Published online Apr 26, 2021. doi: 10.12998/wjcc.v9.i12.2884
Peer-review started: December 4, 2020
First decision: January 24, 2021
Revised: February 2, 2021
Accepted: March 3, 2021
Article in press: March 3, 2021
Published online: April 26, 2021
Processing time: 132 Days and 1.1 Hours
Gastric cancer is the fifth most diagnosed cancer worldwide and the third most common cause of cancer-related death. In recent decades, increasing application of next-generation sequencing has enabled detection of molecular aberrations, including fusions. In cases where tissue is difficult to obtain, cell-free DNA (cfDNA) is used for detecting mutations to identify the molecular profile of cancer. Here, we report a rare case of EGFR-SEPT14 fusion detected from cfDNA analysis in a patient with gastric cancer.
A 49-year-old female diagnosed with advanced gastric cancer in July 2019 received capecitabine and then combination chemotherapy of ramucirumab and paclitaxel, but ascites was detected. The therapy was switched to nivolumab, but disease progression was observed on a positron emission tomography/computed tomography scan in May 2020. Therapy was discontinued, and cfDNA next-generation sequencing was immediately evaluated. All genomic variants, including fusions, were analyzed from cfDNA. The following somatic alterations were detected from the patient’s cfDNA: an APC frameshift mutation (NM_000038.5:c.6579del, p.V2194fs) with variant allele frequency of 0.5%, an EGFR amplification with a copy number of 17.3, and an EGFR-SEPT14 fusion with variant allele frequency of 45.3%. The site of the fusion was exon 24 of EGFR fused to exon 10 of SEPT14. The fusion was in-frame and considered to be protooncogenic. Although the patient refused to continue therapy, we suggest that EGFR-targeted therapies be tried in such future cases.
The expanded applications of the cfDNA assay may open a new horizon in treatment of patients with advanced gastric cancer.
Core Tip: In recent decades, increasing application of next-generation sequencing has enabled detection of molecular aberrations, including fusions. In cases where tissue is not easily obtainable, cell-free DNA is used for detecting mutations to determine the molecular profile of cancer. In this study, we report the first case of EGFR-SEPT14 fusion detected from next-generation sequencing analysis of cell-free DNA from a patient with advanced gastric cancer. We suggest expanded applications of the cell-free DNA assay regardless of cancer type, which may open a new horizon in treatment of patients with advanced gastric cancer.
- Citation: Kim B, Kim Y, Park I, Cho JY, Lee KA. Detection of EGFR-SEPT14 fusion in cell-free DNA of a patient with advanced gastric cancer: A case report . World J Clin Cases 2021; 9(12): 2884-2889
- URL: https://www.wjgnet.com/2307-8960/full/v9/i12/2884.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i12.2884
Gastric cancer is the fifth most diagnosed cancer worldwide with a particularly high incidence in East Asia and the third most common cause of cancer-related death[1]. Curative surgery is the primary treatment of choice, but systemic chemotherapies are used for patients with metastatic or unresectable advanced or recurrent gastric cancer. Because systemic chemotherapies are nonspecific and can cause serious adverse effects, development of molecular targeted drugs has been attempted to improve outcomes in patients with gastric cancer.
In recent decades, increasing application of next-generation sequencing (NGS) has enabled detection of molecular aberrations such as copy number gains or losses, somatic mutations, and gene fusions. For cases where tissue is not easily obtainable, cell-free DNA (cfDNA) is used for detecting mutations to determine the molecular profile of cancer[2]. Successful identification of oncogenic gene fusions can aid in diagnosis and molecular treatment of patients[3]. Here, we report a rare case of EGFR-SEPT14 fusion detected from cfDNA analysis in a patient with gastric cancer.
A 49-year-old female patient had been treated for advanced gastric cancer (AGC) with chemotherapy. After therapy, she expressed whole body pain, especially on the left side of the pelvis.
This patient had been diagnosed with AGC in July 2019. The pathological diagnosis indicated signet ring cell carcinoma. While receiving her first round of chemotherapy with capecitabine, the patient developed acute pyelonephritis and hydronephrosis in both kidneys, leading to a suspicion of periureteral metastases. Therefore, the patient started a new regimen of combination chemotherapy with ramucirumab and paclitaxel. However, ascites was observed after two cycles of chemotherapy. The treatment was switched to nivolumab. After five cycles, an abdominopelvic computed tomography scan was performed in April 2020 that showed improvement in peritoneal carcinomatosis compared to an image from February 2020. She received seven cycles of nivolumab, but progressive disease was observed by the positron emission tomography/computed tomography scan, and other therapeutic options were needed to be discussed.
