Published online Dec 26, 2020. doi: 10.12998/wjcc.v8.i24.6213
Peer-review started: July 13, 2020
First decision: September 30, 2020
Revised: October 13, 2020
Accepted: November 2, 2020
Article in press: November 2, 2020
Published online: December 26, 2020
Processing time: 159 Days and 13.5 Hours
The prevalence of colorectal cancer (CRC) and type 2 diabetes mellitus (T2DM) is increasing globally. It is rarely noticed that the incidence of CRC is higher in patients with T2DM. What needs to be mentioned is that metformin, a commonly used clinical drug for T2DM, attracts scholars’ attention because of its benefits in lowering the risk of developing CRC. Hence, we try to find the common grounds of initiation of T2DM and CRC and the reason why metformin reduces the risk of CRC in patients with T2DM. We noticed consistent changes of gut microbiota, such as elevated Bacteroides, Prevotella and Bifidobacterium and depressed Firmicutes and Lactobacillus. Furthermore, many studies in recent years have proved that the efficacy of metformin, such as improving blood glucose, depends on the gut microbiota. Coincidentally, the progression of CRC is inseparable from the contributions of gut microbiota. Therefore, we first proposed the concept of the metformin-gut microbiota–CRC (in T2DM) axis to explain the effect of metformin in reducing CRC in patients with T2DM. In this review, we elaborated the new concept and its potential clinical application value.
Core Tip: Metformin has been found to reduce colorectal cancer in patients with type 2 diabetes, but the mechanism is unknown. Studies have confirmed that gut microbiota is not only closely related to type 2 diabetes and colorectal cancer, but also mediates the effects of metformin. Therefore, we proposed the concept of the metformin-gut microbiota-colorectal cancer axis in type 2 diabetes mellitus. The concept also provides new ideas for clinical drug use, clinical cancer treatment, and clinical application of gut microbiota.
- Citation: Huang QY, Yao F, Zhou CR, Huang XY, Wang Q, Long H, Wu QM. Role of gut microbiome in regulating the effectiveness of metformin in reducing colorectal cancer in type 2 diabetes. World J Clin Cases 2020; 8(24): 6213-6228
- URL: https://www.wjgnet.com/2307-8960/full/v8/i24/6213.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v8.i24.6213
Several decades ago, colorectal cancer (CRC) was rarely diagnosed. To date, it has become the fourth deadliest cancer in the world, killing an average of 90000 people every year. The accumulation of environmental and genetic factors leading to genetic mutations and epigenetic changes are the factors in the carcinogenesis of intestinal epithelial cells[1]. Nevertheless, recent studies have shown that the occurrence of CRC is closely related to the gut microbiota, and the alterations of gut microbiota are inconsistent at different stages[2,3]. Table 1 summarizes the main alterations of gut microbiota in human studies related to CRC from 2014 to 2019.
| Tumor types | Status | Alteration of gut microbiota |
| Adenomas | Increased1 | Families: Ruminococcaceae, Porphyromonadaceae |
| Genera: Clostridium, Pseudomonas | ||
| Species: Fusobacterium nucleatum, Atopobium parvulum, Actinomyces odontolyticus | ||
| Decreased1 | Order: Clostridiales | |
| Family: Lachnospiraceae | ||
| Genera: Bacteroides, Clostridium | ||
| Species: Bifidobactium animalis, Streptococcus thermophilus | ||
| Carcinoma | Increased1 | Classes: Bacilli, Gammaproteobacteria |
| Order: Enterobacteriales | ||
| Genera: Bacteroides, Parabacteroides, Fusobacterium, Alistipes, Escherichia, Parvimonas, Bilophila. Porphyromonas, Actinomyces, Streptococcus | ||
| Species: Alistipes putredinis, Bilophila wadsworthia, Lachnospiraceae bacterium, Enterobacteriaceae. Fusobacterium nucleatum spp., Actinomyces odontolyticus, Atopobium parvulum | ||
| Decreased1 | Classes: Erysipelotrichi, Clostridia | |
| Order: Clostridiales | ||
| Families: Lachnospiraceae, Ruminococcaceae, Clostridiaceae | ||
| Genera: Bacteroides | ||
| Species: Bifidobactium animalis, Streptococcus thermophilus | ||
| CRC | Increased1 | Phyla: Firmicutes, Fusobacteria, Bacteroidetes |
| Families: Erysipelotrichaceae, Prevotellaceae, Coriobacteriaceae | ||
| Genus: Peptostreptococcus | ||
| Species: Colinsella aerofaciens, Dorea longicatena, Porphyromonas uenonis, Selenomonas sputigena, Streptococcus anginosus, Desulfovibrio vietnamensis, Bilophila wadsworthia, Fusobacterium nucleatum, Parvimonas micra, Peptostreptococcus anaerobius, Solobacterium moorei, Eubacterium ventriosum, Clostridium hathewayi, Bacteroides clarus, Roseburia intestinalis | ||
| Others: clbA+ bacteria, Clostridium symbiosum | ||
| Decreased1 | Genus: Bifidobacterium | |
| Species: Lachnospira multipara, Eubacterium eligens |
There are more than 547 million patients with diabetes mellitus in the world, of which 373 million have been diagnosed. The number of young patients aged 20-39 exceeded 60 million in 2013[4]. Recent studies have shown that type 2 diabetes mellitus (T2DM) is also closely related to changes in gut microbiota. Table 2 summarizes the main alterations of gut microbiota in patients with T2DM. Interestingly, metformin (systematic name: 1-carbamimidamido-N,N-dimethylmethanimidamide, C4H11N5), as one of the most well-known drugs to treat T2DM, has been shown to have a positive pharmacological effect on restraining CRC in T2DM patients. Moreover, more and more studies have proved that its pharmacological function is mediated by gut microbiota.
| Status | Classification | T2DM | T2DM and CRC |
| Increased1 | Phyla | Verrucomicrobia, Proteobacteria, Actinobacteria, Euryarchaeota, Firmicutes | Firmicutes |
| Order | Lactobacillales, Bacteroidales | ||
| Families | Lachnospiraceae, Erysipelotrichaceae | Lachnospiraceae, Erysipelotrichaceae | |
| Genera | Ruminococcus, Enterobacteriaceae, Eggerthella, Desulfovibrio, Escherichia, Haemophilus, Clostridium, Eubacterium, Subdoligranulum, Bacteroides, Parabacteroides, Bifidobacterium spp., Lactobacillus | Enterobacteriaceae, Desulfovibrio, Escherichia, Clostridium, Eubacterium, Bacteroides, Parabacteroides, Bifidobacterium spp.Species: Alistipes, | |
| Species | Akkermansia, Alistipes, Parabacteroides | ||
| Decreased1 | Phyla | Tenericutes, Fusobacteria, Firmicutes, Bacteroidetes, Verrucomicrobia, Proteobacteria, Elusimicrobia | |
| Families | Lachnospiraceae, Peptostreptococcaceae, Flavobacteriaceae, Clostridiaceae | Lachnospiraceae, Clostridiaceae | |
| Genera | Bacteroides, Anaerostipes, Roseburia, Faecalibacterium, Bifidobacterium | Bacteroides, Bifidobacterium | |
| Species | A. muciniphila, F. prausnitzii |
Epidemiological evidence suggests that CRC is predisposed in patients with T2DM[5]. However, after comparing and analyzing the data of current research, the consistent changes of some gut microbiota were observed in patients with T2DM and patients with CRC. Table 2 summarizes the similarities in changing gut microbiota in patients with T2DM and patients with CRC. Therefore, based on the facts above, we set up a new concept called the metformin-gut microbiota-CRC axis (in T2DM) to explain that metformin reduces the incidence of CRC in patients with T2DM. More importantly, the new concept can be extended to be the drug-gut microbiota-diseases axis. If this theory is proven to be effective, it will provide new ideas for the use of clinical drugs and the application of gut microbiota for the treatment of clinical diseases.
To date, epidemiological evidence suggests that T2DM is one of the risk factors for CRC. However, the latest meta-analysis of observational studies displayed insulin therapy significantly increased the risk of CRC [risk ratio (95% confidence interval (CI)): 1.69 (1.25, 2.27)][6]. Compared to this, the latest meta-analysis from observational studies demonstrated that metformin therapy was associated with a significantly lower overall survival [pooled risk ratio = 0.75, 95%CI: 0.66-0.86][7], lower CRC-specific survival [hazard ratio (95%CI): 0.66 (0.50-0.87)][8], and a lower risk of colorectal neoplasm [risk ratio (95%CI): 0.63 (0.50-0.79)] in patients with T2DM[9]. In vivo, Tomimoto et al[10] suggested administration of metformin significantly reduced the number of tumors larger than 2 mm in diameter in Apc (Min/+) mice. Subsequent studies have shown that metformin suppressed colorectal aberrant crypt foci in humans[11,12] and an azoxymethane-induced animal mode of CRC[13]. In patients with T2DM, it has been reported that metformin has a positive prophylactic effect on colorectal adenomas[14] and CRC[15]. In a multicenter double-blind, placebo-controlled, randomized phase 3 trial, Higurashi et al[16] proved the administration of low-dose metformin for 1 year to patients was safe and reduced the prevalence and number of metachronous adenomas or polyps after polypectomy.
