Published online Dec 6, 2020. doi: 10.12998/wjcc.v8.i23.5935
Peer-review started: July 18, 2020
First decision: August 21, 2020
Revised: August 24, 2020
Accepted: September 25, 2020
Article in press: September 25, 2020
Published online: December 6, 2020
Processing time: 138 Days and 19.2 Hours
CD155 is an immune checkpoint protein in cancers and interacts with ligands to regulate the immune microenvironment. The expression of CD155 is correlated with the prognosis and pathological features of breast cancer.
To investigate the expression status of CD155 and the association with exhausted CD4+ helper and CD8+ cytotoxic tumor infiltrating lymphocytes (TILs) and PD-L1 in the breast cancer microenvironment.
One hundred and twenty-six breast cancer patients with invasive ductal breast cancer were consecutively recruited into this study. Immunohistochemistry was used to detect the expression CD155, PD-L1 and PD-1 on tumor-infiltrating immune cells and tumor cells in the microenvironment.
The proportion of patients with CD155 expression was higher in triple negative breast cancer (72.7%) than in Luminal A patients (22.2%, P < 0.05). Patients with positive CD155 expression had a higher percentage of CD4+/PD-1+ helper TILs (30%) than patients with negative CD155 expression (21%, P < 0.05). Patients with positive CD155 expression also had higher cell counts of exhausted CD4+ TILs [47 vs 20/high-power fields (HPF)] and unexhausted CD8+ TILs (30 vs 17/HPF) than patients with negative expression (P < 0.05). CD155 expression was correlated with increased PD-L1 expression in immune cells, 0.8% and 0.02% immune cells expressed PD-L1 in patients with positive and negative CD155 expression, respectively (P < 0.05).
CD155 was related to an inhibitory immune breast cancer microenvironment. CD155 was associated with a high proportion of exhausted CD4+ and unexhausted CD8+ TILs and high PD-L1 expression in immune cells.
Core Tip: In this study, we showed that overexpression of CD155 in the breast cancer microenvironment had a significant association with a high level of programmed cell death ligand 1 expression, exhausted CD4+ helper T cells and unexhausted CD8+ cytotoxic T cells. CD155 expression was related to the inhibitory immune microenvironment and may be an immunotherapeutic target in breast cancer.
- Citation: Wang RB, Li YC, Zhou Q, Lv SZ, Yuan KY, Wu JP, Zhao YJ, Song QK, Zhu B. Overexpression of CD155 is associated with PD-1 and PD-L1 expression on immune cells, rather than tumor cells in the breast cancer microenvironment. World J Clin Cases 2020; 8(23): 5935-5943
- URL: https://www.wjgnet.com/2307-8960/full/v8/i23/5935.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v8.i23.5935
Breast cancer (BC) is the most common malignant tumor among Chinese women, and new cases of breast cancer accounted for 15% of all female cancer patients in 2015[1]. The clinicopathological characteristics of Chinese women with BC are different to those of western women, with a lower expression rate of hormone receptors and higher expression rate of human epidermal growth factor receptor 2[2].
CD155 is one ligand of the T cell immunoglobulin and immunoreceptor tyrosine-based inhibitory motifs (ITIM) domain (TIGIT) expressed in various cell types, including antigen-presenting cells and tumor cells[3] and the interaction limits cell function through feedback inhibition[4]. Normal tissues have no or low expression of CD155 but malignancies have upregulated expression[5,6], which plays a key role in tumor cell invasion and migration.
PD-L1 is mainly expressed on the membrane surface of mature immune cells and various tumor cells[7]. PD-1 is an immune checkpoint molecule and the interaction inhibits biological functions of effector T-cells. PD-1 expression on BC tumor infiltrating lymphocytes (TILs) was shown to be related with different clinicopathological characteristics[8,9]. However, whether the exhausted phenotypes of effector TILs and PD-L1 are related to CD155 expression in BC has not been reported. Therefore, this study was performed to investigate the distribution of CD155 expression and its relationship with PD-L1 and phenotypes of exhausted CD4+ and CD8+ effector TILs to illustrate the effect of CD155 expression on the immune microenvironment of BC in Chinese patients.
All procedures performed in this study involving human participants were approved by the ethical committee of Beijing Shijitan Hospital, Capital Medical University, in accordance with the ethical standards of the 1964 Helsinki Declaration and its later amendments. This was a retrospective study and formal consent was waived.
A total of 126 patients with invasive ductal BC were consecutively recruited into this cohort study from January 1, 2012 to December 31, 2013. Patients were diagnosed with operable BC and received surgical treatment at the Department of Breast Surgery, Beijing Shijitan Hospital, Capital Medical University. All patients were diagnosed with primary invasive BC following pathology testing.
The surgical specimens from all patients were fixed using 4% neutral formaldehyde, embedded in paraffin (FFPE), and stained with hematoxylin and eosin. A series of 4-μm thick sections from each specimen were used to determine the histopathological features. The Nottingham modification of the Bloom–Richardson system was used to classify histological grading of BC.
The expression of CD155, PD-L1 and PD-1 was detected by immunohistochemistry (IHC) on 4 μm-thick FFPE sections. Monoclonal antibody against CD155 (rabbit anti-human, # 81254) was purchased from Cell Signaling Technology. Monoclonal antibody against PD-L1 (rabbit anti-human, # SP142) was from Roche. Monoclonal antibody against PD-1 (mouse antihuman, # UMAB199) CD4 (rabbit anti-human, # EP204) and CD8 (# SP16) were purchased from Beijing Zhong Shan Golden Bridge Biotechnology Co., Ltd. Sections were dehydrated in an oven at 60°C for 60 min, dewaxed for 20 min and washed in 100%, 100%, 95% and 75% alcohol for 2 min, respectively. The sections were then washed in phosphate buffered saline (PBS) 5 times for 2 min each time. EnVision TM FLEX Target Retrieval Solutions were used for antigen retrieval for 2 min 30 s. The sections were left at room temperature for 20 min, and washed in PBS 5 times for 2 min each time. The sections were incubated in 3% H2O2 at room temperature for 15 min; washed in PBS 5 times for 2 min each time and then sealed with 5% serum at 37℃ for 15 min. The supernatant was discarded and the primary antibody was added at 4℃ and left overnight. The samples were washed with PBS 5 times for 2 min each time, DAB was added and reacted for 5-10 min. PD-L1, PD-1 and CD155 were visualized with DAB, whereas CD4 and CD8 were visualized with AP-red. The slides were counterstained with hematoxylin.
TILs located within the borders of the invasive tumor, excluding tumor zones with crush artifacts, necrosis, regressive hyalinization and biopsy sites were evaluated by two pathologists to estimate the average level. All mononuclear cells (including lymphocytes and plasma cells) were scored, and polymorphonuclear leukocytes were excluded. The average number of TILs was counted in 10 high-power fields (HPF, × 400) in randomly selected IHC sections.
Positive CD155 expression was recorded as brown membrane in tumor cells. Negative CD155 tumor cells were defined as having complete weak or incomplete strong staining on the cell membrane. Positive PD-L1 expression was recorded as brown cytoplasm and/or cytomembrane in immune and tumor cells. Positive PD-1 expression was recorded as brown cytoplasm in lymphocytes. CD4 and CD8 were expressed on the cytomembrane of lymphocytes and were red in color. Double staining of CD4/PD-1 and CD8/PD-1 showed red cytomembrane and brown cytoplasm in lymphocytes. PD-1, CD4 or CD8 positive cells in 100 TILs were counted and the expression rate was calculated.
All analyses were conducted with SPSS software (version 17.0). The correlation of age and CD155 expression was analyzed by the Wilcoxon rank sum test. Histological grade and tumor node metastasis (TNM) stage were analyzed with CD155 expression by the Spearman correlation test. The relationship between CD155 expression and molecular subtype was estimated using the Chi-square test. Percentage and cell counts of phenotypic CD4+ and CD8+ effector TILs with CD155 expression were analyzed by the Wilcoxon rank sum test. The percentage of tumor and immune cells expressing PD-L1 with CD155 expression were analyzed by the Wilcoxon rank sum test. All analyses were two sided and the significance level was 0.05.
Patient age was not related to CD155 expression (Table 1). BC patients classified by histological grades and TNM stages had comparable expression of CD155 (P > 0.05, Table 1). Molecular subtypes were correlated with CD155 expression, as 22% of Luminal A BC patients were found to have positive CD155 expression, compared with 73% of triple-negative breast cancer (TNBC) patients (P < 0.05, Table 1).
| CD155 expression | P value | ||
| Negative (n = 78) | Positive (n = 48) | ||
| Age1, mean ± SD (yr) | 58.2 ± 13.87 | 57.8 ± 13.26 | 0.914 |
| Histological grade2, n (%) | 0.112 | ||
| I | 11 (15.3) | 2 (4.3) | |
| II | 47 (65.3) | 32 (69.6) | |
| III | 14 (19.4) | 12 (26.1) | |
| TNM stage2, n (%) | 0.662 | ||
| I | 20 (27.0) | 10 (21.3) | |
| II | 39 (52.7) | 28 (59.6) | |
| III | 12 (16.2) | 6 (12.8) | |
| IV | 3 (4.1) | 3 (6.4) | |
| Molecular subtype3, n (%) | 0.002 | ||
| Luminal A | 49 (77.8) | 14 (22.2) | |
| Luminal B | 16 (51.6) | 15 (48.4) | |
| HER2 over-expression | 5 (50.0) | 5 (50.0) | |
| Triple negative | 3 (27.3) | 8 (72.7) | |
CD155 expression was not associated with percentage of CD4+ helper TILs (Table 2). However, patients with positive CD155 expression had a higher level of CD4+/PD-1+ TILs and a lower level of CD4+/PD-1- TILs (P < 0.05, Table 2). CD155 expression was not related to the percentage of phenotypic CD8+ TILs (Table 2).
| CD155 expression | P value | ||
| TILs phenotypes | Negative (n = 78) | Positive (n = 48) | |
| CD4+ TILs, mean ± SD | 60% ± 22% | 61% ± 22% | 0.788 |
| CD4+/PD-1+ TILs, mean ± SD | 21% ± 20% | 30% ± 19% | 0.004 |
| CD4+/PD-1- TILs, mean ± SD | 39% ± 20% | 31% ± 19% | 0.032 |
| CD8+ TILs, mean ± SD | 23% ± 13% | 24% ± 11% | 0.342 |
| CD8+/PD-1+ TILs, mean ± SD | 4% ± 5% | 5% ± 5% | 0.280 |
| CD8+/PD-1- TILs, mean ± SD | 19% ± 10% | 20% ± 9% | 0.437 |
The expression of CD155 was related to higher cell counts of CD4+ helper TILs (87 vs 54/HPF, Table 3). The increase in cell counts of exhausted, but not unexhausted CD4+ helper TILs was related to CD155 expression (47 vs 20/HPF, Table 3). CD155 expression was associated with higher cell counts of CD8+ TILs and unexhausted CD8+ TILs were increased by 76% in patients with positive CD155 expression (P < 0.05, Table 3).
| TILs phenotypes | CD155 expression | P value | |
| Negative (n = 78) | Positive (n = 48) | ||
| CD4+ TILs, mean ± SD | 54 ± 46 | 87 ± 93 | 0.041 |
| CD4+/PD-1+ TILs, mean ± SD | 20 ± 24 | 47 ± 57 | 0.002 |
| CD4+/PD-1- TILs, mean ± SD | 34 ± 30 | 41 ± 45 | 0.658 |
| CD8+ TILs, mean ± SD | 21 ± 20 | 37 ± 44 | 0.040 |
| CD8+/PD-1+ TILs, mean ± SD | 4 ± 5 | 7 ± 13 | 0.069 |
| CD8+/PD-1- TILs, mean ± SD | 17 ± 17 | 30 ± 35 | 0.040 |
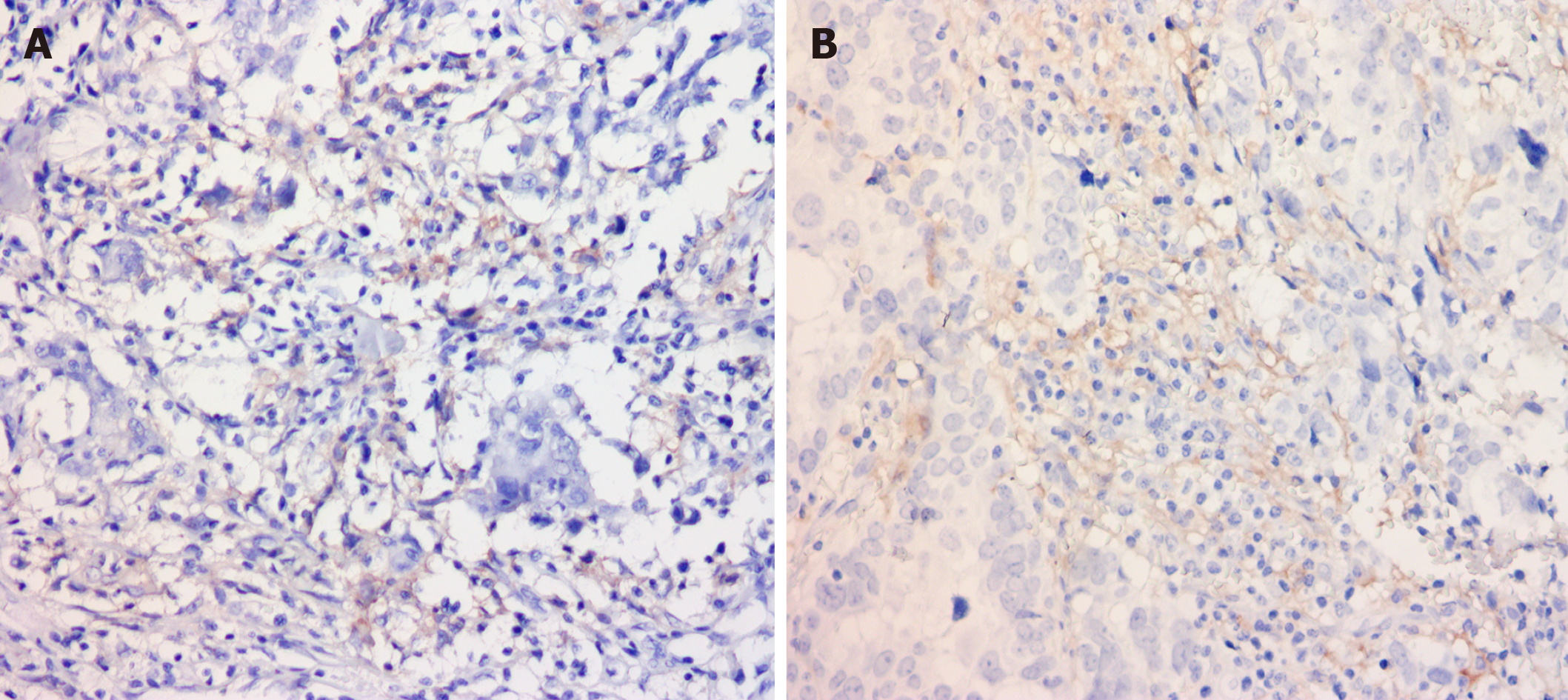
CD155 expression was correlated with a higher proportion of immune cells expressing PD-L1 (Figure 1). The rate of immune cells with PD-L1 expression was 0.02% and 0.8% in patients with negative and positive CD155 expression, respectively (P < 0.05, Figure 1A and B). PD-L1 expression rates were 0.6% and 0.8% in tumor cells with negative and positive CD155 expression, and no significant relationship was observed.
CD155, originally identified as a poliovirus receptor, has similar characteristics of conserved amino acids and domain with the immunoglobulin superfamily[10]. Due to the similar domain to nectin, CD155 is designated as the fifth member of the nectin-like molecular family, and is referred to as necl-5[11]. Up-regulated expression of CD155 can promote cell migration and enhances growth factor-induced cell proliferation[12].
CD155 expression is increased in malignant tumor tissues. In this study, CD155 expression was correlated with molecular subtypes of BC, and the positive rate in TNBC was higher than that in Luminal A patients. Studies[6] have shown that CD155 is less expressed in normal tissues, but is significantly increased in various malignant tumor tissues, and its overexpression was associated with tumor progression and poor prognosis. In addition, plasma soluble CD155 was significantly higher in cancer patients than that in healthy people, and the level in patients with advanced stage cancer was even higher than that in patients with early stage disease[13]. These studies suggest that CD155 may serve as a biomarker for tumor progression and prognosis.
CD155 expression is reported to be regulated by the activation of signaling pathways such as Raf-MEK-ERK-AP1[14], Sonic hedgehog[15], and Toll-like receptor 4 (TLR4)[16]. Overexpression of CD155 inhibited tumor cell apoptosis through the AKT/bcl-2 signaling pathway in colon cancer[17]. In addition, DNA damage is one of the important mechanisms in the induction of CD155 expression. Reactive oxygen species or reactive nitrogen species can induce the expression of CD155 in multiple myeloma cells[18]. Therefore, CD155 expression in tumor tissues is increased under the influence of multiple factors.
T cell activation is initiated after T cell receptor (TCR) recognition of antigens, and the co-signaling molecules affect T cell activation, subsets differentiation and survival[19]. Co-stimulatory and co-inhibitory receptors determine the functional outcome of TCR signaling[20]. TIGIT, like PD-1 and CTLA-4, is a co-inhibitory receptor that can be expressed by CD4+T cells, CD8+T cells, natural killer (NK) cells and other immune cells[21]. CD155 can regulate the function of immune cells. In this study, patients with CD155 overexpression had a higher level of CD4+/PD-1+ TILs and higher cell counts of CD4+, CD8+ TILs. Lymphocytes, T-cells, B-cells, macrophages or NK cells, which moved from the vasculature and localized in tumor stroma are called TILs[22]. The immune system, especially TILs in the epithelium, plays a major role in controlling the growth of virtually all solid tumors[23]. TILs in the microenvironment reportedly affect cancer development, prognosis, and treatment efficacy. The existence of TILs has been determined to be a positive prognostic factor in a number of solid cancers including, but not limited to, colon cancer[24] and BC[25]. Although CD8+ or CD4+ T lymphocytes have been shown to recognize cancer antigens and inhibit the development of cancer, some cancer cells can thwart immune recognition and response[26]. CD155 can interact with its receptors on immune cells to regulate immune function. TIGIT, CD96 and CD226 are common receptors for CD155. When CD155 binds with the co-stimulatory molecule CD226 on the surface of T cells or NK cells, these immune cells are activated to secrete cytokines and kill tumor cells; however, when CD155 interacts with co-inhibitory molecule TIGIT or CD96, the function of immune cells is inhibited[27]. The interaction between CD155 and CD226 down-regulated the expression of CD226 in T cells and NK cells[28]. In contrast to CD226, TIGIT is significantly upregulated on TILs, and its expression parallels that of other co-inhibitory receptors, most notably PD-1[29]. It is now clear that co-signaling molecules have a crucial role in regulating T cell activation, subset differentiation, effector function and survival.
In this study, the proportion of immune cells with PD-L1 expression was correlated with CD155 expression in tumor cells. Studies have confirmed that during the activation process of T cells, interferon-γ (IFN-γ) molecules are secreted to up-regulate the expression of PD-L1 on DC cells, and its binding with PD-1 on T cells will generate inhibitory signals and inhibit the proliferation of T cells[30]. Moreover, in tumor tissues, IFN-γ secretion induced by activation of the TLR4 signaling pathway induced CD155 expression[16]. The common IFN-γ pathway shared by PD-L1 expression in immune cells and CD155 expression in tumor cells might explain this high co-expression.
In this study, although CD155 was observed to be correlated with the molecular phenotype of BC, and there was a significant correlation with TILs and PD-L1, the mechanism is still unclear. The unclear expression of TIGIT, CD96 and CD226 on TILs was the main limitation in this study. The relevant signaling pathways are not discussed in this paper.
CD155 was related to an inhibitory immune microenvironment in breast cancer patients. High CD155 expression was associated with a high level of exhausted CD4+ helper TILs and PD-L1 expression in immune cells. Further studies are warranted.
CD155 is an immune checkpoint protein in cancers and interacts with ligands to regulate the immune microenvironment. The expression of CD155 is correlated with the prognosis and pathological features of breast cancer.
To define whether the expression of CD155 is correlated with the phenotype of tumor infiltrating lymphocytes (TILs) in the breast cancer microenvironment.
To investigate the expression status of CD155 and the association with exhausted CD4+ helper and CD8+ cytotoxic TILs and PD-L1 in the breast cancer microenvironment.
This was a retrospective study of 126 breast cancer patients. Immunohistochemistry was used to detect the expression CD155, PD-L1 and PD-1 on TILs. Univariate and multivariable tests were performed for statistical analysis of the data.
The proportion of patients with CD155 expression was higher in triple negative breast cancer than in Luminal A patients. Patients with positive CD155 expression had a higher percentage of CD4+/PD-1+ helper TILs. Patients with positive CD155 expression also had higher cell counts of exhausted CD4+ TILs and unexhausted CD8+ TILs. CD155 expression was correlated with increased PD-L1 expression in immune cells.
CD155 was related to an inhibitory immune microenvironment in breast cancer patients. High CD155 expression was associated with a high level of exhausted CD4+ helper TILs and PD-L1 expression in immune cells.
CD155 overexpression resulted in a worse overall survival and may be a potential immunotherapy target in breast cancer.
Manuscript source: Unsolicited manuscript
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Majewski M, Treepongkaruna S S-Editor: Yan JP L-Editor: Webster JR P-Editor: Li JH
| 1. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11444] [Cited by in RCA: 13206] [Article Influence: 1467.3] [Reference Citation Analysis (3)] |
| 2. | Zheng S, Bai JQ, Li J, Fan JH, Pang Y, Song QK, Huang R, Yang HJ, Xu F, Lu N, Qiao YL. The pathologic characteristics of breast cancer in China and its shift during 1999-2008: a national-wide multicenter cross-sectional image over 10 years. Int J Cancer. 2012;131:2622-2631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 64] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 3. | Lozano E, Dominguez-Villar M, Kuchroo V, Hafler DA. The TIGIT/CD226 axis regulates human T cell function. J Immunol. 2012;188:3869-3875. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 370] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 4. | Anderson AC, Joller N, Kuchroo VK. Lag-3, Tim-3, and TIGIT: Co-inhibitory Receptors with Specialized Functions in Immune Regulation. Immunity. 2016;44:989-1004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1048] [Cited by in RCA: 1578] [Article Influence: 197.3] [Reference Citation Analysis (0)] |
| 5. | Nakai R, Maniwa Y, Tanaka Y, Nishio W, Yoshimura M, Okita Y, Ohbayashi C, Satoh N, Ogita H, Takai Y, Hayashi Y. Overexpression of Necl-5 correlates with unfavorable prognosis in patients with lung adenocarcinoma. Cancer Sci. 2010;101:1326-1330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 114] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 6. | Bevelacqua V, Bevelacqua Y, Candido S, Skarmoutsou E, Amoroso A, Guarneri C, Strazzanti A, Gangemi P, Mazzarino MC, D'Amico F, McCubrey JA, Libra M, Malaponte G. Nectin like-5 overexpression correlates with the malignant phenotype in cutaneous melanoma. Oncotarget. 2012;3:882-892. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 92] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 7. | Liang SC, Latchman YE, Buhlmann JE, Tomczak MF, Horwitz BH, Freeman GJ, Sharpe AH. Regulation of PD-1, PD-L1, and PD-L2 expression during normal and autoimmune responses. Eur J Immunol. 2003;33:2706-2716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 445] [Cited by in RCA: 504] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 8. | Wang R, Shi F, Zhao L, Zhao Y, Wu G, Song QK. High expression of E-cadherin and Ki-67 associated with functional/dysfunctional phenotypes of tumor-infiltrating lymphocytes among Chinese patients with operable breast cancer. J Int Med Res. 2018;46:5219-5227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 9. | Shi F, Chang H, Zhou Q, Zhao YJ, Wu GJ, Song QK. Distribution of CD4+ and CD8+ exhausted tumor-infiltrating lymphocytes in molecular subtypes of Chinese breast cancer patients. Onco Targets Ther. 2018;11:6139-6145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 10. | He Y, Bowman VD, Mueller S, Bator CM, Bella J, Peng X, Baker TS, Wimmer E, Kuhn RJ, Rossmann MG. Interaction of the poliovirus receptor with poliovirus. Proc Natl Acad Sci USA. 2000;97:79-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 172] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 11. | Takai Y, Irie K, Shimizu K, Sakisaka T, Ikeda W. Nectins and nectin-like molecules: roles in cell adhesion, migration, and polarization. Cancer Sci. 2003;94:655-667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 276] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 12. | Kakunaga S, Ikeda W, Shingai T, Fujito T, Yamada A, Minami Y, Imai T, Takai Y. Enhancement of serum- and platelet-derived growth factor-induced cell proliferation by Necl-5/Tage4/poliovirus receptor/CD155 through the Ras-Raf-MEK-ERK signaling. J Biol Chem. 2004;279:36419-36425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 86] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 13. | Iguchi-Manaka A, Okumura G, Kojima H, Cho Y, Hirochika R, Bando H, Sato T, Yoshikawa H, Hara H, Shibuya A, Shibuya K. Increased Soluble CD155 in the Serum of Cancer Patients. PLoS One. 2016;11:e0152982. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 80] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 14. | Hirota T, Irie K, Okamoto R, Ikeda W, Takai Y. Transcriptional activation of the mouse Necl-5/Tage4/PVR/CD155 gene by fibroblast growth factor or oncogenic Ras through the Raf-MEK-ERK-AP-1 pathway. Oncogene. 2005;24:2229-2235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 61] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 15. | Solecki DJ, Gromeier M, Mueller S, Bernhardt G, Wimmer E. Expression of the human poliovirus receptor/CD155 gene is activated by sonic hedgehog. J Biol Chem. 2002;277:25697-25702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 64] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 16. | Kamran N, Takai Y, Miyoshi J, Biswas SK, Wong JS, Gasser S. Toll-like receptor ligands induce expression of the costimulatory molecule CD155 on antigen-presenting cells. PLoS One. 2013;8:e54406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 66] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 17. | Zheng Q, Wang B, Gao J, Xin N, Wang W, Song X, Shao Y, Zhao C. CD155 knockdown promotes apoptosis via AKT/Bcl-2/Bax in colon cancer cells. J Cell Mol Med. 2018;22:131-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 18. | Fionda C, Abruzzese MP, Zingoni A, Soriani A, Ricci B, Molfetta R, Paolini R, Santoni A, Cippitelli M. Nitric oxide donors increase PVR/CD155 DNAM-1 Ligand expression in multiple myeloma cells: role of DNA damage response activation. BMC Cancer. 2015;15:17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 19. | Josefsson SE, Beiske K, Blaker YN, Førsund MS, Holte H, Østenstad B, Kimby E, Köksal H, Wälchli S, Bai B, Smeland EB, Levy R, Kolstad A, Huse K, Myklebust JH. TIGIT and PD-1 Mark Intratumoral T Cells with Reduced Effector Function in B-cell Non-Hodgkin Lymphoma. Cancer Immunol Res. 2019;7:355-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 82] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 20. | Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol. 2013;13:227-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2216] [Cited by in RCA: 2276] [Article Influence: 189.7] [Reference Citation Analysis (0)] |
| 21. | Fuhrman CA, Yeh WI, Seay HR, Saikumar Lakshmi P, Chopra G, Zhang L, Perry DJ, McClymont SA, Yadav M, Lopez MC, Baker HV, Zhang Y, Li Y, Whitley M, von Schack D, Atkinson MA, Bluestone JA, Brusko TM. Divergent Phenotypes of Human Regulatory T Cells Expressing the Receptors TIGIT and CD226. J Immunol. 2015;195:145-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 201] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 22. | Santoiemma PP, Powell DJ Jr. Tumor infiltrating lymphocytes in ovarian cancer. Cancer Biol Ther. 2015;16:807-820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 267] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 23. | Eggermont A, Robert C, Soria JC, Zitvogel L. Harnessing the immune system to provide long-term survival in patients with melanoma and other solid tumors. Oncoimmunology. 2014;3:e27560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 24. | Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, Tosolini M, Camus M, Berger A, Wind P, Zinzindohoué F, Bruneval P, Cugnenc PH, Trajanoski Z, Fridman WH, Pagès F. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960-1964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4318] [Cited by in RCA: 4903] [Article Influence: 258.1] [Reference Citation Analysis (0)] |
| 25. | Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2228] [Cited by in RCA: 3058] [Article Influence: 254.8] [Reference Citation Analysis (0)] |
| 26. | Mittal D, Gubin MM, Schreiber RD, Smyth MJ. New insights into cancer immunoediting and its three component phases--elimination, equilibrium and escape. Curr Opin Immunol. 2014;27:16-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 960] [Cited by in RCA: 1062] [Article Influence: 96.5] [Reference Citation Analysis (0)] |
| 27. | Pende D, Spaggiari GM, Marcenaro S, Martini S, Rivera P, Capobianco A, Falco M, Lanino E, Pierri I, Zambello R, Bacigalupo A, Mingari MC, Moretta A, Moretta L. Analysis of the receptor-ligand interactions in the natural killer-mediated lysis of freshly isolated myeloid or lymphoblastic leukemias: evidence for the involvement of the Poliovirus receptor (CD155) and Nectin-2 (CD112). Blood. 2005;105:2066-2073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 305] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 28. | Carlsten M, Norell H, Bryceson YT, Poschke I, Schedvins K, Ljunggren HG, Kiessling R, Malmberg KJ. Primary human tumor cells expressing CD155 impair tumor targeting by down-regulating DNAM-1 on NK cells. J Immunol. 2009;183:4921-4930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 220] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 29. | Johnston RJ, Comps-Agrar L, Hackney J, Yu X, Huseni M, Yang Y, Park S, Javinal V, Chiu H, Irving B, Eaton DL, Grogan JL. The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell. 2014;26:923-937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 685] [Cited by in RCA: 880] [Article Influence: 80.0] [Reference Citation Analysis (0)] |
| 30. | Boussiotis VA, Chatterjee P, Li L. Biochemical signaling of PD-1 on T cells and its functional implications. Cancer J. 2014;20:265-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 147] [Article Influence: 14.7] [Reference Citation Analysis (0)] |