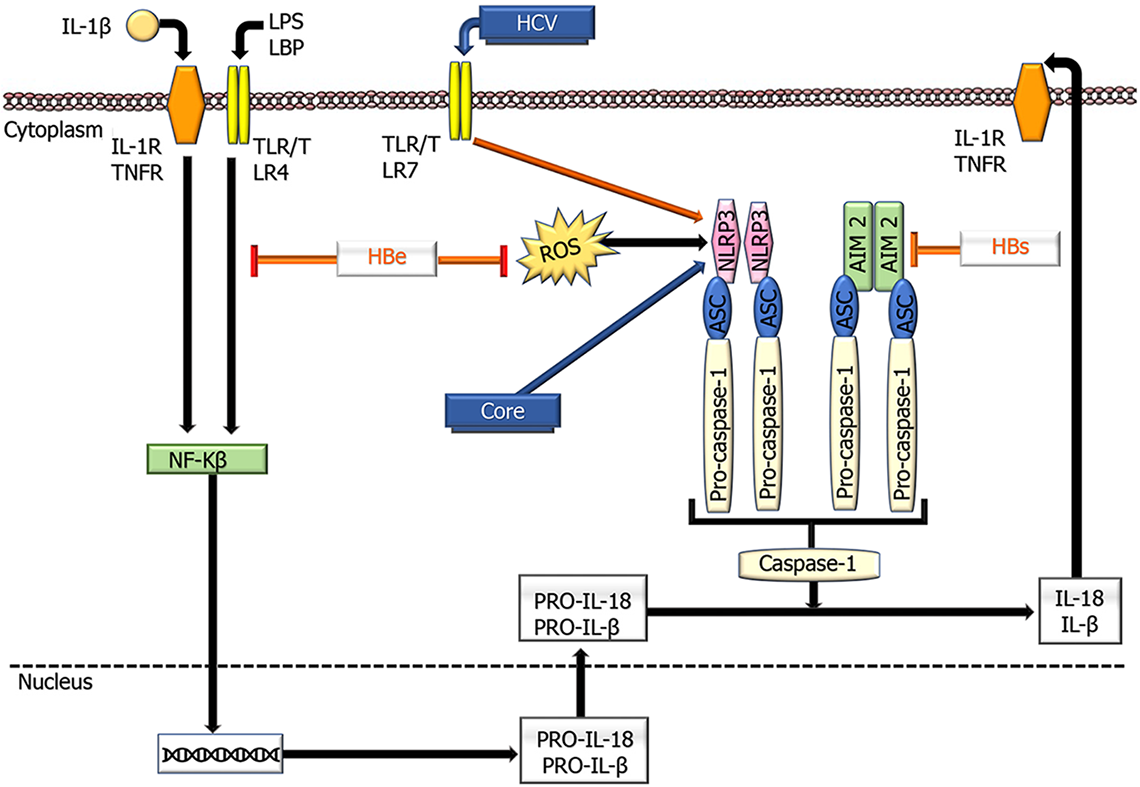
Inflammasomes are important components of the innate immune system that regulate caspase-1 activation and inflammation and have recently been proposed as therapeutic targets for many inflammatory diseases. They consist of large protein complexes that initiate downstream signaling, resulting in the release of type I interferons and proinflammatory cytokines through model recognition receptors (PRRs) that are stimulated by pathogen-related molecular models and DAMPs [125]. Several PRR families are important components in the inflammasome complex, including the nucleotide-binding domain, leucine-rich repeat proteins (also known as NOD-like receptors, NLRs), and melanoma (AIM)-like receptors (also known as ALRs)[204]. After stimulation, the relevant inflammasome PRR (for instance, NLRP3 in hepatocytes) oligomerizes to become a caspase-1 activator, leading to cleavage of proinflammatory IL-1 cytokines into IL-1β and IL-18 and subsequent pyroptosis[205] (Figure 4).
Different findings have been reported on the participation of inflammasomes in oncogenesis in the liver and their interaction with viruses in the development of HCC[206]. Some studies have demonstrated that the NLRP3 inflammasome is significantly reduced in tumors compared to nontumor tissues[207]. It also induces IL-18, resulting in NK cell-mediated suppression of colorectal cancer metastasis to the liver[208]. In addition, 17β-estradiol (E2), which plays a protective role in hepatocarcinogenesis, decreases tumor progression by increasing the NLRP3 inflammasome by the E2/ERβ/MAPK signaling pathway[209]. These findings support the NLRP3 inflammasome's preventive role in HCC. In contrast, it has been observed that during hypoxia, HMGB1, through activation of NLRP3, enhances caspase-1, IL-1β, and IL-18 to promote tumor invasion[210]. HCV activates NLRP3, leading to its accumulation in lipid droplets, where it coordinates with sterol-regulatory-element-binding proteins to promote liver disease[211]. HCV also induces hepatic macrophages to produce IL-1β through the NLRP3 inflammasome, leading to disease progression[212].
Another inflammasome, AIM2, induces IL-18 expression in human Kupffer cells, which stimulates NK cells to produce interferon (IFN)-γ in the regulation of innate immunity[213]. In patients with HBV, increased AIM2 expression was observed in the high HBV replication group compared to the low HBV replication group, with high AIM2 Levels being positively associated with IL-1β and IL-18 expression, suggesting that this inflammasome may be involved in viral elimination[214]. However, another finding indicated that AIM2 prevents the recognition of DDS expressed by HBV to assist in immune evasion[213]. These data also demonstrate that inflammasomes are differentially regulated by HBV proteins (Figure 4).
In the liver, Kupffer cells play a critical role in IL-β and IL-18 release in the inflammasome-mediated inflammatory response. However, there are different reports regarding IL-1β inhibiting HBV infection in liver cells. For instance, priming with IL-1β reduces host cell susceptibility to HBV infection through activation-induced cytidine deaminase[215] and oxidative stress[216]. Although HCV-infected monocytes induce IL-18 through inflammasomes that activate IFN-γ-producing NK cells, patients with chronic HCV infection exhibit reduced monocyte function and low IFN-γ levels[217]. These findings are explained by changes in membrane protein composition on monocytes derived from chronic HCV patients who present depleted levels of IFN-γ due to decreased numbers of CD14+ monocytes. Taken together, these findings show that inflammasomes derived from monocytes, NK cells, and macrophage-derived inflammasomes perform differential functions in hepatitis.
However, all of these data necessitate further investigations to decipher the role of inflammasomes in HCC.
The effect of HBV and HCV on the inflammation-mediated contribution of NF-κB to viral carcinogenesis in HCC has been discussed above.
STAT3 signaling plays an important role in the contribution of inflammation to the development and formation of HCC[143,218]. This pathway can be activated by many cytokines and growth factors, including IL-6, TNF, the EGF family, and hepatocyte growth factor[219,220]. As indicated by Wu et al[143], STAT3 activation leads to strong inhibition of SHP phosphatases and suppressor of cytokine signaling 3, which is inhibited in tumor cells. The accumulation of oxidative stress in both HBV and HCV infection can activate STAT3 by inducing JNK expression, allowing phosphorylation of a critical tyrosine kinase residue (Tyr705). Therefore, viral inflammation causes overexpression of proinflammatory cytokines, including IL-6, IL-10, IL-11, and TGF-α, which regulate the liver microenvironment to support oncogenic conditions, such as inhibition of apoptosis. The multiple roles of STAT3 activation in proliferation and anti-apoptosis (Cyclin D, Bcl-xL, Bcl-2, Caspase), migration and invasion (MMP-4, MMP-9, Slug, Twist), angiogenesis (VEGF, bFGF, HIF-α), and cancer stemness (CD131, NANOG, Notch) make its inhibition an attractive therapeutic target[218]. More recently, in an elegant study, Qin et al[221] demonstrated that in patients with advanced HBV-HCC, the use of icaritin, a small molecule that displays anticancer activities through the IL-6/JAK/STAT3 pathway, could be used as an alternative immune-modulatory regimen to treat advanced HCC patients with poor prognosis.
Hepatocytes have the capacity to develop a tolerogenic immune milieu in which innate and acquired immunity play important roles in preventing permanent damage[222]. Unfortunately, these mechanisms also allow tumor cells to escape from the immune system during oncogenesis. Indeed, the reduction in antigen presentation activity, evidenced by decreased expression of HLA class I molecules and suppression of CD8+ T cells with an increasing number of Tregs, contribute to oncogenesis[223-225]. Similar findings were also noted for NK T cells, myeloid-derived suppressor cells, tumor-associated macrophages (TAMs), and decreased CD4+ T cells[226,227]. In this context, the transition between proinflammatory (M1) and anti-inflammatory (M2) phenotypes of macrophages is very important. The transformation of macrophages into the M2 phenotype, which is effective in exhibiting anti-inflammatory functions by releasing Th2 cytokines (IL-4, IL-10, and IL-13), favors hepatocarcinogenesis[125,228]. In addition, it has been reported that HSCs exhibiting loss of p53 may contribute to the increase in macrophages with the M2 phenotype[143]. In HCC, positive correlations among the increase in anti-inflammatory cytokines, distant metastasis, and poor prognosis demonstrate that immune escape during inflammation promotes carcinogenesis and tumor behavior[229]. Moreover, inflammatory cells and cytokines demonstrably support cancer stem cells by the IL-6/STAT3 pathway. In vitro, the release of IL-6 may induce the expansion of CD133-positive cancer progenitor cells[230]. The IL-6/STAT3 paracrine signaling pathway from TAMs also triggers the proliferation of cancer progenitor cells to facilitate hepatocarcinogenesis[222]. Recently, the results of a study by Song et al[231] delineated that HBV/HCV-related HCC includes new types of immune cells that may play a role in immune escape and highlighted the potential of this condition to play a role in HCC formation and the progression of viruses in uncovering new targets for tumor therapy.
Autophagy is an evolutionarily conserved cellular pathway in which long-lived cytoplasmic proteins and organelles are engulfed into double-membrane vesicles known as autophagosomes that subsequently fuse with lysosomes and are regulated by a series of autophagy-related genes (Atgs)[232]. Atg5, Atg7, and Beclin 1 (BECN1) are essential genes in this process, and silencing any of these genes leads to blockage of autophagy. Previous studies suggest that autophagy acts as a double-edged sword in carcinogenesis and tumor behavior[233]. It can function as a tumor suppressor in the early stage of cancer development by inhibiting inflammation and promoting genomic stability. In contrast, autophagy confers a survival advantage to tumor cells by supplying nutrients and energy and promoting angiogenesis. Many studies have evidenced the role of autophagy in HCC initiation and development. The results indicate that autophagy plays a suppressive role in the onset of HCC development and acts as a promoter after tumor development in advanced stages. For more detailed information on this topic, the elegant reviews by Cui et al[234] and Lee et al[235] are recommended.