At the cellular level, the effect of metformin is mainly manifested in inhibiting the proliferation of CRC cells as a result of AMPK activation and increased reactive oxygen species production[17,18] on the one hand and accelerating the apoptosis of CRC cells induced by immune cells and cytokines on the other hand[19]. Complementarily, metformin may go through a variety of pathways or mechanisms, such as by alteration of cellular responses to oxidative stress, protection of mitochondrial structures via suppressing reactive oxygen species production and NF-kB activity[20-22], and repression of epithelial-mesenchymal transition induced by IL-6 or TGF-β[19,23]. At the genetic level, it has been shown that the upregulation of adenosine A1 receptor[24], blocking of encoding DNA replication proteins and protooncogene protein synthesis[25,26], and regulation of the SNAIL/miR-34:ZEB/miR-200 system[27] are the targets of metformin. In addition, key sites for metformin action have been shown in multiple signaling pathways, such as attenuation of cell stemness via inhibiting the Wnt3a/β-catenin[28] and AMPK/PI3K/Akt pathways[29].
To sum up, in both humans and animals or at both the individual and cellular level, metformin has shown an effect of inhibiting the development of CRC. However, as a treatment for T2DM, the underlying mechanism has not been fully elucidated.
The changes of gut microbiota have been investigated to be associated with metformin administration, contributing to the beneficial therapeutic effects[30,31]. Decreased profiles of Intestinibacter spp. and Clostridium spp. were observed upon administration of metformin to health young men with a corresponding increase of Escherichia/Shigella spp. and Bilophila wadsworthia[32]. To date, there are many studies focusing on the role of the gut microbiota that mediate the effect of metformin in regard to improving the metabolic phenotype.
In diet-induced obese mice, an increase in the population of Akkermansia, a mucin degradation bacteria, induced the improved glucose homeostasis in the cases of metformin administration[33]. Subsequent work focused on the effects of improving metabolism and delaying T2DM progression in animal models[34,35]. The study addressed by Forslund et al[36] supported the opinion from the perspective of the human gut genome that metformin could reduce the consumption of short-chain fatty acid-producing groups in T2DM and improve the transfer of functional microbial groups. Through the implementation of 16S rRNA gene sequencing, it was observed that the participating microbiota were mainly Akkermansia muciniphila and short-chain fatty acid-producing bacteria, including Butyrivibrio, Bifidobacterium bifidum, Megasphaera, and Prevotella after the metformin intervention[37,38].
Overall, at the phylum level, the main shifts by metformin in patients with T2DM are reflected in the increase of Bacteroidetes, Actinobacteria, and Proteobacteria with a corresponding decrease of Firmicutes and Verrucomicrobia. At the genus level, the increased bacteria were mainly Bacteroides, Streptococcus, Collinsella, Escherichia, Clostridium, and Subdoligranulum, while the decreased bacteria were Faecalibacterium and Ruminococcus[39,40]. Finally, through a comparative analysis of currently available data, we found that the alteration of gut microbiota involved in the inhibition of CRC by metformin in T2DM may be mainly the increase of Firmicutes and decrease of Bacteroidetes, Fusobacteria, and Bacteroidetes at the phyla levels. At the genus level, it is mainly manifested as the increase of Bifidobacterium and decrease of Fusobacterium (F.) nucleatum.
In addition, Bauer et al[41], Sun et al[42], and Pryor et al[43] confirmed that metformin improved metabolic dysfunction, such as hyperglycemia, via the glucose-SGLT1-sensing glucoregulatory pathway, B. fragilis–GUDCA–intestinal FXR axis, and the model of host-microbe-drug-nutrient interactions, respectively. These three new findings are profound to help understand how metformin and gut bacteria together affect disease progression. Table 3 lists their details and the involved types of gut microbiota. Not only that, with the participation of gut microbiota, other fields have gradually revealed the role of metformin, including anti-inflammatory effects, downregulation of interleukin expression, and ameliorating polycystic ovary syndrome in an animal model[44]. Moreover, metformin accelerated fatty acid oxidation by regulating host metabolism and longevity under the action of regulating a multipressure metabolic system[43].
| Status | Classification | Metformin-related | CRC-related | ||||
| Multiple polypoid | Stage 0/pTis | Stage SI/II | Stage SIII/IV | ||||
| Increased1 | Phyla | Bacteroidetes, Actinobacteria, Proteobacteria | Proteobacteria | Firmicutes, Fusobacteria Bacteroidetes | Firmicutes, Fusobacteria Bacteroidetes | Firmicutes, Fusobacteria Bacteroidetes | |
| Genus | Bacteroides, Streptococcus, Collinsella, Escherichia, | F. nucleatum | F. nucleatum | F. nucleatum | |||
| Species | Escherichia spp., Anaerostipes, Blautia, Akkermansia muciniphila | Atopobium parvulum | Solobacterium moorei, Lactobacillus sanfranciscensis, Gemella morbillorum, Actinomyces odontolyticus, Desulfovibrio longreachensis, Phascolarctobacterium succinatutens | Solobacterium moorei, Peptostreptococcus stomatis, Peptostreptococcus anaerobius, Parvimonas micra, Gemella morbillorum | Solobacterium moorei, Peptostreptococcus stomatis, Peptostreptococcus anaerobius, Lactobacillus sanfranciscensis, Parvimonas micra, Gemella morbillorum | ||
| Decreased1 | Phyla | Firmicutes, Verrucomicrobia | |||||
| Genus | Faecalibacterium, Ruminococcus | Bifidobacterium | |||||
| Species | Intestinibacter spp., Intestinibacter bartlettii., Alistipes, Oscillibacter, un-Ruminococcaceae | ||||||
| Other axes | Increased1 | Lactobacillus (Glucose-SGLT1-Sensing Glucoregulatory Pathway) | |||||
| E. coli, Proteobacteria (Host-GUDCA-intestinal FXR axis) | |||||||
| Decreased1 | Bacteroides fragilis (B. fragilis–GUDCA–intestinal FXR axis) | ||||||
Therefore, it can be confidently said that the gut microbiota, such as Akkermansia, B. fragilis and E. coli, mediate the pharmacological effects of metformin in different fields separately.
At present, many studies have confirmed that gut microbiota affect the occurrence and development of CRC. In humans, Yachida et al[2] used metagenomics analysis techniques to analyze the changes of gut microbiota in different stages of CRC on samples from a large cohort of 616 participants. They found the relative abundance of Fusobacterium nucleatum spp. was significantly elevated continuously from intramucosal carcinoma to more advanced stages. In addition, Atopobium parvulum and Actinomyces odontolyticus, which co-occurred in intramucosal carcinomas, were significantly increased only in multiple polypoid adenomas and/or intramucosal carcinomas. Moreover, Yu et al[45] has shown that F. nucleatum was abundant in CRC tissues in patients with recurrence post chemotherapy. F. nucleatum promoted CRC resistance to chemotherapy by targeting TLR4 and MYD88 innate immune signaling and specific microRNAs to activate the autophagy pathway. By 16S rRNA gene sequencing of stool samples in taxon-based analysis, stool of conventional adenoma patients was depleted in a network of Clostridia operational taxonomic units from families Ruminococcaceae, Clostridiaceae, and Lachnospiraceae and enriched in the classes Bacilli and Gammaproteobacteria, order Enterobacteriales, and genera Actinomyces and Streptococcus[46].
In animal models through fecal bacteria transplantation, the intestinal microbiota of CRC patients promoted the progression of intestinal adenomas in Apcmin/+ mice[47]. Donohoe et al[48] proved that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner by a gnotobiotic mouse model. Zhu et al[49] found that editing of the gut microbiota reduced carcinogenesis in colitis-associated CRC. Qin et al[50] suggested that the gut microbiome, under specific dietary exposures, stimulates a reprogramming of the enhancer landscape in the colon with downstream effects on transcription factors, which may be associated with CRC development. In addition, the immune system plays an important role in the effect of gut microbiota on CRC. Cremonesi et al[51] demonstrated that gut microbiota modulated T cell trafficking in human CRC. Wang et al[52] has shown that a purified membrane protein from Akkermansia muciniphila or pasteurized bacterium blunts colitis-associated tumorigenesis by modulation of CD8+ T cells. Meanwhile, Long et al[53] has also shown that Peptostreptococcus anaerobius promoted the occurrence of CRC and regulated tumor-related immunity. At the cellular level, Belcheva et al[54] demonstrated that gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells.