The patient did not have any other medical history beyond AGC.
The patient reported a family history of gastric cancer in her grandfather.
Physical examination revealed pain on the left side of the pelvis.
Blood analysis revealed mild leukocytosis (14 × 109/L) with low hemoglobin (10.3 g/dL). Platelet count was in the normal range. Serum C-reactive protein was increased at 181 mg/L (normal range, 0.1-6.0 mg/L).
A positron emission tomography/computed tomography scan obtained in May 2020 revealed bone, multiple nodal, and right lateral abdominal wall soft tissue metastases after the patient had received seven cycles of nivolumab. The therapy was discontinued, and cfDNA NGS was performed immediately.
For genetic testing, the patient provided informed written consent for specimen collection and genetic analysis. This study was approved with a waiver of informed consent by the Institutional Review Board of Gangnam Severance Hospital, Seoul, Korea (IRB No. 3-2020-0268).
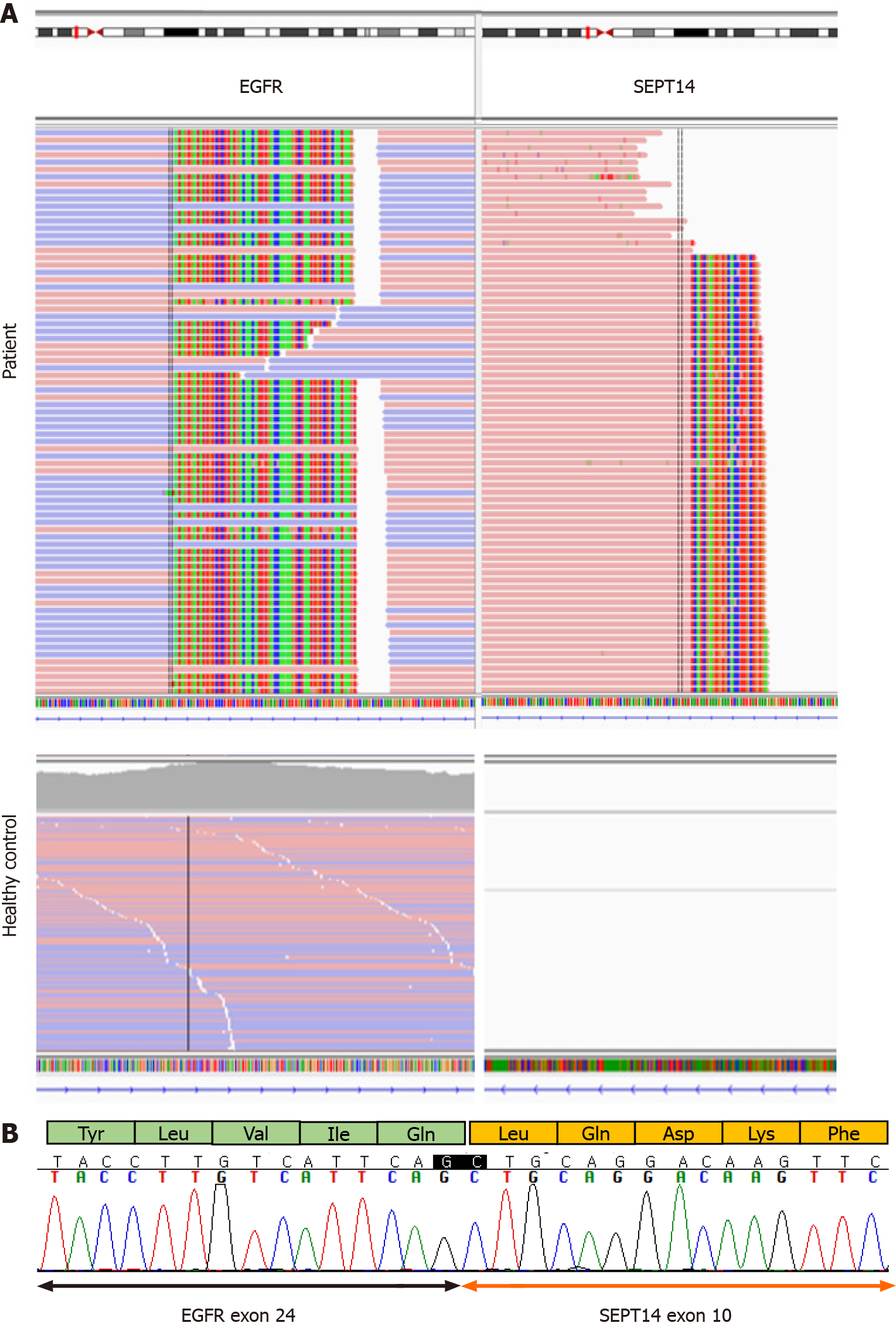
cfDNA was extracted using the MagMAX Cell-Free Total Nucleic Acid Kit (Thermo Fisher Scientific, Waltham, MA, United States). A DNA library was constructed with the AlphaLiquid®100 kit (IMBDx Inc., Seoul, Korea), which was designed to include intronic regions of target genes. Hybrid-capture-selected libraries were sequenced to a mean coverage of 14237x (cfDNA) and 735x (DNA) on an Illumina NextSeq-550 (Illumina, San Diego, CA, United States). GeneFuse was used to detect fusions[4], and a Genome Reference Consortium Human Build 38 was used for variant interpretation. All genomic variants, including fusions, were analyzed from cfDNA. Because of the patient’s family history, the presence of germline mutation was tested in parallel for the following genes: APC, ATM, BRCA1, BRCA2, CDH1, CDK4, CDKN2A, and MLH1. No germline mutations were detected from the genomic DNA. Somatic alterations detected from the cfDNA were an APC frameshift mutation (NM_000038.5:c.6579del, p.V2194fs) with variant allele frequency of 0.5%, an EGFR amplification with a copy number of 17.3, and an EGFR-SEPT14 fusion with variant allele frequency of 45.3% (Figure 1A). Because the EGFR and SEPT14 genes are closely located on chromosome 7, we tested 50 normal healthy controls with the same panel and confirmed that the fusion detected in the patient was a true positive. We also confirmed EGFR-SEPT14 fusion by complementary DNA sequencing, which was processed using the patient’s cell-free RNA extracted by MagMAX Cell-Free Total Nucleic Acid Kit. The site of fusion was exon 24 of EGFR fused to exon 10 of SEPT14 (Figure 1B). The fusion was in-frame and considered to be proto-oncogenic.
The final diagnosis of the present case was EGFR-SEPT14 fusion in AGC.
The patient refused further treatment.