Recent studies have shown that HBV induces autophagy, particularly with HBx and HBs proteins, to improve its survival and replication[236]. However, the role of autophagy in HBV-associated tumorigenesis is not entirely clarified. Autophagy downregulated HBV-HCC in both humans and mouse models[237]. HBV-induced liver cell neoplasia progression stimulated in BECN1-null mice suggests that increased malignant transformation risk is inversely correlated with autophagy induction[238]. In HBV-HCC tissues, the expression of BECN1 and Atg5 is decreased compared to that in chronic hepatitis[239].
Similarly, although the exact mechanisms of downregulation of autophagy in HBV-HCC remain unclear, it is suggested that autophagy can increase the antitumor immune response[240], induce cell death[241], and lead to oncogenic microRNA degradation[242]. These results imply that autophagy, which plays a tumor-suppressive role in hepatocarcinogenesis, is inhibited in HBV-associated tumorigenesis. However, the mechanism by which high autophagy during HBV infection shifts to low autophagy in HBV-related tumorigenesis remains to be elucidated.
HCV infection can also promote inflammation by leading to the activation of inflammasomes and the production of proinflammatory cytokines, including IL-1α and IL-1β, thereby promoting fibrosis carcinogenesis[212]. NLRP3 inflammasome activation through calcium mobilization linked to phospholipase-C through HCV core protein is defined during progressive liver disease, and HCC is also noted[243]. Together with the finding that this activation can be counteracted by autophagy activation under certain circumstances, these data support HCV disease's contribution to progression by autophagy[244]. HCV-induced endoplasmic reticulum ER stress is evidenced by the observation of increased elevation of ER stress markers in HCV-related cirrhosis[245]. Moreover, in HCV cell cultures, Aydın et al[246] observed that excessive ER stress activates NRF2-mediated autophagy switching to promote cell survival. Considering that NRF2 plays a role in the development of HCC through the accumulation of p62, HCV may also contribute to carcinogenesis through autophagy regulation mechanisms[247]. Recently, the contribution of immunity-related GTPases (IRGs), a family of IFN-inducible GTPases, to autophagy has been implicated[248]. IRGM is a critical negative regulator of NLRP3 inflammasome activation by interacting with NLRP3 and ASC and subsequently inhibiting inflammasome assembly[249]. A recent study pointed out that HCV-induced IRGM-mediated phosphorylation of ULK1 induces autophagy[250]. It is proposed that IRGM-mediated autophagy after HCV infection might contribute to the tumor-promoting effects of HCV.
Currently, further studies are needed to fully understand the effect of HCV on carcinogenesis and tumor behavior through autophagy.
Finally, it should also be noted that the association of autophagy with the pathways mentioned above involved in HCC development also contributes to both HBV and HCV-associated carcinogenesis.
Although its role in HCC is not fully clarified, there is evidence that the intestinal flora is involved in carcinogenesis[251-253]. For instance, a human study has shown that Helicobacter hepaticus is present in the livers of patients with HCC compared to the livers of controls without carcinoma[251]. A remarkable observation is the absence of this microorganism in HBV-HCC and HCV-HCC, which necessitates further studies to be clarified entirely. On the other hand, in an elegant study, Huang et al[253] found that Bacteroides, Lachnospiraceae incertae sedis, and Clostridium XIVa were enriched in HBV-HCC patients with a high tumor burden. Therefore, extensive research in this context could reveal more comprehensive information about the use of microbiota changes as a treatment target.
Despite extensive studies on its development and behavior, HCC remains a major health problem worldwide. The main therapeutic strategies that are currently being used in the treatment of HCC include angiogenesis inhibitors, multikinase inhibitors, pathway-targeted therapies (especially the Wnt-β-Catenin and NF-κB pathways), immunotargeting (for immune escape), inflammasome-targeting therapies, and immunotherapy[143]. Compared to other common cancers, such as breast and colon cancers, the numerous variations caused by this tumor's very complex molecular profile constitute a critical limitation for clinical trials. Therefore, uncovering the molecular mechanisms associated with HCC development and tumor behavior is very significant for both decreasing tumor prevalence and identifying new treatment targets. In this context, it is crucial to clarify the effects of HBV and HCV on the molecular events involved in oncogenesis and prognosis due to their close relationship with HCC. Unfortunately, unlike other viruses, it is not possible to identify a single oncogene or mechanism in terms of carcinogenesis and progression. In contrast, complex mechanisms reflect concerted and cumulative effects of chronic inflammation-related alterations, modification of oncogenic pathways (especially tumor suppression, proliferation, apoptosis), and epigenetic dysregulation in response to both viruses. In addition, the integration of HBV into host DNA may also affect tumor development and behavior.
Therefore, further studies to characterize new mechanisms of HBV and HCV-associated carcinogenesis and tumor progression may reduce the prevalence of HCC and lead to the discovery of new treatment targets to overcome the grim prognosis of this disease.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Turkey
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gao YT, Kimkong I S-Editor: Wang JL L-Editor: A P-Editor: Wang LL
| 1. | Ward EM, Sherman RL, Henley SJ, Jemal A, Siegel DA, Feuer EJ, Firth AU, Kohler BA, Scott S, Ma J, Anderson RN, Benard V, Cronin KA. Annual Report to the Nation on the Status of Cancer, Featuring Cancer in Men and Women Age 20-49 Years. J Natl Cancer Inst. 2019;111:1279-1297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 229] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 2. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55833] [Article Influence: 7976.1] [Reference Citation Analysis (132)] |
| 3. | Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2184] [Cited by in RCA: 2907] [Article Influence: 484.5] [Reference Citation Analysis (17)] |
| 4. | Ding XX, Zhu QG, Zhang SM, Guan L, Li T, Zhang L, Wang SY, Ren WL, Chen XM, Zhao J, Lin S, Liu ZZ, Bai YX, He B, Zhang HQ. Precision medicine for hepatocellular carcinoma: driver mutations and targeted therapy. Oncotarget. 2017;8:55715-55730. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 69] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 5. | Lee DH, Lee JM. Primary malignant tumours in the non-cirrhotic liver. Eur J Radiol. 2017;95:349-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 6. | El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 2012; 142: 1264-1273. e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2183] [Cited by in RCA: 2508] [Article Influence: 192.9] [Reference Citation Analysis (2)] |
| 7. | Miao Z, Zhang S, Ou X, Li S, Ma Z, Wang W, Peppelenbosch MP, Liu J, Pan Q. Estimating the Global Prevalence, Disease Progression, and Clinical Outcome of Hepatitis Delta Virus Infection. J Infect Dis. 2020;221:1677-1687. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 227] [Article Influence: 45.4] [Reference Citation Analysis (0)] |
| 8. | World Health Organization. Hepatitis D, 2019. Available from: https://www.who.int/news-room/fact-sheets/detail/hepatitis-d. |
| 9. | World Health Organization. Global hepatitis report, 2017. Available from: https://www.who.int/hepatitis/publications/global-hepatitis-report2017. |
| 10. | Su TH, Kao JH, Liu CJ. Molecular mechanism and treatment of viral hepatitis-related liver fibrosis. Int J Mol Sci. 2014;15:10578-10604. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 11. | Tu T, Budzinska MA, Shackel NA, Urban S. HBV DNA Integration: Molecular Mechanisms and Clinical Implications. Viruses. 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 200] [Cited by in RCA: 300] [Article Influence: 37.5] [Reference Citation Analysis (1)] |
| 12. | Virlogeux V, Trépo C. Extrahepatic manifestations of chronic hepatitis B Infection. Curr Hepatology Rep. 2018;17:156-165. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 13. | Lau KC, Joshi SS, Gao S, Giles E, Swidinsky K, van Marle G, Bathe OF, Urbanski SJ, Terrault NA, Burak KW, Osiowy C, Coffin CS. Oncogenic HBV variants and integration are present in hepatic and lymphoid cells derived from chronic HBV patients. Cancer Lett. 2020;480:39-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 14. | Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1094] [Cited by in RCA: 1115] [Article Influence: 44.6] [Reference Citation Analysis (0)] |
| 15. | Livingston SE, Simonetti JP, McMahon BJ, Bulkow LR, Hurlburt KJ, Homan CE, Snowball MM, Cagle HH, Williams JL, Chulanov VP. Hepatitis B virus genotypes in Alaska Native people with hepatocellular carcinoma: preponderance of genotype F. J Infect Dis. 2007;195:5-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 193] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 16. | Kao JH, Chen PJ, Lai MY, Chen DS. Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology. 2003;124:327-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 414] [Cited by in RCA: 428] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 17. | Liu S, Zhang H, Gu C, Yin J, He Y, Xie J, Cao G. Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: a meta-analysis. J Natl Cancer Inst. 2009;101:1066-1082. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 279] [Cited by in RCA: 328] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 18. | Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933-938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 602] [Cited by in RCA: 601] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 19. | Ploss A, Evans MJ. Hepatitis C virus host cell entry. Curr Opin Virol. 2012;2:14-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 20. | Ding Q, von Schaewen M, Ploss A. The impact of hepatitis C virus entry on viral tropism. Cell Host Microbe. 2014;16:562-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 59] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 21. | Akuta N, Suzuki F, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Arase Y, Ikeda K, Kumada H. Amino acid substitutions in the hepatitis C virus core region are the important predictor of hepatocarcinogenesis. Hepatology. 2007;46:1357-1364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 96] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 22. | Akuta N, Suzuki F, Hirakawa M, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Saitoh S, Arase Y, Ikeda K, Kumada H. Amino acid substitutions in the hepatitis C virus core region of genotype 1b are the important predictor of severe insulin resistance in patients without cirrhosis and diabetes mellitus. J Med Virol. 2009;81:1032-1039. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 23. | Morozov VA, Lagaye S. Hepatitis C virus: Morphogenesis, infection and therapy. World J Hepatol. 2018;10:186-212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 90] [Article Influence: 12.9] [Reference Citation Analysis (13)] |