To sum up, in both humans and animals, gut microbiota is closely related to CRC, and this relationship refers to the immune system. In humans, studies have elaborated changes in the types of gut microbiota, and animal and cell experiments have further elaborated the possible mechanisms by which gut microbiota affects CRC. In Wong et al[55]’s review, the gut microbiota is thought to influence colorectal carcinogenesis via microbial-derived factors such as metabolites or genotoxins, promotion of cancer, or proliferating as opportunistic microorganisms in the tumor-associated microenvironment and the activation of procarcinogenic signaling pathways.
As we described before, the changes in the gut microbiota are inconsistent at different stages of development of CRC. According to the interaction between metformin and gut microbiota, does metformin mediate the changes of different gut microbiota at different stages of the development of CRC?
Based on the eighth Union for International Cancer Control Tumor Node Metastasis Classification of Malignant Tumors, at the phylum level in stage 0/pTis, the enriched bacteria are mainly Actinobacteria accompanied by a certain amount of Proteobacteria, Firmicutes, and Fusobacteria. In stage SI/II, the bacteria that are enriched are Proteobacteri, Firmicutes, Fusobacteria, and Bacteroidetes, and they show roughly the same abundance. In the stage SIII/IV, the enrichment of Firmicutes and Bacteroidetes is more obvious. Proteobacteri, Actinobacteria, and Fusobacteria are also present, but their abundance is not very high and is roughly equal[2].
In a nonblinded, one-armed intervention study, the relative abundance of 11 bacterial genera significantly changed during metformin intervention but returned to baseline levels after treatment cessation[32], which demonstrated that the effects of metformin on the gut microbiota are transient. However, for patients with T2DM, this condition is nothing to worry about because they require long-term medication. Moreover, there was a significant decrease of the phylum Firmicutes and of the ratio of Firmicutes to Bacteroidetes after taking metformin[39,56]. The other result of the microbiome analyses indicated a shift in the bacterial distribution in fish exposed to metformin, leading to an increase of Proteobacteria and a reduction of Firmicutes and Actinobacteria[57]. Compared with the previous data, metformin use from intramucosal cancer to advanced stage is likely to mediate its anticancer effect by provoking different gut microbiota. In stage 0/pTis, Actinobacteria may be the main target of the action of metformin, which is specifically manifested by attenuating its abundance. In stage SI/II and SIII/IV, the decrease of the phylum Firmicutes and of the ratio of Firmicutes to Bacteroidetes may be the main function of metformin. Of course, these processes may also be accompanied by the regulation of other flora. Table 3 details the changes. Therefore, the use of metformin to interfere with different types of gut microbiota at different stages of the development of CRC may be a means to improve the survival rate and prognosis of CRC in the future.
Patients with stage III colon cancer have a risk of recurrence ranging between 15% and 50%[58]. Interestingly, the administration of metformin is associated with decreased recurrence of carcinoma[59], covering the colorectal adenoma (hazard ratio 0.572, 95%CI 0.385-0.852) in diabetic patients with previous colorectal adenoma[60].
So, how does the gut microbiota influence CRC recurrence? A recent study showed that higher fear of cancer recurrence was associated with lower relative abundance of Firmicutes and higher relative abundance of Bacteroidetes at the phylum level and higher relative abundance of Bacteroides and lower relative abundance of Lachnospiraceae and Ruminococcus at the genus level. The chemotherapy-induced changes in gut microbiota influence the fear of cancer recurrence[61]. In addition, colorectal adenoma resection gave rise to a significant increase of Parabacteroides postoperatively. The microbiota signature of Parabacteroides, Streptococcus, and Ruminococcus showed an optimal discriminating performance of postoperative status[62]. These changes suggested that gut microbiota may be used in the future to prevent postoperative recurrence of CRC. Finally, it has been stressed that the intervention of metformin can directly affect the gut microbiota profile. In other words, metformin may inhibit the recurrence of CRC by changing the abundance structure of the corresponding gut microbiota.
The recurrence of CRC is resistant to routine chemotherapy and thought to be due to the enrichment of cancer stem cells. It has been reported that long term treatment of metformin impedes development of chemoresistance by regulating cancer stem cell differentiation in cancer cells[63]. Combination metformin with chemotherapeutics showed an overall modest but intriguing activity in patients with refractory CRC[64] and a synergistic inhibitory cytotoxicity in human CRC cells[65]. Moreover, metformin can act as an alternative radiosensitizing agent to 5-fluorouracil during neoadjuvant treatment for rectal cancer[66]. In addition, a large number of studies have confirmed that metformin, as a cancer stem cell-targeting agent in ovarian cancer, has a direct inhibitory effect on cancer stem cells[63,67]. The metformin intervention reduced expression of cancer stem cell markers and stemness-related genes in primary oral cancer cells[68].
Interestingly, microbiota also have a substantial role in the effectiveness of chemotherapy, chemoresistance, and related side effects[69]. For instance, the presence of specific bacterial species such as Slackia and Blautia obeum may act as microbial markers associated with drug resistance monitoring[70]. The inhibition of the growth of F. nucleatum significantly augments the efficiency of first-line chemotherapy treatments of CRC[71]. In addition, chemotherapy-induced changes in gut microbiota impact chemotherapy via influencing fear of cancer recurrence[61]. Moreover, in terms of cancer stem cells, gut microbiota regulate tumor metastasis via impacting circular RNA expression to regulate levels of corresponding miRNAs[72].
All in all, in the recurrence of CRC due to both the chemoresistance and the enrichment of cancer stem cells the gut microbiota plays a vital role, which may be one of the reasons why metformin can inhibit the recurrence of CRC in T2DM patients.
Metformin, known as a “magic drug”, reduces the incidence of CRC in people with T2DM. Moreover, the gut microbiota has been shown to mediate the pharmacological effects of metformin and the progression of CRC. Finally, the shift of gut microbiota has similarity in the occurrence and development of T2DM and CRC. Therefore, a network of metformin-gut microbiota-CRC (in type 2 diabetes) axis could be assumed. In the network, the gut microbiota plays a key role as a bridge that affects not only the pharmacological effects of metformin but also the occurrence of CRC in T2DM patients.
We have hypothesized the potential mechanism of action of this axis around the gut microbiota. On the one hand, these mechanisms will pave the way for us to understand how gut microbiota can participate in drug therapy. On the other hand, they will provide ideas for the prospective clinical application and transformation of gut microbiota. More importantly, this axis can be expanded into a profound network of gut microbiota-drug-disease to explain the crosstalk relationship between gut microbiota, drugs, and diseases.
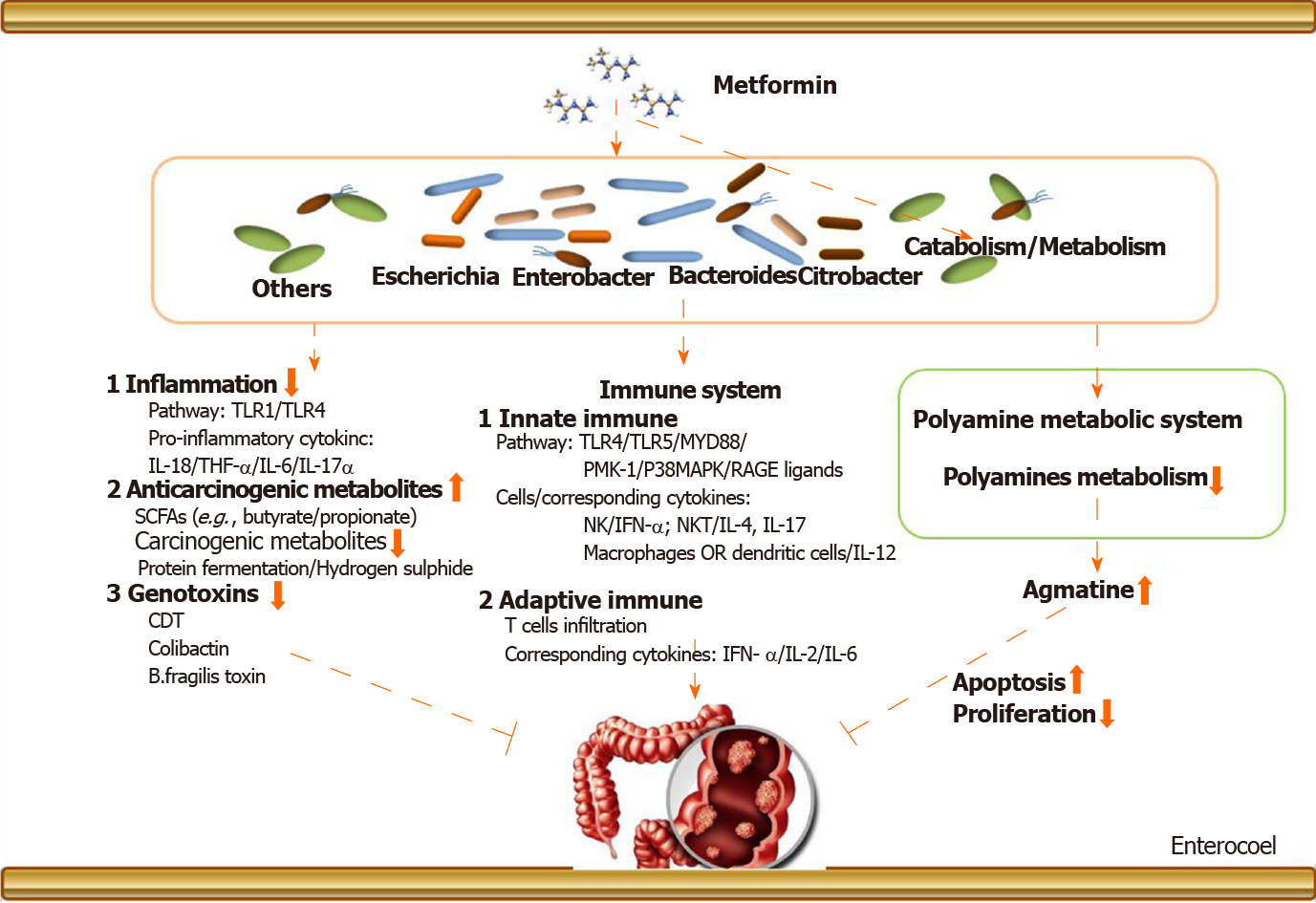
The above narrations provide an elaborate explanation about the metformin-gut microbiota-CRC axis (in T2DM), but we need to know more details about its underlying mechanisms. Figure 1 provides an exhaustive overview. There are two aspects: (1) Metformin may change the gut microbiota after entering the intestine. The changed gut microbiota has several cascade reactions to affect tumorigenesis, including reduction of inflammation, production of metabolites, and regulation of immunity; and (2) Metformin may play a pharmacological role to influence cancer-related systems in colorectal epithelial cells in the presence of gut microbiota.
Noticeably, before oral drugs are absorbed into our bloodstream, they must pass through our intestines for catabolism. In this process, the drugs are likely to first react with the intestinal microorganisms colonized in the intestinal epithelium, one of which is gut microbiota. Therefore, the drug may cause a cascade-like reaction related to gut microbiota by changing the composition and abundance of the gut microbiota. The drug may also be metabolized into other secondary products under the action of gut microbiota to exert subsequent efficacy. But the causation and deep interaction mechanism still remain exclusive. We have mentioned above that the gut microbiota involved in the inhibition of CRC by metformin in T2DM may be mainly Bacteroides, Ruminococcus, Clostridium, Firmicute, Lactobacillus, and E. coli. After these gut microbiota are changed, it may cause subsequent cancer suppression via regulation of inflammation and immunity and reduction of the production of genotoxic metabolites, etc.
Intestinal inflammation and related pathways are closely related to the initiation of CRC. In terms of regulating inflammation, after the mice were treated with metformin, the level of inflammatory markers TNF-α, IL-6, and IL-17α in plasma was reduced with a corresponding increase of the level of IL-10, which was accompanied by a reduction in Helicobacter pylori. Lee et al[44] suggested that fecal microbiota transplantation using metformin-treated mouse fecal material can upregulate the expression of GLP-1 and pattern recognition receptors TLR1 and TLR4. It has been shown that CRC-enriched genotoxic polyketide synthase (pks) + E. coli, E. faecalis, and A. finegoldii and TLR2 and/or TLR4 pathway-related bacteria, such as F. nucleatum and Peptostreptococcus anaerobius, are related closely to intestinal inflammation[55]. In our previous discussion, these bacteria are likely to be the targets of metformin, which provides a theoretical basis for our conjecture.
In terms of innate immune response, it has been proven that metformin specifically affects the accumulation of IFN-γ producing NK cells, IL-4, and IL-17 producing NKT cells and IL-12 producing macrophages/dendritic cells in the intestine in animal models[73]. In addition, the latest studies have shown that metformin could upregulate protein molecules involved in the classical immune pathway such as PMK-1/p38MAPK[74], RAGE ligands[75], and TLRs[76]. It should be mentioned that this process is likely to be mediated by gut microbiota. F. nucleatum has been shown to activate autophagy via the TLR4 and MYD88 pathways closely related to innate immunity[45].
In terms of adaptive immune response, it has been mentioned that metformin regulates adaptive immune cell infiltration[77,78], the expression of related immune factors, such as IFN-γ[79], IL-2[80], and IL-6[23,81,82], and T cell metabolic reprogramming in tumor tissues[83]. Nevertheless, it must be pointed out that these effects may also be manifested in the presence of gut microbiota. Cremonesi et al[51] proved that gut microbiota modulated T cell trafficking into human CRC including CCL5, CXCL9, and CXCL10 for cytotoxic T lymphocytes and T helper (Th) 1 cells, CCL17, CCL22, and CXCL12 for Th1 and regulatory T cells, CXCl3 for follicular Th cells, and CCL20 and CCL17 for IL-17-producing Th cells. Sethi et al[84] showed that gut microbiota depletion significantly reduced tumor burden in subcutaneous and liver metastasis models of pancreatic cancer, colon cancer, and melanoma. However, the effect was counteracted in Rag1-knockout mice that lacked mature T and B cells. In fact, gut microbiota is likely more complicated in the mechanisms of drug-mediated immunity affecting the development of CRC. A lot of work needs to be done to supplement this field.
In addition, it should be mentioned that after metformin changes the abundance of gut microbiota, the metabolites of the gut microbiota such as carcinogenic-related metabolites and genotoxicity will also change accordingly. Some of these metabolites are cancer-promoting, such as products of protein fermentation, hydrogen sulfide, bile acid metabolism, and ethanol, but some show protective effects. For example, short-chain fatty acids (butyrate and propionate) are anti-inflammatory molecules. Butyrate can inhibit histone deacetylase in colon epithelial cells and immune cells to downregulate proinflammatory cytokines and induce apoptosis in CRC cell lines. Therefore, the reduction of gut microbiota that produces procarcinogenic metabolites and the increase of gut microbiota that produces anti-inflammatory metabolites are likely to be the reason of metformin’s action. Similarly, metformin can directly slow down the abundance of E. coli, restraining to the intestinal bacteria’s carcinogenic effects. On the other hand, metformin may also promote some floras that are beneficial for anticancer effects to become a dominant flora. This is consistent with the changes in gut microbiota caused by the metformin intervention mentioned in Table 3.
Polyamines are a class of compounds containing two or more amino groups. The raw materials for their synthesis are ornithine and arginine. The key enzymes are ornithine decarboxylase and arginine decarboxylase. It has been proven that they can regulate cell proliferation and apoptosis, and their biosynthesis is closely related to the formation and metastasis of cancer[85]. There is a correlation between the expression of ornithine decarboxylase and the prediction of cancer risk and treatment response in certain epithelial cancers. However, it must be pointed out that the function of the polyamine system is affected by metformin in the presence of several pathogens, including Shigella flexneri, Streptococcus pneumoniae, Salmonella enterica subsp., enterica serovar Typhimurium, and Helicobacter pylori[86]. Therefore, metformin may inhibit the development of CRC by regulating the polyamine system under the action of certain gut microbiota.
Agmatine, a biogenic amine, is a polyamine that is converted from arginine by the action of arginine decarboxylase on the mitochondrial membrane of cells, and it has vasomotor implication and regulation of anti-inflammation[87]. Interestingly, the ability of bacteria to produce agmatine was enhanced under the administration of metformin in T2DM in a nutrient-dependent manner[43]. In addition, it must be emphasized that agmatine has a directly inhibitory effect on the proliferation of tumor cells with the corresponding acceleration of apoptosis[88]. As for CRC, agmatine has been confirmed to inhibit the proliferation of six types of human intestinal tumor cell lines in a concentration-dependent manner[89]. It is able to arrest proliferation in cell lines by depleting intracellular polyamine levels and enter mammalian cells via the polyamine transport system.
To date, there are few reports on the use of agmatine metabolism as a direct therapeutic target in clinical research, but there are studies affirming that the agmatine metabolism system has the potential to treat tumors. The role of metformin and gut microbiota is like a fuse in the antitumor effect mediated by the agmatine metabolic system. Moreover, Pryor et al[43] suggested that the Escherichia, Bacteroides, Enterobacter, and Citrobacter genera produce the most agmatine in the intestine, and these bacteria were abundant in the gut of patients after metformin intervention. Furthermore, this inhibitory effect of agmatine on tumors was probably attributable to an interaction between agmatine and the intracellular polyamine metabolism system[90,91]. For example, agmatine regulates the functional gene delivery system of spermidine/spermine acetyltransferase in cancer cell[90]. Therefore, the exploration of a metformin-gut microbiota-cell polyamine metabolism system-cancer chain helps us to understand how metformin inhibits CRC by regulating the agmatine metabolism system in the presence of gut microbiota. From the perspective of drugs-gut microbiota-pharmacological effects-diseases, this will be an interesting and profound finding.
To sum up, metformin may indirectly restrain clinical colorectal tumors by regulating the agmatine metabolism system in the presence of gut microbiota. In future studies, more attention should be attached to the drug’s interaction with gut microbiota to the intracellular polyamine system because the intervention of the polyamine system may be one of the important methods for clinical cancerous treatment.
One clear goal moving forward is to explore how to better apply the gut microbiota in clinical treatment. Gut microbiota may directly induce metformin’s anti-CRC effect via the structure of its own composition, the production of secondary metabolites, and the regulation of the immune system after interplay with metformin. As a consequence, accumulated information has proven that metformin influences colorectal tissue carcinogenesis, and this effect is dependent on gut microbiota. Then we need to figure it out that what types of gut microbiota play key roles in this process.
Herein, we advocate to build a complete network of relationships of drugs-gut microbiota-diseases. This concept will provide a new treatment strategy for the diagnosis and treatment of clinical diseases by using gut microbiota. However, in order to achieve this goal, the relationship between changes in the structure and function of the gut microbiota and the pharmacological effects of drugs needs to be thoroughly explained. For example, composition of gut microbiota is a determinant for development of gastrointestinal adverse effects following metformin intake[32]. Wu et al[92] investigated the pharmacodynamic and pharmacokinetic effects of metformin mediated by the gut microbiota in vivo. The pharmacodynamic indexes were evaluated, and metformin concentrations were measured with a validated liquid chromatography-tandem mass spectrometry method after oral administration. They described the pharmacodynamic differences between metformin in sterile diabetic rats and conventional diabetic rats. Compared to conventional diabetic rats, fasting blood glucose and cmax in pseudosterile diabetic rats were significantly increased with a corresponding reduction in oral glucose and t1/2α. All of these elaborated that these differences in pharmacodynamics and pharmacokinetics may be due to the decrease in the expression of Oct1 in the liver, contributing to the changes in liver uptake of metformin[92]. Napolitano et al[93] suggested that metformin has complex effects that alter bile acid recycling and gut microbiota (positive correlation between the microbiota abundance of the phylum Firmicutes and changes in cholic acid and conjugates, negative correlation in Bacteroidetes) due to gut-based pharmacology, which might provide insights into novel therapeutic approaches to treat T2DM and associated metabolic diseases. But the explanation of these pharmacodynamics and pharmacokinetic mechanisms is not enough for the determination of clinical drugs. In addition, the current majority of research focuses on the relationship between the changes in the profiles of gut microbiota and the effects of drugs on diseases (e.g., metformin on CRC).
To date, the research in respect to the role of metformin to promote the formation of CRC in T2DM were mainly carried out in the observational studies of populations. In future work, more experiments need to be conducted to explore deeper mechanisms in individuals and in animal models, which can also verify the effect of an antihyperglycemia pharmaceutical for anticancer treatment. Furthermore, metformin, a biguanide derivative, has pleiotropic effects beyond glucose reduction, including antitumor, antihaze-induced pneumonia, and anti-aging. Other pharmacological effects and corresponding mechanisms of metformin need to be further discovered. For instance, we elaborated that metformin elicits its role in tumors via regulating the function of the immune system and how the effect is performed in the presence of gut microbiota based on the fact that the occurrence of tumors is closely related to the body’s immunity.
Overall, CRC is an urgent research topic undertaken by many researchers all over the world. Nevertheless, the diagnosis and treatment of CRC still has many unresolved issues, but the research on the gut microbiota has opened a new window. Through this window, we could see the emergence of gut microbiota as a novel way to fight tumors. However, the current research is limited to the changes in its quantity and distribution due to the limitations of current scientific research technologies and methods. When metformin is regarded as an anticancer drug in the intestine, the intestinal microbiota will change or affect the production of certain anticancer substances. Hence, it must be stressed again that establishing a complete drug-gut microbiota-disease relationship network is necessary for the potential medical application of gut microbiota. In addition, novel methods for microbiological research should be systematically carried out in order to explore the function and structure of gut microbiota in the future: structure functions, genomics, transcriptomics, proteomics, and metabolomics. These efforts will benefit the prevention, diagnosis, and treatment of clinical CRC. Advances in this field will also promote the progress of other microbial-related diseases and provide new ideas for the use of clinical drugs.
In summary, metformin can inhibit the initiation of CRC in T2DM patients by changing the abundance of gut microbiota or under the participation of gut microbiota. Further research is needed to confirm this hypothesis and explore the mechanisms behind it. Moreover, in order to understand thoroughly the function and structure of gut microbiota, more cutting-edge scientific research methods and technologies are needed to implement.
Manuscript source: Unsolicited manuscript
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Majumdar APN S-Editor: Huang P L-Editor: Filipodia P-Editor: Liu JH
| 1. | Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467-1480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1570] [Cited by in RCA: 3015] [Article Influence: 502.5] [Reference Citation Analysis (3)] |
| 2. | Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M, Hosoda F, Rokutan H, Matsumoto M, Takamaru H, Yamada M, Matsuda T, Iwasaki M, Yamaji T, Yachida T, Soga T, Kurokawa K, Toyoda A, Ogura Y, Hayashi T, Hatakeyama M, Nakagama H, Saito Y, Fukuda S, Shibata T, Yamada T. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25:968-976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 404] [Cited by in RCA: 875] [Article Influence: 145.8] [Reference Citation Analysis (0)] |
| 3. | Allen J, Sears CL. Impact of the gut microbiome on the genome and epigenome of colon epithelial cells: contributions to colorectal cancer development. Genome Med. 2019;11:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 132] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 4. | Lascar N, Brown J, Pattison H, Barnett AH, Bailey CJ, Bellary S. Type 2 diabetes in adolescents and young adults. Lancet Diabetes Endocrinol. 2018;6:69-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 367] [Cited by in RCA: 531] [Article Influence: 75.9] [Reference Citation Analysis (1)] |
| 5. | Soltani G, Poursheikhani A, Yassi M, Hayatbakhsh A, Kerachian M, Kerachian MA. Obesity, diabetes and the risk of colorectal adenoma and cancer. BMC Endocr Disord. 2019;19:113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 6. | Bu WJ, Song L, Zhao DY, Guo B, Liu J. Insulin therapy and the risk of colorectal cancer in patients with type 2 diabetes: a meta-analysis of observational studies. Br J Clin Pharmacol. 2014;78:301-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 7. | Nie Z, Zhu H, Gu M. Reduced colorectal cancer incidence in type 2 diabetic patients treated with metformin: a meta-analysis. Pharm Biol. 2016;54:2636-2642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 8. | Mei ZB, Zhang ZJ, Liu CY, Liu Y, Cui A, Liang ZL, Wang GH, Cui L. Survival benefits of metformin for colorectal cancer patients with diabetes: a systematic review and meta-analysis. PLoS One. 2014;9:e91818. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 75] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 9. | Zhang ZJ, Zheng ZJ, Kan H, Song Y, Cui W, Zhao G, Kip KE. Reduced risk of colorectal cancer with metformin therapy in patients with type 2 diabetes: a meta-analysis. Diabetes Care. 2011;34:2323-2328. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 205] [Cited by in RCA: 228] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 10. | Tomimoto A, Endo H, Sugiyama M, Fujisawa T, Hosono K, Takahashi H, Nakajima N, Nagashima Y, Wada K, Nakagama H, Nakajima A. Metformin suppresses intestinal polyp growth in ApcMin/+ mice. Cancer Sci. 2008;99:2136-2141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 153] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 11. | Bordini HP, Kremer JL, Fagundes TR, Melo GP, Conchon-Costa I, da Silva SS, Cecchini AL, Panis C, Luiz RC. Protective effect of metformin in an aberrant crypt foci model induced by 1,2-dimethylhydrazine: Modulation of oxidative stress and inflammatory process. Mol Carcinog. 2017;56:913-922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 12. | Hosono K, Endo H, Takahashi H, Sugiyama M, Sakai E, Uchiyama T, Suzuki K, Iida H, Sakamoto Y, Yoneda K, Koide T, Tokoro C, Abe Y, Inamori M, Nakagama H, Nakajima A. Metformin suppresses colorectal aberrant crypt foci in a short-term clinical trial. Cancer Prev Res (Phila). 2010;3:1077-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 244] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 13. | Hosono K, Endo H, Takahashi H, Sugiyama M, Uchiyama T, Suzuki K, Nozaki Y, Yoneda K, Fujita K, Yoneda M, Inamori M, Tomatsu A, Chihara T, Shimpo K, Nakagama H, Nakajima A. Metformin suppresses azoxymethane-induced colorectal aberrant crypt foci by activating AMP-activated protein kinase. Mol Carcinog. 2010;49:662-671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 152] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 14. | Kanadiya MK, Gohel TD, Sanaka MR, Thota PN, Shubrook JH Jr. Relationship between type-2 diabetes and use of metformin with risk of colorectal adenoma in an American population receiving colonoscopy. J Diabetes Complications. 2013;27:463-466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 15. | Demb J, Yaseyyedi A, Liu L, Bustamante R, Earles A, Ghosh P, Gutkind JS, Gawron AJ, Kaltenbach TR, Martinez ME, Gupta S. Metformin Is Associated With Reduced Odds for Colorectal Cancer Among Persons With Diabetes. Clin Transl Gastroenterol. 2019;10:e00092. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 16. | Higurashi T, Hosono K, Takahashi H, Komiya Y, Umezawa S, Sakai E, Uchiyama T, Taniguchi L, Hata Y, Uchiyama S, Hattori A, Nagase H, Kessoku T, Arimoto J, Matsuhashi N, Inayama Y, Yamanaka S, Taguri M, Nakajima A. Metformin for chemoprevention of metachronous colorectal adenoma or polyps in post-polypectomy patients without diabetes: a multicentre double-blind, placebo-controlled, randomised phase 3 trial. Lancet Oncol. 2016;17:475-483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 239] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 17. | Mogavero A, Maiorana MV, Zanutto S, Varinelli L, Bozzi F, Belfiore A, Volpi CC, Gloghini A, Pierotti MA, Gariboldi M. Metformin transiently inhibits colorectal cancer cell proliferation as a result of either AMPK activation or increased ROS production. Sci Rep. 2017;7:15992. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 106] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 18. | Mohamed Suhaimi NA, Phyo WM, Yap HY, Choy SHY, Wei X, Choudhury Y, Tan WJ, Tan LAPY, Foo RSY, Tan SHS, Tiang Z, Wong CF, Koh PK, Tan MH. Metformin Inhibits Cellular Proliferation and Bioenergetics in Colorectal Cancer Patient-Derived Xenografts. Mol Cancer Ther. 2017;16:2035-2044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 19. | Park JH, Kim YH, Park EH, Lee SJ, Kim H, Kim A, Lee SB, Shim S, Jang H, Myung JK, Park S, Lee SJ, Kim MJ. Effects of metformin and phenformin on apoptosis and epithelial-mesenchymal transition in chemoresistant rectal cancer. Cancer Sci. 2019;110:2834-2845. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 20. | Wang SQ, Cui SX, Qu XJ. Metformin inhibited colitis and colitis-associated cancer (CAC) through protecting mitochondrial structures of colorectal epithelial cells in mice. Cancer Biol Ther. 2019;20:338-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 21. | Sena P, Mancini S, Benincasa M, Mariani F, Palumbo C, Roncucci L. Metformin Induces Apoptosis and Alters Cellular Responses to Oxidative Stress in Ht29 Colon Cancer Cells: Preliminary Findings. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 22. | Nguyen TT, Ung TT, Li S, Lian S, Xia Y, Park SY, Do Jung Y. Metformin inhibits lithocholic acid-induced interleukin 8 upregulation in colorectal cancer cells by suppressing ROS production and NF-kB activity. Sci Rep. 2019;9:2003. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 23. | Kang S, Kim BR, Kang MH, Kim DY, Lee DH, Oh SC, Min BW, Um JW. Anti-metastatic effect of metformin via repression of interleukin 6-induced epithelial-mesenchymal transition in human colon cancer cells. PLoS One. 2018;13:e0205449. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 24. | Lan B, Zhang J, Zhang P, Zhang W, Yang S, Lu D, Li W, Dai Q. Metformin suppresses CRC growth by inducing apoptosis via ADORA1. Front Biosci (Landmark Ed). 2017;22:248-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 25. | Kim SH, Kim SC, Ku JL. Metformin increases chemo-sensitivity via gene downregulation encoding DNA replication proteins in 5-Fu resistant colorectal cancer cells. Oncotarget. 2017;8:56546-56557. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 26. | Shen P, Reineke LC, Knutsen E, Chen M, Pichler M, Ling H, Calin GA. Metformin blocks MYC protein synthesis in colorectal cancer via mTOR-4EBP-eIF4E and MNK1-eIF4G-eIF4E signaling. Mol Oncol. 2018;12:1856-1870. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 27. | Wang Y, Wu Z, Hu L. The regulatory effects of metformin on the [SNAIL/miR-34]:[ZEB/miR-200] system in the epithelial-mesenchymal transition(EMT) for colorectal cancer(CRC). Eur J Pharmacol. 2018;834:45-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 28. | Zhang C, Wang Y. Metformin attenuates cells stemness and epithelial-mesenchymal transition in colorectal cancer cells by inhibiting the Wnt3a/β-catenin pathway. Mol Med Rep. 2019;19:1203-1209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 29. | Amable G, Martínez-León E, Picco ME, Di Siervi N, Davio C, Rozengurt E, Rey O. Metformin inhibits β-catenin phosphorylation on Ser-552 through an AMPK/PI3K/Akt pathway in colorectal cancer cells. Int J Biochem Cell Biol. 2019;112:88-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 30. | Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Mannerås-Holm L, Ståhlman M, Olsson LM, Serino M, Planas-Fèlix M, Xifra G, Mercader JM, Torrents D, Burcelin R, Ricart W, Perkins R, Fernàndez-Real JM, Bäckhed F. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017;23:850-858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 824] [Cited by in RCA: 1135] [Article Influence: 141.9] [Reference Citation Analysis (0)] |
| 31. | Tong X, Xu J, Lian F, Yu X, Zhao Y, Xu L, Zhang M, Zhao X, Shen J, Wu S, Pang X, Tian J, Zhang C, Zhou Q, Wang L, Pang B, Chen F, Peng Z, Wang J, Zhen Z, Fang C, Li M, Chen L, Zhao L. Structural Alteration of Gut Microbiota during the Amelioration of Human Type 2 Diabetes with Hyperlipidemia by Metformin and a Traditional Chinese Herbal Formula: a Multicenter, Randomized, Open Label Clinical Trial. mBio. 2018;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 282] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 32. | Bryrup T, Thomsen CW, Kern T, Allin KH, Brandslund I, Jørgensen NR, Vestergaard H, Hansen T, Hansen TH, Pedersen O, Nielsen T. Metformin-induced changes of the gut microbiota in healthy young men: results of a non-blinded, one-armed intervention study. Diabetologia. 2019;62:1024-1035. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 147] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 33. | Shin NR, Lee JC, Lee HY, Kim MS, Whon TW, Lee MS, Bae JW. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2014;63:727-735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1031] [Cited by in RCA: 1185] [Article Influence: 107.7] [Reference Citation Analysis (0)] |
| 34. | Lee H, Ko G. Effect of metformin on metabolic improvement and gut microbiota. Appl Environ Microbiol. 2014;80:5935-5943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 292] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 35. | Reimer RA, Grover GJ, Koetzner L, Gahler RJ, Lyon MR, Wood S. Combining sitagliptin/metformin with a functional fiber delays diabetes progression in Zucker rats. J Endocrinol. 2014;220:361-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 36. | Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, Prifti E, Vieira-Silva S, Gudmundsdottir V, Pedersen HK, Arumugam M, Kristiansen K, Voigt AY, Vestergaard H, Hercog R, Costea PI, Kultima JR, Li J, Jørgensen T, Levenez F, Dore J; MetaHIT consortium; Nielsen HB; Brunak S; Raes J; Hansen T; Wang J; Ehrlich SD; Bork P; Pedersen O. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262-266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1208] [Cited by in RCA: 1511] [Article Influence: 151.1] [Reference Citation Analysis (0)] |
| 37. | Zhang X, Zhao Y, Xu J, Xue Z, Zhang M, Pang X, Zhang X, Zhao L. Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci Rep. 2015;5:14405. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 370] [Cited by in RCA: 475] [Article Influence: 47.5] [Reference Citation Analysis (0)] |
| 38. | de la Cuesta-Zuluaga J, Mueller NT, Corrales-Agudelo V, Velásquez-Mejía EP, Carmona JA, Abad JM, Escobar JS. Metformin Is Associated With Higher Relative Abundance of Mucin-Degrading Akkermansia muciniphila and Several Short-Chain Fatty Acid-Producing Microbiota in the Gut. Diabetes Care. 2017;40:54-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 397] [Cited by in RCA: 522] [Article Influence: 65.3] [Reference Citation Analysis (0)] |