The patient could have tried EGFR targeted therapy such as erlotinib, which has been used in other types of carcinomas with EGFR-SEPT14 fusion[5], but she refused further treatment and passed away about 1 month after discontinuation of nivolumab.
EGFR1 (EGFR; ErbB1; HER1) is one of four transmembrane growth factor receptor proteins that constitute the EGFR family of receptor tyrosine kinases[6]. Activation of EGFR leads to cell proliferation, differentiation, motility, and metastasis[7]. SEPT14 is a member of a highly conserved septin family of guanosine 5’-triphosphate-binding cytoskeletal proteins with multiple cellular functions, such as membrane transport, apoptosis, cell polarity, cell cycle regulation, cytokinesis, and oncogenesis[8]. Among all septins, SEPT14 shows the highest mutation frequency in skin cancer followed by SEPT9 exhibiting high mutation frequency in stomach cancer[9].
The EGFR-SEPT14 fusion was first reported in glioblastoma in which the site of fusion was the tyrosine kinase domain of EGFR and the coiled-coil domain of SEPT14. The EGFR-SEPT14 fusion is the most frequent functional gene fusion in human glioblastoma[10]. The EGFR-SEPT14 fusion was also identified in tissue from salivary gland secretory carcinoma using fluorescence in situ hybridization. That previous case indicated that a tumor harboring this fusion would be sensitive to EGFR inhibitors[11]. Recently, the EGFR-SEPT14 fusion was reported in colorectal adenocarcinoma by using a comprehensive NGS assay on tumor samples[5].
In the present study, the tissue biopsy of the patient was difficult. Therefore, we used a comprehensive NGS assay with a sample of cfDNA from the patient. We identified an EGFR-SEPT14 fusion in AGC. To our knowledge, this is the first case of EGFR-SEPT14 fusion identified in a cfDNA sample from an AGC patient. The patient went through unusually rapid disease progression, and this progression might have been caused by the fusion mutation. Unfortunately, because the patient refused to continue therapy, we could not determine whether the EGFR-SEPT14 fusion responded to EGFR targeted therapies, such as tyrosine kinase inhibitors. However, the use of such therapies might have been effective in AGC with an EGFR-SEPT14 fusion because there was a report of a patient with colorectal cancer with an EGFR-SEPT14 fusion treated with erlotinib therapy. The fusion site reported in that study is the same as that in the present study, and the patient was administered erlotinib therapy to which the EGFR-SEPT14 fusion is known to be sensitive[10]. However, soon after treatment, an EGFR variant III was detected and can result in resistance to erlotinib[5]. To confirm the treatment effect and disease progression in AGC, further studies are needed.