| 24. | Grassi G, Di Caprio G, Fimia GM, Ippolito G, Tripodi M, Alonzi T. Hepatitis C virus relies on lipoproteins for its life cycle. World J Gastroenterol. 2016;22:1953-1965. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 40] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (2)] |
| 25. | Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013;19:837-849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 445] [Cited by in RCA: 421] [Article Influence: 35.1] [Reference Citation Analysis (0)] |
| 26. | Kanwal F, Kramer JR, Ilyas J, Duan Z, El-Serag HB. HCV genotype 3 is associated with an increased risk of cirrhosis and hepatocellular cancer in a national sample of U.S. Veterans with HCV. Hepatology. 2014;60:98-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 241] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 27. | Lee MH, Hsiao TI, Subramaniam SR, Le AK, Vu VD, Trinh HN, Zhang J, Jin M, Wong VW, Wong GL, Nguyen MH. HCV Genotype 6 Increased the Risk for Hepatocellular Carcinoma Among Asian Patients With Liver Cirrhosis. Am J Gastroenterol. 2017;112:1111-1119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 28. | Bréchot C. Pathogenesis of hepatitis B virus-related hepatocellular carcinoma: old and new paradigms. Gastroenterology. 2004;127:S56-S61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 330] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 29. | Zhao LH, Liu X, Yan HX, Li WY, Zeng X, Yang Y, Zhao J, Liu SP, Zhuang XH, Lin C, Qin CJ, Zhao Y, Pan ZY, Huang G, Liu H, Zhang J, Wang RY, Wen W, Lv GS, Zhang HL, Wu H, Huang S, Wang MD, Tang L, Cao HZ, Wang L, Lee TL, Jiang H, Tan YX, Yuan SX, Hou GJ, Tao QF, Xu QG, Zhang XQ, Wu MC, Xu X, Wang J, Yang HM, Zhou WP, Wang HY. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat Commun. 2016;7:12992. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 217] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 30. | Toh ST, Jin Y, Liu L, Wang J, Babrzadeh F, Gharizadeh B, Ronaghi M, Toh HC, Chow PK, Chung AY, Ooi LL, Lee CG. Deep sequencing of the hepatitis B virus in hepatocellular carcinoma patients reveals enriched integration events, structural alterations and sequence variations. Carcinogenesis. 2013;34:787-798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 86] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 31. | Lee J, Tsai KN, Ou JJ. Mechanisms of Hepatitis B Virus-Induced Hepatocarcinogenesis. Recent Results Cancer Res. 2021;217:47-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 32. | Murakami Y, Saigo K, Takashima H, Minami M, Okanoue T, Bréchot C, Paterlini-Bréchot P. Large scaled analysis of hepatitis B virus (HBV) DNA integration in HBV related hepatocellular carcinomas. Gut. 2005;54:1162-1168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 241] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 33. | Tamori A, Yamanishi Y, Kawashima S, Kanehisa M, Enomoto M, Tanaka H, Kubo S, Shiomi S, Nishiguchi S. Alteration of gene expression in human hepatocellular carcinoma with integrated hepatitis B virus DNA. Clin Cancer Res. 2005;11:5821-5826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 60] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 34. | Ding D, Lou X, Hua D, Yu W, Li L, Wang J, Gao F, Zhao N, Ren G, Lin B. Recurrent targeted genes of hepatitis B virus in the liver cancer genomes identified by a next-generation sequencing-based approach. PLoS Genet. 2012;8:e1003065. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 135] [Cited by in RCA: 153] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 35. | Jiang Z, Jhunjhunwala S, Liu J, Haverty PM, Kennemer MI, Guan Y, Lee W, Carnevali P, Stinson J, Johnson S, Diao J, Yeung S, Jubb A, Ye W, Wu TD, Kapadia SB, de Sauvage FJ, Gentleman RC, Stern HM, Seshagiri S, Pant KP, Modrusan Z, Ballinger DG, Zhang Z. The effects of hepatitis B virus integration into the genomes of hepatocellular carcinoma patients. Genome Res. 2012;22:593-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 234] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 36. | Sung WK, Zheng H, Li S, Chen R, Liu X, Li Y, Lee NP, Lee WH, Ariyaratne PN, Tennakoon C, Mulawadi FH, Wong KF, Liu AM, Poon RT, Fan ST, Chan KL, Gong Z, Hu Y, Lin Z, Wang G, Zhang Q, Barber TD, Chou WC, Aggarwal A, Hao K, Zhou W, Zhang C, Hardwick J, Buser C, Xu J, Kan Z, Dai H, Mao M, Reinhard C, Wang J, Luk JM. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet. 2012;44:765-769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 619] [Cited by in RCA: 724] [Article Influence: 55.7] [Reference Citation Analysis (0)] |
| 37. | Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, Arai Y, Takahashi H, Shirakihara T, Nagasaki M, Shibuya T, Nakano K, Watanabe-Makino K, Tanaka H, Nakamura H, Kusuda J, Ojima H, Shimada K, Okusaka T, Ueno M, Shigekawa Y, Kawakami Y, Arihiro K, Ohdan H, Gotoh K, Ishikawa O, Ariizumi S, Yamamoto M, Yamada T, Chayama K, Kosuge T, Yamaue H, Kamatani N, Miyano S, Nakagama H, Nakamura Y, Tsunoda T, Shibata T, Nakagawa H. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760-764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 631] [Cited by in RCA: 714] [Article Influence: 54.9] [Reference Citation Analysis (0)] |
| 38. | Wan S, Civan J, Rossi S, Yang H. Profiling HBV integrations in hepatocellular carcinoma. Hepatobiliary Surg Nutr. 2013;2:124-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 39. | Li W, Zeng X, Lee NP, Liu X, Chen S, Guo B, Yi S, Zhuang X, Chen F, Wang G, Poon RT, Fan ST, Mao M, Li Y, Li S, Wang J, Jianwang, Xu X, Jiang H, Zhang X. HIVID: an efficient method to detect HBV integration using low coverage sequencing. Genomics. 2013;102:338-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 88] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 40. | Hai H, Tamori A, Kawada N. Role of hepatitis B virus DNA integration in human hepatocarcinogenesis. World J Gastroenterol. 2014;20:6236-6243. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 71] [Cited by in RCA: 71] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 41. | Zhang X, Wang L, Yan Y. Identification of potential key genes and pathways in hepatitis B virus-associated hepatocellular carcinoma by bioinformatics analyses. Oncol Lett. 2020;19:3477-3486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 42. | Ou J, Rutter WJ. Hybrid hepatitis B virus-host transcripts in a human hepatoma cell. Proc Natl Acad Sci USA. 1985;82:83-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 56] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 43. | Ma NF, Lau SH, Hu L, Xie D, Wu J, Yang J, Wang Y, Wu MC, Fung J, Bai X, Tzang CH, Fu L, Yang M, Su YA, Guan XY. COOH-terminal truncated HBV X protein plays key role in hepatocarcinogenesis. Clin Cancer Res. 2008;14:5061-5068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 138] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 44. | Peng Z, Zhang Y, Gu W, Wang Z, Li D, Zhang F, Qiu G, Xie K. Integration of the hepatitis B virus X fragment in hepatocellular carcinoma and its effects on the expression of multiple molecules: a key to the cell cycle and apoptosis. Int J Oncol. 2005;26:467-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 45. | Liu X, Wang L, Zhang S, Lin J, Feitelson MA, Gao H, Zhu M. Mutations in the C-terminus of the X protein of hepatitis B virus regulate Wnt-5a expression in hepatoma Huh7 cells: cDNA microarray and proteomic analyses. Carcinogenesis. 2008;29:1207-1214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 46] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 46. | Yip WK, Cheng AS, Zhu R, Lung RW, Tsang DP, Lau SS, Chen Y, Sung JG, Lai PB, Ng EK, Yu J, Wong N, To KF, Wong VW, Sung JJ, Chan HL. Carboxyl-terminal truncated HBx regulates a distinct microRNA transcription program in hepatocellular carcinoma development. PLoS One. 2011;6:e22888. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 64] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 47. | Li W, Li M, Liao D, Lu X, Gu X, Zhang Q, Zhang Z, Li H. Carboxyl-terminal truncated HBx contributes to invasion and metastasis via deregulating metastasis suppressors in hepatocellular carcinoma. Oncotarget. 2016;7:55110-55127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 48. | Jung SY, Kim YJ. C-terminal region of HBx is crucial for mitochondrial DNA damage. Cancer Lett. 2013;331:76-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 49. | Sze KM, Chu GK, Lee JM, Ng IO. C-terminal truncated hepatitis B virus x protein is associated with metastasis and enhances invasiveness by C-Jun/matrix metalloproteinase protein 10 activation in hepatocellular carcinoma. Hepatology. 2013;57:131-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 120] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 50. | Ng KY, Chai S, Tong M, Guan XY, Lin CH, Ching YP, Xie D, Cheng AS, Ma S. C-terminal truncated hepatitis B virus X protein promotes hepatocellular carcinogenesis through induction of cancer and stem cell-like properties. Oncotarget. 2016;7:24005-24017. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 51. | Tu H, Bonura C, Giannini C, Mouly H, Soussan P, Kew M, Paterlini-Bréchot P, Bréchot C, Kremsdorf D. Biological impact of natural COOH-terminal deletions of hepatitis B virus X protein in hepatocellular carcinoma tissues. Cancer Res. 2001;61:7803-7810. [PubMed] |
| 52. | Xu R, Zhang X, Zhang W, Fang Y, Zheng S, Yu XF. Association of human APOBEC3 cytidine deaminases with the generation of hepatitis virus B x antigen mutants and hepatocellular carcinoma. Hepatology. 2007;46:1810-1820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 82] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 53. | Lau CC, Sun T, Ching AK, He M, Li JW, Wong AM, Co NN, Chan AW, Li PS, Lung RW, Tong JH, Lai PB, Chan HL, To KF, Chan TF, Wong N. Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell. 2014;25:335-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 227] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 54. | Lee SM, Lee YG, Bae JB, Choi JK, Tayama C, Hata K, Yun Y, Seong JK, Kim YJ. HBx induces hypomethylation of distal intragenic CpG islands required for active expression of developmental regulators. Proc Natl Acad Sci USA. 2014;111:9555-9560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 55. | Rongrui L, Na H, Zongfang L, Fanpu J, Shiwen J. Epigenetic mechanism involved in the HBV/HCV-related hepatocellular carcinoma tumorigenesis. Curr Pharm Des. 2014;20:1715-1725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 56. | Yu G, Bing Y, Li W, Xia L, Liu Z. Hepatitis B virus inhibits the expression of CD82 through hypermethylation of its promoter in hepatoma cells. Mol Med Rep. 2014;10:2580-2586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 57. | Lee MH, Na H, Na TY, Shin YK, Seong JK, Lee MO. Epigenetic control of metastasis-associated protein 1 gene expression by hepatitis B virus X protein during hepatocarcinogenesis. Oncogenesis. 2014;3:e88. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 58. | Fu X, Song X, Li Y, Tan D, Liu G. Hepatitis B virus X protein upregulates DNA methyltransferase 3A/3B and enhances SOCS-1CpG island methylation. Mol Med Rep. 2016;13:301-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 59. | Ying J, Li H, Seng TJ, Langford C, Srivastava G, Tsao SW, Putti T, Murray P, Chan AT, Tao Q. Functional epigenetics identifies a protocadherin PCDH10 as a candidate tumor suppressor for nasopharyngeal, esophageal and multiple other carcinomas with frequent methylation. Oncogene. 2006;25:1070-1080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 217] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 60. | Arzumanyan A, Friedman T, Kotei E, Ng IO, Lian Z, Feitelson MA. Epigenetic repression of E-cadherin expression by hepatitis B virus x antigen in liver cancer. Oncogene. 2012;31:563-572. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 91] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 61. | Fang S, Huang SF, Cao J, Wen YA, Zhang LP, Ren GS. Silencing of PCDH10 in hepatocellular carcinoma via de novo DNA methylation independent of HBV infection or HBX expression. Clin Exp Med. 2013;13:127-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 62. | Qiu X, Zhang L, Lu S, Song Y, Lao Y, Hu J, Fan H. Upregulation of DNMT1 mediated by HBx suppresses RASSF1A expression independent of DNA methylation. Oncol Rep. 2014;31:202-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 63. | Weichert W. HDAC expression and clinical prognosis in human malignancies. Cancer Lett. 2009;280:168-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 322] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 64. | Xie HJ, Noh JH, Kim JK, Jung KH, Eun JW, Bae HJ, Kim MG, Chang YG, Lee JY, Park H, Nam SW. HDAC1 inactivation induces mitotic defect and caspase-independent autophagic cell death in liver cancer. PLoS One. 2012;7:e34265. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 84] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 65. | Liu XY, Tang SH, Wu SL, Luo YH, Cao MR, Zhou HK, Jiang XW, Shu JC, Bie CQ, Huang SM, Zheng ZH, Gao F. Epigenetic modulation of insulin-like growth factor-II overexpression by hepatitis B virus X protein in hepatocellular carcinoma. Am J Cancer Res. 2015;5:956-978. [PubMed] |
| 66. | Zhao Q, Li T, Qi J, Liu J, Qin C. The miR-545/374a cluster encoded in the Ftx lncRNA is overexpressed in HBV-related hepatocellular carcinoma and promotes tumorigenesis and tumor progression. PLoS One. 2014;9:e109782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 75] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 67. | Gao P, Wong CC, Tung EK, Lee JM, Wong CM, Ng IO. Deregulation of microRNA expression occurs early and accumulates in early stages of HBV-associated multistep hepatocarcinogenesis. J Hepatol. 2011;54:1177-1184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 122] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 68. | Connolly E, Melegari M, Landgraf P, Tchaikovskaya T, Tennant BC, Slagle BL, Rogler LE, Zavolan M, Tuschl T, Rogler CE. Elevated expression of the miR-17-92 polycistron and miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. Am J Pathol. 2008;173:856-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 207] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 69. | Xu J, An P, Winkler CA, Yu Y. Dysregulated microRNAs in Hepatitis B Virus-Related Hepatocellular Carcinoma: Potential as Biomarkers and Therapeutic Targets. Front Oncol. 2020;10:1271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 70. | Huang J, Wang Y, Guo Y, Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 2010;52:60-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 274] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 71. | Xu L, Beckebaum S, Iacob S, Wu G, Kaiser GM, Radtke A, Liu C, Kabar I, Schmidt HH, Zhang X, Lu M, Cicinnati VR. MicroRNA-101 inhibits human hepatocellular carcinoma progression through EZH2 downregulation and increased cytostatic drug sensitivity. J Hepatol. 2014;60:590-598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 137] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 72. | Guo X, Lv X, Ma Y, Chen L, Chen Y. Circulating miR-21 serves as a serum biomarker for hepatocellular carcinoma and correlated with distant metastasis. Oncotarget. 2017;8:44050-44058. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 74] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 73. | Damania P, Sen B, Dar SB, Kumar S, Kumari A, Gupta E, Sarin SK, Venugopal SK. Hepatitis B virus induces cell proliferation via HBx-induced microRNA-21 in hepatocellular carcinoma by targeting programmed cell death protein4 (PDCD4) and phosphatase and tensin homologue (PTEN). PLoS One. 2014;9:e91745. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 69] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 74. | Li CH, Xu F, Chow S, Feng L, Yin D, Ng TB, Chen Y. Hepatitis B virus X protein promotes hepatocellular carcinoma transformation through interleukin-6 activation of microRNA-21 expression. Eur J Cancer. 2014;50:2560-2569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 75. | Cui M, Wang Y, Sun B, Xiao Z, Ye L, Zhang X. MiR-205 modulates abnormal lipid metabolism of hepatoma cells via targeting acyl-CoA synthetase long-chain family member 1 (ACSL1) mRNA. Biochem Biophys Res Commun. 2014;444:270-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 89] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 76. | Tang Y, Zhou J, Hooi SC, Jiang YM, Lu GD. Fatty acid activation in carcinogenesis and cancer development: Essential roles of long-chain acyl-CoA synthetases. Oncol Lett. 2018;16:1390-1396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 100] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 77. | Wang G, Dong F, Xu Z, Sharma S, Hu X, Chen D, Zhang L, Zhang J, Dong Q. MicroRNA profile in HBV-induced infection and hepatocellular carcinoma. BMC Cancer. 2017;17:805. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 67] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 78. | Caligiuri P, Cerruti R, Icardi G, Bruzzone B. Overview of hepatitis B virus mutations and their implications in the management of infection. World J Gastroenterol. 2016;22:145-154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 102] [Cited by in RCA: 109] [Article Influence: 12.1] [Reference Citation Analysis (2)] |
| 79. | Salpini R, Colagrossi L, Bellocchi MC, Surdo M, Becker C, Alteri C, Aragri M, Ricciardi A, Armenia D, Pollicita M, Di Santo F, Carioti L, Louzoun Y, Mastroianni CM, Lichtner M, Paoloni M, Esposito M, D'Amore C, Marrone A, Marignani M, Sarrecchia C, Sarmati L, Andreoni M, Angelico M, Verheyen J, Perno CF, Svicher V. Hepatitis B surface antigen genetic elements critical for immune escape correlate with hepatitis B virus reactivation upon immunosuppression. Hepatology. 2015;61:823-833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 106] [Article Influence: 10.6] [Reference Citation Analysis (1)] |
| 80. | Aragri M, Alteri C, Battisti A, Di Carlo D, Minichini C, Sagnelli C, Bellocchi MC, Pisaturo MA, Starace M, Armenia D, Carioti L, Pollicita M, Salpini R, Sagnelli E, Perno CF, Coppola N, Svicher V. Multiple Hepatitis B Virus (HBV) Quasispecies and Immune-Escape Mutations Are Present in HBV Surface Antigen and Reverse Transcriptase of Patients With Acute Hepatitis B. J Infect Dis. 2016;213:1897-1905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 81. | Pollicino T, Cacciola I, Saffioti F, Raimondo G. Hepatitis B virus PreS/S gene variants: pathobiology and clinical implications. J Hepatol. 2014;61:408-417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 211] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 82. | Wang HC, Huang W, Lai MD, Su IJ. Hepatitis B virus pre-S mutants, endoplasmic reticulum stress and hepatocarcinogenesis. Cancer Sci. 2006;97:683-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 224] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 83. | Ou JH, Laub O, Rutter WJ. Hepatitis B virus gene function: the precore region targets the core antigen to cellular membranes and causes the secretion of the e antigen. Proc Natl Acad Sci USA. 1986;83:1578-1582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 221] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 84. | Su IJ, Wang LH, Hsieh WC, Wu HC, Teng CF, Tsai HW, Huang W. The emerging role of hepatitis B virus pre-S2 deletion mutant proteins in HBV tumorigenesis. J Biomed Sci. 2014;21:98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 85. | Chen CJ, Yang HI, Su J, Jen CL, You SL, Lu SN, Huang GT, Iloeje UH; REVEAL-HBV Study Group. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA. 2006;295:65-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2309] [Cited by in RCA: 2365] [Article Influence: 124.5] [Reference Citation Analysis (0)] |
| 86. | Su IJ, Wang HC, Wu HC, Huang WY. Ground glass hepatocytes contain pre-S mutants and represent preneoplastic lesions in chronic hepatitis B virus infection. J Gastroenterol Hepatol. 2008;23:1169-1174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 115] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 87. | Na B, Huang Z, Wang Q, Qi Z, Tian Y, Lu CC, Yu J, Hanes MA, Kakar S, Huang EJ, Ou JH, Liu L, Yen TS. Transgenic expression of entire hepatitis B virus in mice induces hepatocarcinogenesis independent of chronic liver injury. PLoS One. 2011;6:e26240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 88. | Lee SA, Kim KJ, Kim DW, Kim BJ. Male-specific W4P/R mutation in the pre-S1 region of hepatitis B virus, increasing the risk of progression of liver diseases in chronic patients. J Clin Microbiol. 2013;51:3928-3936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 89. | Hsieh YH, Su IJ, Wang HC, Chang WW, Lei HY, Lai MD, Chang WT, Huang W. Pre-S mutant surface antigens in chronic hepatitis B virus infection induce oxidative stress and DNA damage. Carcinogenesis. 2004;25:2023-2032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 256] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 90. | Buckwold VE, Xu Z, Chen M, Yen TS, Ou JH. Effects of a naturally occurring mutation in the hepatitis B virus basal core promoter on precore gene expression and viral replication. J Virol. 1996;70:5845-5851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 374] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 91. | Locarnini S, McMillan J, Bartholomeusz A. The hepatitis B virus and common mutants. Semin Liver Dis. 2003;23:5-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 103] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 92. | Parekh S, Zoulim F, Ahn SH, Tsai A, Li J, Kawai S, Khan N, Trépo C, Wands J, Tong S. Genome replication, virion secretion, and e antigen expression of naturally occurring hepatitis B virus core promoter mutants. J Virol. 2003;77:6601-6612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 210] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 93. | Kosaka Y, Takase K, Kojima M, Shimizu M, Inoue K, Yoshiba M, Tanaka S, Akahane Y, Okamoto H, Tsuda F. Fulminant hepatitis B: induction by hepatitis B virus mutants defective in the precore region and incapable of encoding e antigen. Gastroenterology. 1991;100:1087-1094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 180] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 94. | Chiang PW, Jeng KS, Hu CP, Chang CM. Characterization of a cis element required for packaging and replication of the human hepatitis B virus. Virology. 1992;186:701-711. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 22] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 95. | Rodriguez-Frias F, Buti M, Jardi R, Cotrina M, Viladomiu L, Esteban R, Guardia J. Hepatitis B virus infection: precore mutants and its relation to viral genotypes and core mutations. Hepatology. 1995;22:1641-1647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 122] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 96. | Yeh CT, So M, Ng J, Yang HW, Chang ML, Lai MW, Chen TC, Lin CY, Yeh TS, Lee WC. Hepatitis B virus-DNA level and basal core promoter A1762T/G1764A mutation in liver tissue independently predict postoperative survival in hepatocellular carcinoma. Hepatology. 2010;52:1922-1933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 78] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 97. | Datta S, Chatterjee S, Veer V, Chakravarty R. Molecular biology of the hepatitis B virus for clinicians. J Clin Exp Hepatol. 2012;2:353-365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 98. | Croagh CM, Desmond PV, Bell SJ. Genotypes and viral variants in chronic hepatitis B: A review of epidemiology and clinical relevance. World J Hepatol. 2015;7:289-303. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 101] [Article Influence: 10.1] [Reference Citation Analysis (1)] |
| 99. | Yan J, Yao Z, Hu K, Zhong Y, Li M, Xiong Z, Deng M. Hepatitis B Virus Core Promoter A1762T/G1764A (TA)/T1753A/T1768A Mutations Contribute to Hepatocarcinogenesis by Deregulating Skp2 and P53. Dig Dis Sci. 2015;60:1315-1324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 100. | Hensel KO, Rendon JC, Navas MC, Rots MG, Postberg J. Virus-host interplay in hepatitis B virus infection and epigenetic treatment strategies. FEBS J. 2017;284:3550-3572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 101. | Keasler VV, Hodgson AJ, Madden CR, Slagle BL. Hepatitis B virus HBx protein localized to the nucleus restores HBx-deficient virus replication in HepG2 cells and in vivo in hydrodynamically-injected mice. Virology. 2009;390:122-129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 57] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 102. | Belloni L, Pollicino T, De Nicola F, Guerrieri F, Raffa G, Fanciulli M, Raimondo G, Levrero M. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci USA. 2009;106:19975-19979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 400] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 103. | Kim S, Kim HY, Lee S, Kim SW, Sohn S, Kim K, Cho H. Hepatitis B virus x protein induces perinuclear mitochondrial clustering in microtubule- and Dynein-dependent manners. J Virol. 2007;81:1714-1726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 75] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 104. | Bouchard MJ, Wang LH, Schneider RJ. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science. 2001;294:2376-2378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 327] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 105. | Ali A, Abdel-Hafiz H, Suhail M, Al-Mars A, Zakaria MK, Fatima K, Ahmad S, Azhar E, Chaudhary A, Qadri I. Hepatitis B virus, HBx mutants and their role in hepatocellular carcinoma. World J Gastroenterol. 2014;20:10238-10248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 113] [Cited by in RCA: 114] [Article Influence: 10.4] [Reference Citation Analysis (1)] |
| 106. | Cougot D, Allemand E, Rivière L, Benhenda S, Duroure K, Levillayer F, Muchardt C, Buendia MA, Neuveut C. Inhibition of PP1 phosphatase activity by HBx: a mechanism for the activation of hepatitis B virus transcription. Sci Signal. 2012;5:ra1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 107. | Srisuttee R, Koh SS, Kim SJ, Malilas W, Boonying W, Cho IR, Jhun BH, Ito M, Horio Y, Seto E, Oh S, Chung YH. Hepatitis B virus X (HBX) protein upregulates β-Catenin in a human hepatic cell line by sequestering SIRT1 deacetylase. Oncol Rep. 2012;28:276-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 108. | Liu S, Koh SS, Lee CG. Hepatitis B Virus X Protein and Hepatocarcinogenesis. Int J Mol Sci. 2016;17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 109. | Xie Q, Chen L, Shan X, Tang J, Zhou F, Chen Q, Quan H, Nie D, Zhang W, Huang AL, Tang N. Epigenetic silencing of SFRP1 and SFRP5 by hepatitis B virus X protein enhances hepatoma cell tumorigenicity through Wnt signaling pathway. Int J Cancer. 2014;135:635-646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 91] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 110. | Peng C, Xiao X, Kang B, He S, Li J. Serum secreted frizzled-related protein 5 Levels differentially decrease in patients with hepatitis B virus-associated chronic infection and hepatocellular carcinoma. Oncol Lett. 2014;8:1340-1344. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 111. | Zhang H, Diab A, Fan H, Mani SK, Hullinger R, Merle P, Andrisani O. PLK1 and HOTAIR Accelerate Proteasomal Degradation of SUZ12 and ZNF198 during Hepatitis B Virus-Induced Liver Carcinogenesis. Cancer Res. 2015;75:2363-2374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 119] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 112. | Zhang H, Xing Z, Mani SK, Bancel B, Durantel D, Zoulim F, Tran EJ, Merle P, Andrisani O. RNA helicase DEAD box protein 5 regulates Polycomb repressive complex 2/Hox transcript antisense intergenic RNA function in hepatitis B virus infection and hepatocarcinogenesis. Hepatology. 2016;64:1033-1048. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 117] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 113. | Qadri I, Fatima K, AbdeL-Hafiz H. Hepatitis B virus X protein impedes the DNA repair via its association with transcription factor, TFIIH. BMC Microbiol. 2011;11:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 114. | van de Klundert MA, van Hemert FJ, Zaaijer HL, Kootstra NA. The hepatitis B virus x protein inhibits thymine DNA glycosylase initiated base excision repair. PLoS One. 2012;7:e48940. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 115. | Daud M, Rana MA, Husnain T, Ijaz B. Modulation of Wnt signaling pathway by hepatitis B virus. Arch Virol. 2017;162:2937-2947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 116. | Jain S, Chang TT, Hamilton JP, Lin SY, Lin YJ, Evans AA, Selaru FM, Lin PW, Chen SH, Block TM, Hu CT, Song W, Meltzer SJ, Su YH. Methylation of the CpG sites only on the sense strand of the APC gene is specific for hepatocellular carcinoma. PLoS One. 2011;6:e26799. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 117. | D'souza S, Lau KC, Coffin CS, Patel TR. Molecular mechanisms of viral hepatitis induced hepatocellular carcinoma. World J Gastroenterol. 2020;26:5759-5783. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 94] [Cited by in RCA: 161] [Article Influence: 32.2] [Reference Citation Analysis (8)] |
| 118. | Kim M, Lee HC, Tsedensodnom O, Hartley R, Lim YS, Yu E, Merle P, Wands JR. Functional interaction between Wnt3 and Frizzled-7 Leads to activation of the Wnt/beta-catenin signaling pathway in hepatocellular carcinoma cells. J Hepatol. 2008;48:780-791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 145] [Cited by in RCA: 148] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 119. | Lin X, Wang Q, Cao Z, Geng M, Cao Y, Liu X. Differential Expression of Wnt Pathway Genes in Sporadic Hepatocellular Carcinomas Infected With Hepatitis B Virus Identified With OligoGE Arrays. Hepat Mon. 2013;13:e6192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 120. | Lian Z, Liu J, Li L, Li X, Clayton M, Wu MC, Wang HY, Arbuthnot P, Kew M, Fan D, Feitelson MA. Enhanced cell survival of Hep3B cells by the hepatitis B x antigen effector, URG11, is associated with upregulation of beta-catenin. Hepatology. 2006;43:415-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 53] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 121. | Sun Q, Wang R, Luo J, Wang P, Xiong S, Liu M, Cheng B. Notch1 promotes hepatitis B virus X protein-induced hepatocarcinogenesis via Wnt/β-Catenin pathway. Int J Oncol. 2014;45:1638-1648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 122. | Tian X, Li J, Ma ZM, Zhao C, Wan DF, Wen YM. Role of hepatitis B surface antigen in the development of hepatocellular carcinoma: regulation of lymphoid enhancer-binding factor 1. J Exp Clin Cancer Res. 2009;28:58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 123. | Shokri S, Mahmoudvand S, Taherkhani R, Farshadpour F, Jalalian FA. Complexity on modulation of NF-κB pathways by hepatitis B and C: A double-edged sword in hepatocarcinogenesis. J Cell Physiol. 2019;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 124. | Ivanov AV, Valuev-Elliston VT, Tyurina DA, Ivanova ON, Kochetkov SN, Bartosch B, Isaguliants MG. Oxidative stress, a trigger of hepatitis C and B virus-induced liver carcinogenesis. Oncotarget. 2017;8:3895-3932. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 128] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 125. | Yu LX, Ling Y, Wang HY. Role of nonresolving inflammation in hepatocellular carcinoma development and progression. NPJ Precis Oncol. 2018;2:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 215] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 126. | Weil R, Sirma H, Giannini C, Kremsdorf D, Bessia C, Dargemont C, Bréchot C, Israël A. Direct association and nuclear import of the hepatitis B virus X protein with the NF-kappaB inhibitor IkappaBalpha. Mol Cell Biol. 1999;19:6345-6354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 108] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 127. | Bui-Nguyen TM, Pakala SB, Sirigiri RD, Xia W, Hung MC, Sarin SK, Kumar V, Slagle BL, Kumar R. NF-kappaB signaling mediates the induction of MTA1 by hepatitis B virus transactivator protein HBx. Oncogene. 2010;29:1179-1189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 62] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 128. | Ma J, Sun T, Park S, Shen G, Liu J. The role of hepatitis B virus X protein is related to its differential intracellular localization. Acta Biochim Biophys Sin (Shanghai). 2011;43:583-588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 129. | Kim SY, Kim JC, Kim JK, Kim HJ, Lee HM, Choi MS, Maeng PJ, Ahn JK. Hepatitis B virus X protein enhances NFkappaB activity through cooperating with VBP1. BMB Rep. 2008;41:158-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 130. | Liu Y, Tong Z, Li T, Chen Q, Zhuo L, Li W, Wu RC, Yu C. Hepatitis B virus X protein stabilizes amplified in breast cancer 1 protein and cooperates with it to promote human hepatocellular carcinoma cell invasiveness. Hepatology. 2012;56:1015-1024. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 131. | Moon H, Cho K, Shin S, Kim DY, Han KH, Ro SW. High Risk of Hepatocellular Carcinoma Development in Fibrotic Liver: Role of the Hippo-YAP/TAZ Signaling Pathway. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 132. | Zhang T, Zhang J, You X, Liu Q, Du Y, Gao Y, Shan C, Kong G, Wang Y, Yang X, Ye L, Zhang X. Hepatitis B virus X protein modulates oncogene Yes-associated protein by CREB to promote growth of hepatoma cells. Hepatology. 2012;56:2051-2059. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 158] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 133. | Wu Y, Zhang J, Zhang H, Zhai Y. Hepatitis B virus X protein mediates yes-associated protein 1 upregulation in hepatocellular carcinoma. Oncol Lett. 2016;12:1971-1974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 134. | Gao Y, Feng J, Yang G, Zhang S, Liu Y, Bu Y, Sun M, Zhao M, Chen F, Zhang W, Ye L, Zhang X. Hepatitis B virus X protein-elevated MSL2 modulates hepatitis B virus covalently closed circular DNA by inducing degradation of APOBEC3B to enhance hepatocarcinogenesis. Hepatology. 2017;66:1413-1429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 135. | Liu N, Zhang J, Yang X, Jiao T, Zhao X, Li W, Zhu J, Yang P, Jin J, Peng J, Li Z, Ye X. HDM2 Promotes NEDDylation of Hepatitis B Virus HBx To Enhance Its Stability and Function. J Virol. 2017;91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 136. | Liu P, Zhang H, Liang X, Ma H, Luan F, Wang B, Bai F, Gao L, Ma C. HBV preS2 promotes the expression of TAZ via miRNA-338-3p to enhance the tumorigenesis of hepatocellular carcinoma. Oncotarget. 2015;6:29048-29059. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 137. | Nosaka T, Naito T, Hiramatsu K, Ohtani M, Nemoto T, Marusawa H, Ma N, Hiraku Y, Kawanishi S, Yamashita T, Kaneko S, Nakamoto Y. Gene expression profiling of hepatocarcinogenesis in a mouse model of chronic hepatitis B. PLoS One. 2017;12:e0185442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 138. | Cheng AS, Chan HL, To KF, Leung WK, Chan KK, Liew CT, Sung JJ. Cyclooxygenase-2 pathway correlates with vascular endothelial growth factor expression and tumor angiogenesis in hepatitis B virus-associated hepatocellular carcinoma. Int J Oncol. 2004;24:853-860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 139. | Alkharsah KR. VEGF Upregulation in Viral Infections and Its Possible Therapeutic Implications. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 140. | Vrancken K, Paeshuyse J, Liekens S. Angiogenic activity of hepatitis B and C viruses. Antivir Chem Chemother. 2012;22:159-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (1)] |
| 141. | Liu LP, Liang HF, Chen XP, Zhang WG, Yang SL, Xu T, Ren L. The role of NF-kappaB in Hepatitis b virus X protein-mediated upregulation of VEGF and MMPs. Cancer Invest. 2010;28:443-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 56] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 142. | Xu Q, Gu S, Liang J, Lin Z, Zheng S, Yan J. The Biological Function of Hepatitis B Virus X Protein in Hepatocellular Carcinoma. Oncol Res. 2019;27:509-514. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 143. | Wu MY, Yiang GT, Cheng PW, Chu PY, Li CJ. Molecular Targets in Hepatocarcinogenesis and Implications for Therapy. J Clin Med. 2018;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 144. | Kim YR, Byun MR, Choi JW. Integrin α6 as an invasiveness marker for hepatitis B viral X-driven hepatocellular carcinoma. Cancer Biomark. 2018;23:135-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 145. | Zhu Y, Liu H, Zhang M, Guo GL. Fatty liver diseases, bile acids, and FXR. Acta Pharm Sin B. 2016;6:409-412. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 93] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 146. | Kennedy L, Francis H. Defining the relationship between farsenoid X receptor, hepatitis B virus X protein and hepatocellular carcinoma: It's complicated. Hepatology. 2017;65:774-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 147. | Niu Y, Chen L, Wu M, Huang W, Wu X, Huang D, Xie Y, Shi G. Partial abrogation of FXR-KNG1 signaling by carboxyl-terminal truncated HBx-C30 in hepatitis B virus-associated hepatocellular carcinoma. Virus Res. 2021;293:198264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 148. | Gjoneska E, Pfenning AR, Mathys H, Quon G, Kundaje A, Tsai LH, Kellis M. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer's disease. Nature. 2015;518:365-369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 414] [Cited by in RCA: 413] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 149. | Polak P, Karlić R, Koren A, Thurman R, Sandstrom R, Lawrence M, Reynolds A, Rynes E, Vlahoviček K, Stamatoyannopoulos JA, Sunyaev SR. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015;518:360-364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 405] [Cited by in RCA: 401] [Article Influence: 40.1] [Reference Citation Analysis (0)] |
| 150. | Moore CC, McKillop IH, Huynh T. MicroRNA expression following activated protein C treatment during septic shock. J Surg Res. 2013;182:116-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 151. | Levin A, Neufeldt CJ, Pang D, Wilson K, Loewen-Dobler D, Joyce MA, Wozniak RW, Tyrrell DL. Functional characterization of nuclear localization and export signals in hepatitis C virus proteins and their role in the membranous web. PLoS One. 2014;9:e114629. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 152. | Toh TB, Lim JJ, Chow EK. Epigenetics of hepatocellular carcinoma. Clin Transl Med. 2019;8:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 78] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 153. | Khalaf AM, Fuentes D, Morshid AI, Burke MR, Kaseb AO, Hassan M, Hazle JD, Elsayes KM. Role of Wnt/β-Catenin signaling in hepatocellular carcinoma, pathogenesis, and clinical significance. J Hepatocell Carcinoma. 2018;5:61-73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 170] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 154. | Hayes CN, Zhang P, Zhang Y, Chayama K. Molecular Mechanisms of Hepatocarcinogenesis Following Sustained Virological Response in Patients with Chronic Hepatitis C Virus Infection. Viruses. 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (1)] |
| 155. | Zheng Y, Hlady RA, Joyce BT, Robertson KD, He C, Nannini DR, Kibbe WA, Achenbach CJ, Murphy RL, Roberts LR, Hou L. DNA methylation of individual repetitive elements in hepatitis C virus infection-induced hepatocellular carcinoma. Clin Epigenetics. 2019;11:145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 156. | Hamdane N, Jühling F, Crouchet E, El Saghire H, Thumann C, Oudot MA, Bandiera S, Saviano A, Ponsolles C, Roca Suarez AA, Li S, Fujiwara N, Ono A, Davidson I, Bardeesy N, Schmidl C, Bock C, Schuster C, Lupberger J, Habersetzer F, Doffoël M, Piardi T, Sommacale D, Imamura M, Uchida T, Ohdan H, Aikata H, Chayama K, Boldanova T, Pessaux P, Fuchs BC, Hoshida Y, Zeisel MB, Duong FHT, Baumert TF. HCV-Induced Epigenetic Changes Associated With Liver Cancer Risk Persist After Sustained Virologic Response. Gastroenterology 2019; 156: 2313-2329. e7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 211] [Article Influence: 35.2] [Reference Citation Analysis (0)] |
| 157. | Jühling F, Hamdane N, Crouchet E, Li S, El Saghire H, Mukherji A, Fujiwara N, Oudot MA, Thumann C, Saviano A, Roca Suarez AA, Goto K, Masia R, Sojoodi M, Arora G, Aikata H, Ono A, Tabrizian P, Schwartz M, Polyak SJ, Davidson I, Schmidl C, Bock C, Schuster C, Chayama K, Pessaux P, Tanabe KK, Hoshida Y, Zeisel MB, Duong FH, Fuchs BC, Baumert TF. Targeting clinical epigenetic reprogramming for chemoprevention of metabolic and viral hepatocellular carcinoma. Gut. 2021;70:157-169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 72] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 158. | Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931-21936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2751] [Cited by in RCA: 3108] [Article Influence: 207.2] [Reference Citation Analysis (0)] |
| 159. | Perez S, Kaspi A, Domovitz T, Davidovich A, Lavi-Itzkovitz A, Meirson T, Alison Holmes J, Dai CY, Huang CF, Chung RT, Nimer A, El-Osta A, Yaari G, Stemmer SM, Yu ML, Haviv I, Gal-Tanamy M. Hepatitis C virus leaves an epigenetic signature post cure of infection by direct-acting antivirals. PLoS Genet. 2019;15:e1008181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 126] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 160. | Goto K, Roca Suarez AA, Wrensch F, Baumert TF, Lupberger J. Hepatitis C Virus and Hepatocellular Carcinoma: When the Host Loses Its Grip. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 63] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 161. | Halley-Stott RP, Gurdon JB. Epigenetic memory in the context of nuclear reprogramming and cancer. Brief Funct Genomics. 2013;12:164-173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 162. | Hoshida Y, Fuchs BC, Bardeesy N, Baumert TF, Chung RT. Pathogenesis and prevention of hepatitis C virus-induced hepatocellular carcinoma. J Hepatol. 2014;61:S79-S90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 166] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 163. | Sodroski C, Lowey B, Hertz L, Jake Liang T, Li Q. MicroRNA-135a Modulates Hepatitis C Virus Genome Replication through Downregulation of Host Antiviral Factors. Virol Sin. 2019;34:197-210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 164. | Van Renne N, Roca Suarez AA, Duong FHT, Gondeau C, Calabrese D, Fontaine N, Ababsa A, Bandiera S, Croonenborghs T, Pochet N, De Blasi V, Pessaux P, Piardi T, Sommacale D, Ono A, Chayama K, Fujita M, Nakagawa H, Hoshida Y, Zeisel MB, Heim MH, Baumert TF, Lupberger J. miR-135a-5p-mediated downregulation of protein tyrosine phosphatase receptor delta is a candidate driver of HCV-associated hepatocarcinogenesis. Gut. 2018;67:953-962. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 165. | Bandiera S, Pernot S, El Saghire H, Durand SC, Thumann C, Crouchet E, Ye T, Fofana I, Oudot MA, Barths J, Schuster C, Pessaux P, Heim MH, Baumert TF, Zeisel MB. Hepatitis C Virus-Induced Upregulation of MicroRNA miR-146a-5p in Hepatocytes Promotes Viral Infection and Deregulates Metabolic Pathways Associated with Liver Disease Pathogenesis. J Virol. 2016;90:6387-6400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 83] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 166. | Luna JM, Scheel TK, Danino T, Shaw KS, Mele A, Fak JJ, Nishiuchi E, Takacs CN, Catanese MT, de Jong YP, Jacobson IM, Rice CM, Darnell RB. Hepatitis C virus RNA functionally sequesters miR-122. Cell. 2015;160:1099-1110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 276] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 167. | Plissonnier ML, Herzog K, Levrero M, Zeisel MB. Non-Coding RNAs and Hepatitis C Virus-Induced Hepatocellular Carcinoma. Viruses. 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 168. | Pezzuto F, Buonaguro L, Buonaguro FM, Tornesello ML. Frequency and geographic distribution of TERT promoter mutations in primary hepatocellular carcinoma. Infect Agent Cancer. 2017;12:27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 169. | Krump NA, You J. Molecular mechanisms of viral oncogenesis in humans. Nat Rev Microbiol. 2018;16:684-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 189] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 170. | Yamane D, McGivern DR, Masaki T, Lemon SM. Liver Injury and Disease Pathogenesis in Chronic Hepatitis C. In: Hepatitis C Virus: From Molecular Virology to Antiviral Therapy. Berlin: Springer-Verlag, 2013: 263-288. |
| 171. | Moradpour D, Penin F. Hepatitis C virus proteins: from structure to function. Curr Top Microbiol Immunol. 2013;369:113-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 149] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 172. | Wang W, Pan Q, Fuhler GM, Smits R, Peppelenbosch MP. Action and function of Wnt/β-Catenin signaling in the progression from chronic hepatitis C to hepatocellular carcinoma. J Gastroenterol. 2017;52:419-431. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 65] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 173. | Liu J, Ding X, Tang J, Cao Y, Hu P, Zhou F, Shan X, Cai X, Chen Q, Ling N, Zhang B, Bi Y, Chen K, Ren H, Huang A, He TC, Tang N. Enhancement of canonical Wnt/β-Catenin signaling activity by HCV core protein promotes cell growth of hepatocellular carcinoma cells. PLoS One. 2011;6:e27496. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 100] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 174. | Mahmoudvand S, Shokri S, Taherkhani R, Farshadpour F. Hepatitis C virus core protein modulates several signaling pathways involved in hepatocellular carcinoma. World J Gastroenterol. 2019;25:42-58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 60] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (1)] |
| 175. | Quan H, Zhou F, Nie D, Chen Q, Cai X, Shan X, Zhou Z, Chen K, Huang A, Li S, Tang N. Hepatitis C virus core protein epigenetically silences SFRP1 and enhances HCC aggressiveness by inducing epithelial-mesenchymal transition. Oncogene. 2014;33:2826-2835. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 88] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 176. | Umer M, Qureshi SA, Hashmi ZY, Raza A, Ahmad J, Rahman M, Iqbal M. Promoter hypermethylation of Wnt pathway inhibitors in hepatitis C virus - induced multistep hepatocarcinogenesis. Virol J. 2014;11:117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 177. | Nguyen H, Sankaran S, Dandekar S. Hepatitis C virus core protein induces expression of genes regulating immune evasion and anti-apoptosis in hepatocytes. Virology. 2006;354:58-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 54] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 178. | Chung YM, Park KJ, Choi SY, Hwang SB, Lee SY. Hepatitis C virus core protein potentiates TNF-alpha-induced NF-kappaB activation through TRAF2-IKKbeta-dependent pathway. Biochem Biophys Res Commun. 2001;284:15-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 39] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 179. | Zhu N, Khoshnan A, Schneider R, Matsumoto M, Dennert G, Ware C, Lai MM. Hepatitis C virus core protein binds to the cytoplasmic domain of tumor necrosis factor (TNF) receptor 1 and enhances TNF-induced apoptosis. J Virol. 1998;72:3691-3697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 314] [Cited by in RCA: 301] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 180. | Lee J, Tian Y, Chan ST, Kim JY, Cho C, Ou JH. TNF-α Induced by Hepatitis C Virus via TLR7 and TLR8 in Hepatocytes Supports Interferon Signaling via an Autocrine Mechanism. PLoS Pathog. 2015;11:e1004937. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 181. | Yoshida H, Kato N, Shiratori Y, Otsuka M, Maeda S, Kato J, Omata M. Hepatitis C virus core protein activates nuclear factor kappa B-dependent signaling through tumor necrosis factor receptor-associated factor. J Biol Chem. 2001;276:16399-16405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 80] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 182. | Kondo Y, Ninomiya M, Kimura O, Machida K, Funayama R, Nagashima T, Kobayashi K, Kakazu E, Kato T, Nakayama K, Lai MM, Shimosegawa T. HCV infection enhances Th17 commitment, which could affect the pathogenesis of autoimmune diseases. PLoS One. 2014;9:e98521. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 183. | Masalova OV, Lesnova EI, Permyakova KY, Samokhvalov EI, Ivanov AV, Kochetkov SN, Kushch AA. [Effect of Hepatitis C virus proteins on the production of proinflammatory and profibrotic cytokines in Huh7.5 human hepatoma cells]. Mol Biol (Mosk). 2016;50:486-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 184. | Chen Y, He L, Peng Y, Shi X, Chen J, Zhong J, Chen X, Cheng G, Deng H. The hepatitis C virus protein NS3 suppresses TNF-α-stimulated activation of NF-κB by targeting LUBAC. Sci Signal. 2015;8:ra118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 185. | Ivanov AV, Bartosch B, Smirnova OA, Isaguliants MG, Kochetkov SN. HCV and oxidative stress in the liver. Viruses. 2013;5:439-469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 174] [Cited by in RCA: 160] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 186. | Paracha UZ, Fatima K, Alqahtani M, Chaudhary A, Abuzenadah A, Damanhouri G, Qadri I. Oxidative stress and hepatitis C virus. Virol J. 2013;10:251. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 110] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 187. | Chen WC, Tseng CK, Chen YH, Lin CK, Hsu SH, Wang SN, Lee JC. HCV NS5A Up-Regulates COX-2 Expression via IL-8-Mediated Activation of the ERK/JNK MAPK Pathway. PLoS One. 2015;10:e0133264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 188. | Hu B, Xie S, Hu Y, Chen W, Chen X, Zheng Y, Wu X. Hepatitis C virus NS4B protein induces epithelial-mesenchymal transition by upregulation of Snail. Virol J. 2017;14:83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 189. | Xue Y, Mars WM, Bowen W, Singhi AD, Stoops J, Michalopoulos GK. Hepatitis C Virus Mimics Effects of Glypican-3 on CD81 and Promotes Development of Hepatocellular Carcinomas via Activation of Hippo Pathway in Hepatocytes. Am J Pathol. 2018;188:1469-1477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 190. | Li Y, Chen J, Wu C, Wang L, Lu M, Chen X. Hepatitis B virus/hepatitis C virus upregulate angiopoietin-2 expression through mitogen-activated protein kinase pathway. Hepatol Res. 2010;40:1022-1033. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 191. | Jahan S, Khaliq S, Ijaz B, Ahmad W, Hassan S. Role of HCV Core gene of genotype 1a and 3a and host gene Cox-2 in HCV-induced pathogenesis. Virol J. 2011;8:155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 192. | Luangdilok S, Box C, Harrington K, Rhŷs-Evans P, Eccles S. MAPK and PI3K signalling differentially regulate angiogenic and lymphangiogenic cytokine secretion in squamous cell carcinoma of the head and neck. Eur J Cancer. 2011;47:520-529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 193. | Im YK, La Selva R, Gandin V, Ha JR, Sabourin V, Sonenberg N, Pawson T, Topisirovic I, Ursini-Siegel J. The ShcA adaptor activates AKT signaling to potentiate breast tumor angiogenesis by stimulating VEGF mRNA translation in a 4E-BP-dependent manner. Oncogene. 2015;34:1729-1735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 194. | Kanda T, Steele R, Ray R, Ray RB. Hepatitis C virus core protein augments androgen receptor-mediated signaling. J Virol. 2008;82:11066-11072. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 76] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 195. | Shao YY, Hsieh MS, Wang HY, Li YS, Lin H, Hsu HW, Huang CY, Hsu CH, Cheng AL. Hepatitis C virus core protein potentiates proangiogenic activity of hepatocellular carcinoma cells. Oncotarget. 2017;8:86681-86692. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 196. | Hayashi J, Aoki H, Kajino K, Moriyama M, Arakawa Y, Hino O. Hepatitis C virus core protein activates the MAPK/ERK cascade synergistically with tumor promoter TPA, but not with epidermal growth factor or transforming growth factor alpha. Hepatology. 2000;32:958-961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 107] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 197. | Tsutsumi T, Suzuki T, Moriya K, Yotsuyanagi H, Shintani Y, Fujie H, Matsuura Y, Kimura S, Koike K, Miyamura T. Alteration of intrahepatic cytokine expression and AP-1 activation in transgenic mice expressing hepatitis C virus core protein. Virology. 2002;304:415-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 63] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 198. | Nakamura H, Aoki H, Hino O, Moriyama M. HCV core protein promotes heparin binding EGF-like growth factor expression and activates Akt. Hepatol Res. 2011;41:455-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 199. | Kwon YC, Sasaki R, Meyer K, Ray R. Hepatitis C Virus Core Protein Modulates Endoglin (CD105) Signaling Pathway for Liver Pathogenesis. J Virol. 2017;91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 200. | Lv SY, Cui B, Chen WD, Wang YD. Apelin/APJ system: A key therapeutic target for liver disease. Oncotarget. 2017;8:112145-112151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 201. | Cabiati M, Gaggini M, De Simone P, Del Ry S. Evaluation of Apelin/APJ system expression in hepatocellular carcinoma as a function of clinical severity. Clin Exp Med. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 202. | Benkheil M, Paeshuyse J, Neyts J, Van Haele M, Roskams T, Liekens S. HCV-induced EGFR-ERK signaling promotes a pro-inflammatory and pro-angiogenic signature contributing to liver cancer pathogenesis. Biochem Pharmacol. 2018;155:305-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 203. | Cui J, Chen Y, Wang HY, Wang RF. Mechanisms and pathways of innate immune activation and regulation in health and cancer. Hum Vaccin Immunother. 2014;10:3270-3285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 239] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 204. | Carty M, Guy C, Bowie AG. Detection of Viral Infections by Innate Immunity. Biochem Pharmacol. 2021;183:114316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 261] [Article Influence: 52.2] [Reference Citation Analysis (0)] |
| 205. | Mridha AR, Wree A, Robertson AAB, Yeh MM, Johnson CD, Van Rooyen DM, Haczeyni F, Teoh NC, Savard C, Ioannou GN, Masters SL, Schroder K, Cooper MA, Feldstein AE, Farrell GC. NLRP3 inflammasome blockade reduces liver inflammation and fibrosis in experimental NASH in mice. J Hepatol. 2017;66:1037-1046. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 546] [Cited by in RCA: 829] [Article Influence: 103.6] [Reference Citation Analysis (0)] |
| 206. | He Q, Fu Y, Tian D, Yan W. The contrasting roles of inflammasomes in cancer. Am J Cancer Res. 2018;8:566-583. [PubMed] |
| 207. | Wei Q, Mu K, Li T, Zhang Y, Yang Z, Jia X, Zhao W, Huai W, Guo P, Han L. Deregulation of the NLRP3 inflammasome in hepatic parenchymal cells during liver cancer progression. Lab Invest. 2014;94:52-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 218] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 208. | Dupaul-Chicoine J, Arabzadeh A, Dagenais M, Douglas T, Champagne C, Morizot A, Rodrigue-Gervais IG, Breton V, Colpitts SL, Beauchemin N, Saleh M. The Nlrp3 Inflammasome Suppresses Colorectal Cancer Metastatic Growth in the Liver by Promoting Natural Killer Cell Tumoricidal Activity. Immunity. 2015;43:751-763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 268] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 209. | Wei Q, Guo P, Mu K, Zhang Y, Zhao W, Huai W, Qiu Y, Li T, Ma X, Liu Y, Chen X, Han L. Estrogen suppresses hepatocellular carcinoma cells through ERβ-mediated upregulation of the NLRP3 inflammasome. Lab Invest. 2015;95:804-816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 115] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 210. | Yan W, Chang Y, Liang X, Cardinal JS, Huang H, Thorne SH, Monga SP, Geller DA, Lotze MT, Tsung A. High-mobility group box 1 activates caspase-1 and promotes hepatocellular carcinoma invasiveness and metastases. Hepatology. 2012;55:1863-1875. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 197] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 211. | Negash AA, Olson RM, Griffin S, Gale M Jr. Modulation of calcium signaling pathway by hepatitis C virus core protein stimulates NLRP3 inflammasome activation. PLoS Pathog. 2019;15:e1007593. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 212. | Negash AA, Ramos HJ, Crochet N, Lau DT, Doehle B, Papic N, Delker DA, Jo J, Bertoletti A, Hagedorn CH, Gale M Jr. IL-1β production through the NLRP3 inflammasome by hepatic macrophages links hepatitis C virus infection with liver inflammation and disease. PLoS Pathog. 2013;9:e1003330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 368] [Cited by in RCA: 356] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 213. | Zannetti C, Roblot G, Charrier E, Ainouze M, Tout I, Briat F, Isorce N, Faure-Dupuy S, Michelet M, Marotel M, Kati S, Schulz TF, Rivoire M, Traverse-Glehen A, Luangsay S, Alatiff O, Henry T, Walzer T, Durantel D, Hasan U. Characterization of the Inflammasome in Human Kupffer Cells in Response to Synthetic Agonists and Pathogens. J Immunol. 2016;197:356-367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 51] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 214. | Han Y, Chen Z, Hou R, Yan D, Liu C, Chen S, Li X, Du W. Expression of AIM2 is correlated with increased inflammation in chronic hepatitis B patients. Virol J. 2015;12:129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 215. | Watashi K, Liang G, Iwamoto M, Marusawa H, Uchida N, Daito T, Kitamura K, Muramatsu M, Ohashi H, Kiyohara T, Suzuki R, Li J, Tong S, Tanaka Y, Murata K, Aizaki H, Wakita T. Interleukin-1 and tumor necrosis factor-α trigger restriction of hepatitis B virus infection via a cytidine deaminase activation-induced cytidine deaminase (AID). J Biol Chem. 2013;288:31715-31727. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 134] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 216. | Togashi H, Ohno S, Matsuo T, Watanabe H, Saito T, Shinzawa H, Takahashi T. Interferon-gamma, tumor necrosis factor-alpha, and interleukin 1-beta suppress the replication of hepatitis B virus through oxidative stress. Res Commun Mol Pathol Pharmacol. 2000;107:407-417. [PubMed] |
| 217. | Serti E, Werner JM, Chattergoon M, Cox AL, Lohmann V, Rehermann B. Monocytes activate natural killer cells via inflammasome-induced interleukin 18 in response to hepatitis C virus replication. Gastroenterology 2014; 147: 209-220. e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 76] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 218. | Lee C, Cheung ST. STAT3: An Emerging Therapeutic Target for Hepatocellular Carcinoma. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 99] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 219. | Takeda K, Akira S. STAT family of transcription factors in cytokine-mediated biological responses. Cytokine Growth Factor Rev. 2000;11:199-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 239] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 220. | Hirano T, Ishihara K, Hibi M. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene. 2000;19:2548-2556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 889] [Cited by in RCA: 948] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 221. | Qin SK, Li Q, Ming Xu J, Liang J, Cheng Y, Fan Y, Jiang J, Ye H, Tao H, Li L, Zheng L, Wei Z, Li S, Meng K, Ye B, Sun Y. Icaritin-induced immunomodulatory efficacy in advanced hepatitis B virus-related hepatocellular carcinoma: Immunodynamic biomarkers and overall survival. Cancer Sci. 2020;111:4218-4231. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 222. | Li CJ, Liao WT, Wu MY, Chu PY. New Insights into the Role of Autophagy in Tumor Immune Microenvironment. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 223. | Matsui M, Machida S, Itani-Yohda T, Akatsuka T. Downregulation of the proteasome subunits, transporter, and antigen presentation in hepatocellular carcinoma, and their restoration by interferon-gamma. J Gastroenterol Hepatol. 2002;17:897-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 55] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 224. | Ormandy LA, Hillemann T, Wedemeyer H, Manns MP, Greten TF, Korangy F. Increased populations of regulatory T cells in peripheral blood of patients with hepatocellular carcinoma. Cancer Res. 2005;65:2457-2464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 452] [Cited by in RCA: 486] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 225. | Unitt E, Rushbrook SM, Marshall A, Davies S, Gibbs P, Morris LS, Coleman N, Alexander GJ. Compromised lymphocytes infiltrate hepatocellular carcinoma: the role of T-regulatory cells. Hepatology. 2005;41:722-730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 222] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 226. | Cariani E, Pilli M, Zerbini A, Rota C, Olivani A, Pelosi G, Schianchi C, Soliani P, Campanini N, Silini EM, Trenti T, Ferrari C, Missale G. Immunological and molecular correlates of disease recurrence after liver resection for hepatocellular carcinoma. PLoS One. 2012;7:e32493. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 227. | Yeung OW, Lo CM, Ling CC, Qi X, Geng W, Li CX, Ng KT, Forbes SJ, Guan XY, Poon RT, Fan ST, Man K. Alternatively activated (M2) macrophages promote tumour growth and invasiveness in hepatocellular carcinoma. J Hepatol. 2015;62:607-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 324] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 228. | Wu MY, Li CJ, Hou MF, Chu PY. New Insights into the Role of Inflammation in the Pathogenesis of Atherosclerosis. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 295] [Cited by in RCA: 280] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 229. | Budhu A, Forgues M, Ye QH, Jia HL, He P, Zanetti KA, Kammula US, Chen Y, Qin LX, Tang ZY, Wang XW. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10:99-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 623] [Cited by in RCA: 653] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 230. | Won C, Kim BH, Yi EH, Choi KJ, Kim EK, Jeong JM, Lee JH, Jang JJ, Yoon JH, Jeong WI, Park IC, Kim TW, Bae SS, Factor VM, Ma S, Thorgeirsson SS, Lee YH, Ye SK. Signal transducer and activator of transcription 3-mediated CD133 up-regulation contributes to promotion of hepatocellular carcinoma. Hepatology. 2015;62:1160-1173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 153] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 231. | Song G, Shi Y, Zhang M, Goswami S, Afridi S, Meng L, Ma J, Chen Y, Lin Y, Zhang J, Liu Y, Jin Z, Yang S, Rao D, Zhang S, Ke A, Wang X, Cao Y, Zhou J, Fan J, Zhang X, Xi R, Gao Q. Global immune characterization of HBV/HCV-related hepatocellular carcinoma identifies macrophage and T-cell subsets associated with disease progression. Cell Discov. 2020;6:90. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 116] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 232. | Yun CW, Lee SH. The Roles of Autophagy in Cancer. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 377] [Cited by in RCA: 684] [Article Influence: 97.7] [Reference Citation Analysis (0)] |
| 233. | Choi Y, Bowman JW, Jung JU. Autophagy during viral infection - a double-edged sword. Nat Rev Microbiol. 2018;16:341-354. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 342] [Cited by in RCA: 593] [Article Influence: 98.8] [Reference Citation Analysis (0)] |
| 234. | Cui J, Shen HM, Lim LHK. The Role of Autophagy in Liver Cancer: Crosstalk in Signaling Pathways and Potential Therapeutic Targets. Pharmaceuticals (Basel). 2020;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 235. | Lee YG, Jeon TI. Modulation of the Autophagy-lysosomal Pathway in Hepatocellular Carcinoma Using Small Molecules. Molecules. 2020;25. [PubMed] |
| 236. | Zhang L. Autophagy in hepatitis B or C virus infection: An incubator and a potential therapeutic target. Life Sci. 2020;242:117206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 237. | Xie M, Yang Z, Liu Y, Zheng M. The role of HBV-induced autophagy in HBV replication and HBV related-HCC. Life Sci. 2018;205:107-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 238. | Kotsafti A, Farinati F, Cardin R, Cillo U, Nitti D, Bortolami M. Autophagy and apoptosis-related genes in chronic liver disease and hepatocellular carcinoma. BMC Gastroenterol. 2012;12:118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 239. | Lan SH, Wu SY, Zuchini R, Lin XZ, Su IJ, Tsai TF, Lin YJ, Wu CT, Liu HS. Autophagy suppresses tumorigenesis of hepatitis B virus-associated hepatocellular carcinoma through degradation of microRNA-224. Hepatology. 2014;59:505-517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 163] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 240. | Zhang XD, Wang Y, Ye LH. Hepatitis B virus X protein accelerates the development of hepatoma. Cancer Biol Med. 2014;11:182-190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 56] [Reference Citation Analysis (0)] |
| 241. | Zhang J, Fu LL, Tian M, Liu HQ, Li JJ, Li Y, He J, Huang J, Ouyang L, Gao HY, Wang JH. Design and synthesis of a novel candidate compound NTI-007 targeting sodium taurocholate cotransporting polypeptide [NTCP]-APOA1-HBx-Beclin1-mediated autophagic pathway in HBV therapy. Bioorg Med Chem. 2015;23:976-984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 242. | Rogov V, Dötsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53:167-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 678] [Cited by in RCA: 805] [Article Influence: 73.2] [Reference Citation Analysis (0)] |
| 243. | Del Campo JA, Gallego P, Grande L. Role of inflammatory response in liver diseases: Therapeutic strategies. World J Hepatol. 2018;10:1-7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 251] [Article Influence: 35.9] [Reference Citation Analysis (4)] |
| 244. | Chung C, Seo W, Silwal P, Jo EK. Crosstalks between inflammasome and autophagy in cancer. J Hematol Oncol. 2020;13:100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 99] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 245. | Chandra PK, Gunduz F, Hazari S, Kurt R, Panigrahi R, Poat B, Bruce D, Cohen AJ, Bohorquez HE, Carmody I, Loss G, Balart LA, Wu T, Dash S. Impaired expression of type I and type II interferon receptors in HCV-associated chronic liver disease and liver cirrhosis. PLoS One. 2014;9:e108616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 246. | Aydin Y, Stephens CM, Chava S, Heidari Z, Panigrahi R, Williams DD, Wiltz K, Bell A, Wilson W, Reiss K, Dash S. Chaperone-Mediated Autophagy Promotes Beclin1 Degradation in Persistently Infected Hepatitis C Virus Cell Culture. Am J Pathol. 2018;188:2339-2355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 247. | Bartolini D, Dallaglio K, Torquato P, Piroddi M, Galli F. Nrf2-p62 autophagy pathway and its response to oxidative stress in hepatocellular carcinoma. Transl Res. 2018;193:54-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 173] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 248. | MacMicking JD, Taylor GA, McKinney JD. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science. 2003;302:654-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 517] [Cited by in RCA: 538] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 249. | Mehto S, Jena KK, Nath P, Chauhan S, Kolapalli SP, Das SK, Sahoo PK, Jain A, Taylor GA. The Crohn's Disease Risk Factor IRGM Limits NLRP3 Inflammasome Activation by Impeding Its Assembly and by Mediating Its Selective Autophagy. Mol Cell 2019; 73: 429-445. e7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 166] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 250. | Hansen MD, Johnsen IB, Stiberg KA, Sherstova T, Wakita T, Richard GM, Kandasamy RK, Meurs EF, Anthonsen MW. Hepatitis C virus triggers Golgi fragmentation and autophagy through the immunity-related GTPase M. Proc Natl Acad Sci USA. 2017;114:E3462-E3471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 104] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 251. | Wang L, Wan YY. The role of gut microbiota in liver disease development and treatment. Liver Res. 2019;3:3-18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 252. | Mohamadkhani A. On the potential role of intestinal microbial community in hepatocarcinogenesis in chronic hepatitis B. Cancer Med. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 253. | Huang H, Ren Z, Gao X, Hu X, Zhou Y, Jiang J, Lu H, Yin S, Ji J, Zhou L, Zheng S. Integrated analysis of microbiome and host transcriptome reveals correlations between gut microbiota and clinical outcomes in HBV-related hepatocellular carcinoma. Genome Med. 2020;12:102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 119] [Article Influence: 23.8] [Reference Citation Analysis (0)] |