| 39. | Nakajima H, Takewaki F, Hashimoto Y, Kajiyama S, Majima S, Okada H, Senmaru T, Ushigome E, Nakanishi N, Hamaguchi M, Yamazaki M, Tanaka Y, Oikawa Y, Nakajima S, Ohno H, Fukui M. The Effects of Metformin on the Gut Microbiota of Patients with Type 2 Diabetes: A Two-Center, Quasi-Experimental Study. Life (Basel). 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 40. | Rosario D, Benfeitas R, Bidkhori G, Zhang C, Uhlen M, Shoaie S, Mardinoglu A. Understanding the Representative Gut Microbiota Dysbiosis in Metformin-Treated Type 2 Diabetes Patients Using Genome-Scale Metabolic Modeling. Front Physiol. 2018;9:775. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 41. | Bauer PV, Duca FA, Waise TMZ, Rasmussen BA, Abraham MA, Dranse HJ, Puri A, O'Brien CA, Lam TKT. Metformin Alters Upper Small Intestinal Microbiota that Impact a Glucose-SGLT1-Sensing Glucoregulatory Pathway. Cell Metab 2018; 27: 101-117. e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 195] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 42. | Sun L, Xie C, Wang G, Wu Y, Wu Q, Wang X, Liu J, Deng Y, Xia J, Chen B, Zhang S, Yun C, Lian G, Zhang X, Zhang H, Bisson WH, Shi J, Gao X, Ge P, Liu C, Krausz KW, Nichols RG, Cai J, Rimal B, Patterson AD, Wang X, Gonzalez FJ, Jiang C. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat Med. 2018;24:1919-1929. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 704] [Article Influence: 100.6] [Reference Citation Analysis (0)] |
| 43. | Pryor R, Norvaisas P, Marinos G, Best L, Thingholm LB, Quintaneiro LM, De Haes W, Esser D, Waschina S, Lujan C, Smith RL, Scott TA, Martinez-Martinez D, Woodward O, Bryson K, Laudes M, Lieb W, Houtkooper RH, Franke A, Temmerman L, Bjedov I, Cochemé HM, Kaleta C, Cabreiro F. Host-Microbe-Drug-Nutrient Screen Identifies Bacterial Effectors of Metformin Therapy. Cell 2019; 178: 1299-1312. e29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 187] [Cited by in RCA: 192] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 44. | Lee H, Kim J, An J, Lee S, Choi D, Kong H, Song Y, Park IH, Lee CK, Kim K. Downregulation of IL-18 Expression in the Gut by Metformin-induced Gut Microbiota Modulation. Immune Netw. 2019;19:e28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 45. | Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, Chen Y, Chen H, Hong J, Zou W, Fang JY. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017; 170: 548-563. e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 814] [Cited by in RCA: 1474] [Article Influence: 184.3] [Reference Citation Analysis (0)] |
| 46. | Peters BA, Dominianni C, Shapiro JA, Church TR, Wu J, Miller G, Yuen E, Freiman H, Lustbader I, Salik J, Friedlander C, Hayes RB, Ahn J. The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome. 2016;4:69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 190] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 47. | Li L, Li X, Zhong W, Yang M, Xu M, Sun Y, Ma J, Liu T, Song X, Dong W, Liu X, Chen Y, Liu Y, Abla Z, Liu W, Wang B, Jiang K, Cao H. Gut microbiota from colorectal cancer patients enhances the progression of intestinal adenoma in Apcmin/+ mice. EBioMedicine. 2019;48:301-315. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 99] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 48. | Donohoe DR, Holley D, Collins LB, Montgomery SA, Whitmore AC, Hillhouse A, Curry KP, Renner SW, Greenwalt A, Ryan EP, Godfrey V, Heise MT, Threadgill DS, Han A, Swenberg JA, Threadgill DW, Bultman SJ. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 2014;4:1387-1397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 345] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 49. | Zhu W, Miyata N, Winter MG, Arenales A, Hughes ER, Spiga L, Kim J, Sifuentes-Dominguez L, Starokadomskyy P, Gopal P, Byndloss MX, Santos RL, Burstein E, Winter SE. Editing of the gut microbiota reduces carcinogenesis in mouse models of colitis-associated colorectal cancer. J Exp Med. 2019;216:2378-2393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 95] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 50. | Qin Y, Roberts JD, Grimm SA, Lih FB, Deterding LJ, Li R, Chrysovergis K, Wade PA. An obesity-associated gut microbiome reprograms the intestinal epigenome and leads to altered colonic gene expression. Genome Biol. 2018;19:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 100] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 51. | Cremonesi E, Governa V, Garzon JFG, Mele V, Amicarella F, Muraro MG, Trella E, Galati-Fournier V, Oertli D, Däster SR, Droeser RA, Weixler B, Bolli M, Rosso R, Nitsche U, Khanna N, Egli A, Keck S, Slotta-Huspenina J, Terracciano LM, Zajac P, Spagnoli GC, Eppenberger-Castori S, Janssen KP, Borsig L, Iezzi G. Gut microbiota modulate T cell trafficking into human colorectal cancer. Gut. 2018;67:1984-1994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 195] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 52. | Wang L, Tang L, Feng Y, Zhao S, Han M, Zhang C, Yuan G, Zhu J, Cao S, Wu Q, Li L, Zhang Z. A purified membrane protein from Akkermansia muciniphila or the pasteurised bacterium blunts colitis associated tumourigenesis by modulation of CD8+ T cells in mice. Gut. 2020;69:1988-1997. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 184] [Cited by in RCA: 411] [Article Influence: 82.2] [Reference Citation Analysis (0)] |
| 53. | Long X, Wong CC, Tong L, Chu ESH, Ho Szeto C, Go MYY, Coker OO, Chan AWH, Chan FKL, Sung JJY, Yu J. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat Microbiol. 2019;4:2319-2330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 323] [Article Influence: 53.8] [Reference Citation Analysis (0)] |
| 54. | Belcheva A, Irrazabal T, Robertson SJ, Streutker C, Maughan H, Rubino S, Moriyama EH, Copeland JK, Surendra A, Kumar S, Green B, Geddes K, Pezo RC, Navarre WW, Milosevic M, Wilson BC, Girardin SE, Wolever TMS, Edelmann W, Guttman DS, Philpott DJ, Martin A. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell. 2014;158:288-299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 375] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 55. | Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol. 2019;16:690-704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 384] [Cited by in RCA: 812] [Article Influence: 135.3] [Reference Citation Analysis (0)] |
| 56. | Ryan PM, Patterson E, Carafa I, Mandal R, Wishart DS, Dinan TG, Cryan JF, Tuohy KM, Stanton C, Ross RP. Metformin and Dipeptidyl Peptidase-4 Inhibitor Differentially Modulate the Intestinal Microbiota and Plasma Metabolome of Metabolically Dysfunctional Mice. Can J Diabetes 2020; 44: 146-155. e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 57. | Jacob S, Dötsch A, Knoll S, Köhler HR, Rogall E, Stoll D, Tisler S, Huhn C, Schwartz T, Zwiener C, Triebskorn R. Does the antidiabetic drug metformin affect embryo development and the health of brown trout (Salmo trutta f. fario)? Environ Sci Eur. 2018;30:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 58. | Brody H. Colorectal cancer. Nature. 2015;521:S1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 378] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 59. | Yang Q, Guo X, Yang L. Metformin Enhances the Effect of Regorafenib and Inhibits Recurrence and Metastasis of Hepatic Carcinoma After Liver Resection via Regulating Expression of Hypoxia Inducible Factors 2α (HIF-2α) and 30 kDa HIV Tat-Interacting Protein (TIP30). Med Sci Monit. 2018;24:2225-2234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 60. | Han MS, Lee HJ, Park SJ, Hong SP, Cheon JH, Kim WH, Kim TI. The effect of metformin on the recurrence of colorectal adenoma in diabetic patients with previous colorectal adenoma. Int J Colorectal Dis. 2017;32:1223-1226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 61. | Okubo R, Kinoshita T, Katsumata N, Uezono Y, Xiao J, Matsuoka YJ. Impact of chemotherapy on the association between fear of cancer recurrence and the gut microbiota in breast cancer survivors. Brain Behav Immun. 2020;85:186-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 62. | Yu SY, Xie YH, Qiu YW, Chen YX, Fang JY. Moderate alteration to gut microbiota brought by colorectal adenoma resection. J Gastroenterol Hepatol. 2019;34:1758-1765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 63. | Bishnu A, Sakpal A, Ghosh N, Choudhury P, Chaudhury K, Ray P. Long term treatment of metformin impedes development of chemoresistance by regulating cancer stem cell differentiation through taurine generation in ovarian cancer cells. Int J Biochem Cell Biol. 2019;107:116-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 64. | Miranda VC, Braghiroli MI, Faria LD, Bariani G, Alex A, Bezerra Neto JE, Capareli FC, Sabbaga J, Lobo Dos Santos JF, Hoff PM, Riechelmann RP. Phase 2 Trial of Metformin Combined With 5-Fluorouracil in Patients With Refractory Metastatic Colorectal Cancer. Clin Colorectal Cancer 2016; 15: 321-328. e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 65. | Huang WS, Lin CT, Chen CN, Chang SF, Chang HI, Lee KC. Metformin increases the cytotoxicity of oxaliplatin in human DLD-1 colorectal cancer cells through down-regulating HMGB1 expression. J Cell Biochem. 2018;119:6943-6952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 66. | Fernandes JM, Jandrey EHF, Koyama FC, Leite KRM, Camargo AA, Costa ÉT, Perez RO, Asprino PF. Metformin as an Alternative Radiosensitizing Agent to 5-Fluorouracil During Neoadjuvant Treatment for Rectal Cancer. Dis Colon Rectum. 2020;63:918-926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 67. | Brown JR, Chan DK, Shank JJ, Griffith KA, Fan H, Szulawski R, Yang K, Reynolds RK, Johnston C, McLean K, Uppal S, Liu JR, Cabrera L, Taylor SE, Orr BC, Modugno F, Mehta P, Bregenzer M, Mehta G, Shen H, Coffman LG, Buckanovich RJ. Phase II clinical trial of metformin as a cancer stem cell-targeting agent in ovarian cancer. JCI Insight. 2020;5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 89] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 68. | Patil S. Metformin treatment decreases the expression of cancer stem cell marker CD44 and stemness related gene expression in primary oral cancer cells. Arch Oral Biol. 2020;113:104710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 69. | Bartolini I, Risaliti M, Ringressi MN, Melli F, Nannini G, Amedei A, Muiesan P, Taddei A. Role of gut microbiota-immunity axis in patients undergoing surgery for colorectal cancer: Focus on short and long-term outcomes. World J Gastroenterol. 2020;26:2498-2513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 29] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (1)] |
| 70. | Deleemans JM, Chleilat F, Reimer RA, Henning JW, Baydoun M, Piedalue KA, McLennan A, Carlson LE. The chemo-gut study: investigating the long-term effects of chemotherapy on gut microbiota, metabolic, immune, psychological and cognitive parameters in young adult Cancer survivors; study protocol. BMC Cancer. 2019;19:1243. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 51] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 71. | Zheng DW, Dong X, Pan P, Chen KW, Fan JX, Cheng SX, Zhang XZ. Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nat Biomed Eng. 2019;3:717-728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 300] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 72. | Zhu Z, Huang J, Li X, Xing J, Chen Q, Liu R, Hua F, Qiu Z, Song Y, Bai C, Mo YY, Zhang Z. Gut microbiota regulate tumor metastasis via circRNA/miRNA networks. Gut Microbes. 2020;12:1788891. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 85] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 73. | Volarevic V, Misirkic M, Vucicevic L, Paunovic V, Simovic Markovic B, Stojanovic M, Milovanovic M, Jakovljevic V, Micic D, Arsenijevic N, Trajkovic V, Lukic ML. Metformin aggravates immune-mediated liver injury in mice. Arch Toxicol. 2015;89:437-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 74. | Xiao Y, Liu F, Li S, Jiang N, Yu C, Zhu X, Qin Y, Hui J, Meng L, Song C, Li XF, Liu Y. Metformin promotes innate immunity through a conserved PMK-1/p38 MAPK pathway. Virulence. 2020;11:39-48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 75. | Kumar NP, Moideen K, Nancy A, Viswanathan V, Shruthi BS, Sivakumar S, Hissar S, Kornfeld H, Babu S. Systemic RAGE ligands are upregulated in tuberculosis individuals with diabetes co-morbidity and modulated by anti-tuberculosis treatment and metformin therapy. BMC Infect Dis. 2019;19:1039. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 76. | Vaez H, Najafi M, Rameshrad M, Toutounchi NS, Garjani M, Barar J, Garjani A. AMPK activation by metformin inhibits local innate immune responses in the isolated rat heart by suppression of TLR 4-related pathway. Int Immunopharmacol. 2016;40:501-507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 77. | Curry JM, Johnson J, Mollaee M, Tassone P, Amin D, Knops A, Whitaker-Menezes D, Mahoney MG, South A, Rodeck U, Zhan T, Harshyne L, Philp N, Luginbuhl A, Cognetti D, Tuluc M, Martinez-Outschoorn U. Metformin Clinical Trial in HPV+ and HPV- Head and Neck Squamous Cell Carcinoma: Impact on Cancer Cell Apoptosis and Immune Infiltrate. Front Oncol. 2018;8:436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 78. | Pereira FV, Melo ACL, Low JS, de Castro ÍA, Braga TT, Almeida DC, Batista de Lima AGU, Hiyane MI, Correa-Costa M, Andrade-Oliveira V, Origassa CST, Pereira RM, Kaech SM, Rodrigues EG, Câmara NOS. Metformin exerts antitumor activity via induction of multiple death pathways in tumor cells and activation of a protective immune response. Oncotarget. 2018;9:25808-25825. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 79. | Hirayama T, Nagata Y, Nishida M, Matsuo M, Kobayashi S, Yoneda A, Kanetaka K, Udono H, Eguchi S. Metformin Prevents Peritoneal Dissemination via Immune-suppressive Cells in the Tumor Microenvironment. Anticancer Res. 2019;39:4699-4709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 80. | Eikawa S, Nishida M, Mizukami S, Yamazaki C, Nakayama E, Udono H. Immune-mediated antitumor effect by type 2 diabetes drug, metformin. Proc Natl Acad Sci USA. 2015;112:1809-1814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 339] [Cited by in RCA: 433] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 81. | Xue J, Li X, Liu P, Li K, Sha L, Yang X, Zhu L, Wang Z, Dong Y, Zhang L, Lei H, Zhang X, Dong X, Wang H. Inulin and metformin ameliorate polycystic ovary syndrome via anti-inflammation and modulating gut microbiota in mice. Endocr J. 2019;66:859-870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 77] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 82. | Soberanes S, Misharin AV, Jairaman A, Morales-Nebreda L, McQuattie-Pimentel AC, Cho T, Hamanaka RB, Meliton AY, Reyfman PA, Walter JM, Chen CI, Chi M, Chiu S, Gonzalez-Gonzalez FJ, Antalek M, Abdala-Valencia H, Chiarella SE, Sun KA, Woods PS, Ghio AJ, Jain M, Perlman H, Ridge KM, Morimoto RI, Sznajder JI, Balch WE, Bhorade SM, Bharat A, Prakriya M, Chandel NS, Mutlu GM, Budinger GRS. Metformin Targets Mitochondrial Electron Transport to Reduce Air-Pollution-Induced Thrombosis. Cell Metab 2019; 29: 335-347. e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 84] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 83. | Bahrambeigi S, Shafiei-Irannejad V. Immune-mediated anti-tumor effects of metformin; targeting metabolic reprogramming of T cells as a new possible mechanism for anti-cancer effects of metformin. Biochem Pharmacol. 2020;174:113787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 84. | Sethi V, Kurtom S, Tarique M, Lavania S, Malchiodi Z, Hellmund L, Zhang L, Sharma U, Giri B, Garg B, Ferrantella A, Vickers SM, Banerjee S, Dawra R, Roy S, Ramakrishnan S, Saluja A, Dudeja V. Gut Microbiota Promotes Tumor Growth in Mice by Modulating Immune Response. Gastroenterology 2018; 155: 33-37. e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 299] [Article Influence: 42.7] [Reference Citation Analysis (0)] |
| 85. | Cirenajwis H, Smiljanic S, Honeth G, Hegardt C, Marton LJ, Oredsson SM. Reduction of the putative CD44+CD24- breast cancer stem cell population by targeting the polyamine metabolic pathway with PG11047. Anticancer Drugs. 2010;21:897-906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 86. | Di Martino ML, Campilongo R, Casalino M, Micheli G, Colonna B, Prosseda G. Polyamines: emerging players in bacteria-host interactions. Int J Med Microbiol. 2013;303:484-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 114] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 87. | Zhao D, Ren LM. Non-adrenergic inhibition at prejunctional sites by agmatine of purinergic vasoconstriction in rabbit saphenous artery. Neuropharmacology. 2005;48:597-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 88. | Isome M, Lortie MJ, Murakami Y, Parisi E, Matsufuji S, Satriano J. The antiproliferative effects of agmatine correlate with the rate of cellular proliferation. Am J Physiol Cell Physiol. 2007;293:C705-C711. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 89. | Molderings GJ, Kribben B, Heinen A, Schröder D, Brüss M, Göthert M. Intestinal tumor and agmatine (decarboxylated arginine): low content in colon carcinoma tissue specimens and inhibitory effect on tumor cell proliferation in vitro. Cancer. 2004;101:858-868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 90. | Cui PF, Xing L, Qiao JB, Zhang JL, He YJ, Zhang M, Lyu JY, Luo CQ, Jin L, Jiang HL. Polyamine metabolism-based dual functional gene delivery system to synergistically inhibit the proliferation of cancer. Int J Pharm. 2016;506:79-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 91. | Wang JF, Su RB, Wu N, Xu B, Lu XQ, Liu Y, Li J. Inhibitory effect of agmatine on proliferation of tumor cells by modulation of polyamine metabolism. Acta Pharmacol Sin. 2005;26:616-622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 92. | Wu B, Chen M, Gao Y, Hu J, Liu M, Zhang W, Huang W. In vivo pharmacodynamic and pharmacokinetic effects of metformin mediated by the gut microbiota in rats. Life Sci. 2019;226:185-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 93. | Napolitano A, Miller S, Nicholls AW, Baker D, Van Horn S, Thomas E, Rajpal D, Spivak A, Brown JR, Nunez DJ. Novel gut-based pharmacology of metformin in patients with type 2 diabetes mellitus. PLoS One. 2014;9:e100778. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 168] [Cited by in RCA: 223] [Article Influence: 20.3] [Reference Citation Analysis (0)] |