Nevertheless, detection of genomic fusion by the well-established cfDNA NGS assay confirmed that cfDNA can serve as an alternate source for detecting gene aberrations, including fusions. Furthermore, EGFR-SEPT14 fusion has been reported in various types of cancer. Therefore, expanded applications of cfDNA assays should be considered regardless of cancer type. We also suggest that genomic variants including fusions can be therapeutic targets in AGC, which may open a new horizon in treatment.
To the best of our knowledge, this is the first case of an EGFR-SEPT14 fusion identified in a cfDNA sample from a patient with AGC. Detection of genomic fusion by the well-established cfDNA NGS assay confirmed that cfDNA can serve as an alternate source for detecting gene aberrations, including fusions. Successful identification of genomic variants, including fusions, from cfDNA can aid in diagnosis and molecular treatment of patients with AGC.
Manuscript source: Unsolicited manuscript
Specialty type: Medicine, research and experimental
Country/Territory of origin: South Korea
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Jin X S-Editor: Fan JR L-Editor: Filipodia P-Editor: Yuan YY
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55670] [Article Influence: 7952.9] [Reference Citation Analysis (132)] |
| 2. | Volckmar AL, Sültmann H, Riediger A, Fioretos T, Schirmacher P, Endris V, Stenzinger A, Dietz S. A field guide for cancer diagnostics using cell-free DNA: From principles to practice and clinical applications. Genes Chromosomes Cancer. 2018;57:123-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 121] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 3. | Schram AM, Chang MT, Jonsson P, Drilon A. Fusions in solid tumours: diagnostic strategies, targeted therapy, and acquired resistance. Nat Rev Clin Oncol. 2017;14:735-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 245] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 4. | Chen S, Liu M, Huang T, Liao W, Xu M, Gu J. GeneFuse: detection and visualization of target gene fusions from DNA sequencing data. Int J Biol Sci. 2018;14:843-848. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 5. | Li Y, Zhang HB, Chen X, Yang X, Ye Y, Bekaii-Saab T, Zheng Y, Zhang Y. A Rare EGFR-SEPT14 Fusion in a Patient with Colorectal Adenocarcinoma Responding to Erlotinib. Oncologist. 2020;25:203-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 6. | Herbst RS. Review of epidermal growth factor receptor biology. Int J Radiat Oncol Biol Phys. 2004;59:21-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 825] [Cited by in RCA: 935] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 7. | Sigismund S, Avanzato D, Lanzetti L. Emerging functions of the EGFR in cancer. Mol Oncol. 2018;12:3-20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 609] [Cited by in RCA: 1003] [Article Influence: 143.3] [Reference Citation Analysis (0)] |
| 8. | Peterson EA, Kalikin LM, Steels JD, Estey MP, Trimble WS, Petty EM. Characterization of a SEPT9 interacting protein, SEPT14, a novel testis-specific septin. Mamm Genome. 2007;18:796-807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 42] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 9. | Angelis D, Spiliotis ET. Septin Mutations in Human Cancers. Front Cell Dev Biol. 2016;4:122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 57] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 10. | Frattini V, Trifonov V, Chan JM, Castano A, Lia M, Abate F, Keir ST, Ji AX, Zoppoli P, Niola F, Danussi C, Dolgalev I, Porrati P, Pellegatta S, Heguy A, Gupta G, Pisapia DJ, Canoll P, Bruce JN, McLendon RE, Yan H, Aldape K, Finocchiaro G, Mikkelsen T, Privé GG, Bigner DD, Lasorella A, Rabadan R, Iavarone A. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45:1141-1149. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 373] [Cited by in RCA: 433] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 11. | Black M, Liu CZ, Onozato M, Iafrate AJ, Darvishian F, Jour G, Cotzia P. Concurrent Identification of Novel EGFR-SEPT14 Fusion and ETV6-RET Fusion in Secretory Carcinoma of the Salivary Gland. Head Neck Pathol. 2020;14:817-821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |