Published online Nov 6, 2020. doi: 10.12998/wjcc.v8.i21.5203
Peer-review started: July 21, 2020
First decision: August 8, 2020
Revised: August 12, 2020
Accepted: September 16, 2020
Article in press: September 16, 2020
Published online: November 6, 2020
Processing time: 107 Days and 22.1 Hours
Pulmonary tuberculosis (TB) and lung cancer (LC) are common diseases with a high incidence and similar symptoms, which may be misdiagnosed by radiologists, thus delaying the best treatment opportunity for patients.
To develop and validate radiomics methods for distinguishing pulmonary TB from LC based on computed tomography (CT) images.
We enrolled 478 patients (January 2012 to October 2018), who underwent preoperative CT screening. Radiomics features were extracted and selected from the CT data to establish a logistic regression model. A radiomics nomogram model was constructed, with the receiver operating characteristic, decision and calibration curves plotted to evaluate the discriminative performance.
Radiomics features extracted from lesions with 4 mm radial dilation distances outside the lesion showed the best discriminative performance. The radiomics nomogram model exhibited good discrimination, with an area under the curve of 0.914 (sensitivity = 0.890, specificity = 0.796) in the training cohort, and 0.900 (sensitivity = 0.788, specificity = 0.907) in the validation cohort. The decision curve analysis revealed that the constructed nomogram had clinical usefulness.
These proposed radiomic methods can be used as a noninvasive tool for differentiation of TB and LC based on preoperative CT data.
Core Tip: Pulmonary tuberculosis (TB) often exhibits similarities to lung cancer (LC) on computed tomography (CT) images, which may lead to clinical misdiagnosis. Our study evaluated the discriminative performance of peritumoral regions in differentiating between TB and LC. Radiomics features were extracted and selected from preoperative lung CT images. An eight-feature-combined radiomics signature was constructed as an identifier of TB and LC. A radiomics nomogram model was also plotted and validated with calibration curve and decision curve analyses. The good performance of our model could improve current applications of computer-aided diagnosis for pulmonary TB and LC.
- Citation: Cui EN, Yu T, Shang SJ, Wang XY, Jin YL, Dong Y, Zhao H, Luo YH, Jiang XR. Radiomics model for distinguishing tuberculosis and lung cancer on computed tomography scans. World J Clin Cases 2020; 8(21): 5203-5212
- URL: https://www.wjgnet.com/2307-8960/full/v8/i21/5203.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v8.i21.5203
Pulmonary tuberculosis (TB) is a global public health threat, which represent > 80% of clinical TB cases. Its effects on the lungs involve chronic inflammation that is reported to cause carcinogenesis of lung tissue[1]. Lung cancer (LC) has a poor prognosis, and is one of the most common cause of death due to cancer worldwide[2]. These two diseases are both common, with high prevalence and similar symptoms and clinical presentation. Hence, patients with LC are often misdiagnosed with pulmonary TB, which may delay timely treatment, and even expose patients to inappropriate medication.
Previous studies have examined the association between and diagnosis of pulmonary TB and LC through clinical symptoms and signs, and blood transcriptional profiles[3-5]. Such methods mainly relied on the subjective experiences of clinicians and were therefore unreliable. Imaging examinations, such as computed tomography (CT), are useful tools. However, in clinical practice, due to the radiological similarities between TB and LC, even highly trained radiologists relying on CT data are often prone to misdiagnosis or missed diagnosis. Therefore, the determination of TB or LC is based on histopathological analysis, such as invasive biopsy, with the associated inherent risk of these invasive procedures[6-8]. Thus, noninvasive and computer-aided alternatives are required to improve the discrimination of TB and LC.
In recent years, radiomics has attracted increasing attention due to its high-throughput extraction and selection of discriminative features from medical imaging data, and to construct machine learning classifiers and a radiomics nomogram model to assist in disease diagnosis, prediction of disease status, and response to treatment[9-11]. This has been shown to improve the detection and discrimination performance of medical images compared with those made by radiologists[12-15]. The radiomics approach has been used to predict tumor subtype[16] and metastasis[17] in patients with lung disease. However, to the best of our knowledge, there is still no instance of the application of radiomics in differentiating TB and LC. Thus, the present study aims to establish and validate radiomic methods to distinguish TB from LC, based on pretreatment CT data.
The retrospective analysis conducted on lung CT data was approved by the Institutional Research Ethics Board of our institute. A total of 478 patients were enrolled between January 2012 and October 2018 in the Liaoning Cancer Hospital and Institute. The number of patients with pulmonary TB and LC was 244 and 234, respectively. All patients were pathologically confirmed with pulmonary TB or LC, which is the gold standard. Inclusion criteria were as follows: (1) Patients aged > 18 years; (2) Patients who underwent CT thorax screening before surgery; and (3) Patients who underwent surgical resection with pathological confirmation. Exclusion criteria were as follows: (1) Patients exhibiting other tumors; (2) Patients with a history of lung surgery, or radiotherapy or chemotherapy; and (3) Patients with artifacts in CT images. All patients were randomly divided into the training and validation cohorts at a ratio of 2:1.
All patients were scanned with a 64-slice spiral CT (Syngo 2009A; Siemens, Germany): voltage 120 kV, current 200-350 mAs, slice thickness 5.0 mm, and array 512 × 512. The obtained CT thoracic images with a resolution of 2457 × 1996 were interpreted on a Hologic breast computer-aided diagnosis workstation (SecureView Dx; Hologic) equipped with two 5-megapixel monitors, and stored in the Picture Archiving and Communication System of the hospital in Digital Imaging and Communications in Medicine format.
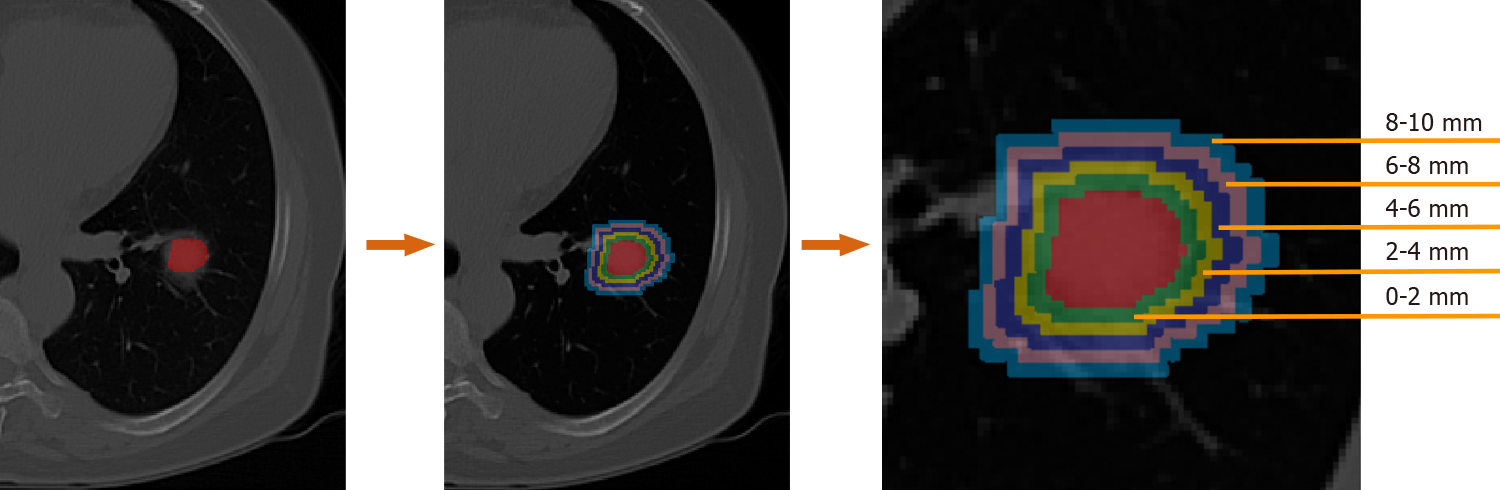
The lesion regions of interests (ROIs) were drawn manually by two radiologists with 12 and 14 years of experience for each patient using the ITK-SNAP software (version 3.6.0, www.itk-snap.org). Other senior radiologists and clinicians were invited to join the decision-making process whenever a divergence occurred during the segmentation. None of the radiologists and clinicians had prior knowledge of the pathological results of these patients. The segmented ROIs were exported into MHA format, and used for image feature extraction. To evaluate the discriminative power of the peritumor tissues. Dilated masks were obtained by dilating the original ROI of each CT slice with 10 different radial distances. The dilated radial distance was up to 10 mm outside the lesion region. The dilated masks are shown in Figure 1. The original ROI segmented by radiologists is colored red. Rings with different colors indicate various radial dilation distances surrounding the lesion.
The imaging features included the following: First order statistics, shape-based, gray-level co-occurrence matrix (GLCM), gray-level size zone matrix, gray-level run length matrix (GLRLM) and neighborhood gray-tone difference matrix[9,18]. These were extracted from the lesions using Python (version 3.6.5). The least absolute shrinkage and selection operator (LASSO) logistic regression was used to exclude features that were redundant, while the predictive features in relation to pulmonary TB and LC remained[19]. The LASSO-selected features were further used to calculate a radiomics score for constructing the radiomics nomogram as a routine radiomics analysis process[20].
The radiomics score was calculated by a linear combination of selected features weighted by the respective LASSO coefficients for each patient[14,21]. A radiomics nomogram model for differentiating LC from TB was constructed based on the multivariable logistic regression analysis using the “rms” package in the R language (v. 3.5.0; available from URL: https://www.r-project.org).
The performance of binary classifications was evaluated using the receiver operating characteristic (ROC) curve analysis for both the nomogram model and machine learning classifiers. The optimal cut-off values of the ROC curves were selected based on the maximum Youden index[22]. The area under the ROC curve (AUC) values were calculated to quantify the discrimination performance. Three comparison metrics, including accuracy, sensitivity and specificity, were also computed following the standard formulas described previously[23]. Calibration curves were plotted to evaluate the calibration of the constructed radiomics nomogram model. A decision curve analysis (DCA) was conducted to assess the clinical utility of the nomogram, by quantifying the net benefits for a range of threshold probabilities in the training and validation groups. All algorithms were run on a 64-bit hexa-core 3.7 GHz Intel i7-6700K CPU with 128 GB of 3000 MHz DDR4 RAM.
To evaluate the discriminative performance of peritumoral tissues, dilations of ten distances were performance from the original ROI. As shown in Table 1, the radiomics features were extracted from the ROI when the dilation was 0. The features were obtained from peritumor tissues when the dilations were from 1 to 10.
| Dilations | Cohorts | AUC | Specificity | Sensitivity |
| 0.0 mm | Training cohort | 0.875 | 0.761 | 0.846 |
| Validation cohort | 0.832 | 0.791 | 0.769 | |
| 1.0 mm | Training cohort | 0.875 | 0.791 | 0.819 |
| Validation cohort | 0.797 | 0.884 | 0.635 | |
| 2.0 mm | Training cohort | 0.889 | 0.751 | 0.879 |
| Validation cohort | 0.810 | 0.907 | 0.635 | |
| 3.0 mm | Training cohort | 0.900 | 0.811 | 0.846 |
| Validation cohort | 0.865 | 0.907 | 0.635 | |
| 4.0 mm | Training cohort | 0.914 | 0.796 | 0.890 |
| Validation cohort | 0.900 | 0.907 | 0.788 | |
| 5.0 mm | Training cohort | 0.888 | 0.818 | 0.816 |
| Validation cohort | 0.779 | 0.652 | 0.837 | |
| 6.0 mm | Training cohort | 0.899 | 0.843 | 0.810 |
| Validation cohort | 0.823 | 0.804 | 0.775 | |
| 7.0 mm | Training cohort | 0.897 | 0.854 | 0.800 |
| Validation cohort | 0.819 | 0.630 | 0.939 | |
| 8.0 mm | Training cohort | 0.906 | 0.737 | 0.924 |
| Validation cohort | 0.840 | 0.739 | 0.837 | |
| 9.0 mm | Training cohort | 0.904 | 0.737 | 0.930 |
| Validation cohort | 0.836 | 0.739 | 0.837 | |
| 10.0 mm | Training cohort | 0.906 | 0.727 | 0.935 |
| Validation cohort | 0.836 | 0.739 | 0.837 |
The model with lowest overfitting was obtained when the dilated radial distance equaled 4.0 mm. At this dilated distance, the highest AUCs of 0.914 and 0.900 on the training and validation cohorts, respectively, were also achieved.
Eight radiomics features were selected by the LASSO process at the best dilation distance. Table 2 shows the selected features with the AUCs and P values in the training and validation cohorts. Figure 2 shows the boxplots of the eight selected radiomics features between the TB and LC groups.
| Radiomics features | Cohorts | AUC | P value |
| Lbp-2D_firstorder_Entropy | Training cohort | 0.627 | 0.000 |
| Validation cohort | 0.618 | 0.033 | |
| Lbp-3D-k_firstorder_10Percentile | Training cohort | 0.633 | 0.022 |
| Validation cohort | 0.568 | 0.026 | |
| Log-sigma-3-0-mm-3D_glcm_Idn | Training cohort | 0.557 | 0.359 |
| Validation cohort | 0.527 | 0.344 | |
| Log-sigma-5-0-mm-3D_glrlm_RunLengthNonUniformity | Training cohort | 0.559 | 0.010 |
| Validation cohort | 0.576 | 0.329 | |
| Squareroot_gldm_DependenceNonUniformity | Training cohort | 0.550 | 0.006 |
| Validation cohort | 0.581 | 0.404 | |
| Wavelet-HLH_glcm_Idn | Training cohort | 0.562 | 0.086 |
| Validation cohort | 0.551 | 0.304 | |
| Wavelet-HLL_glcm_Idn | Training cohort | 0.547 | 0.160 |
| Validation cohort | 0.542 | 0.435 | |
| Wavelet-LLL_glcm_Idmn | Training cohort | 0.658 | 0.000 |
| Validation cohort | 0.663 | 0.008 |
The radiomics signature that consisted of eight features from the best radial dilation distance was obtained by logistic regression, and was as follows: Ct Score = 447.771 - 360.807 × lbp-2D_firstorder_Entropy-4.955 × lbp-3D-k_firstorder_10Percentile + 27.755 × log-sigma-3-0-mm-3D_glcm_Idn + 0.0000143 × log-sigma-5-0-mm-3D_glrlm_RunLengthNonUniformity - 0.0000753 × squareroot_gldm_ DependenceNonUniformity + 33.277 × wavelet-HLH_glcm_Idn + 4.746 × wavelet-HLL_glcm_Idn-195.455 × wavelet-LLL_glcm_Idmn.
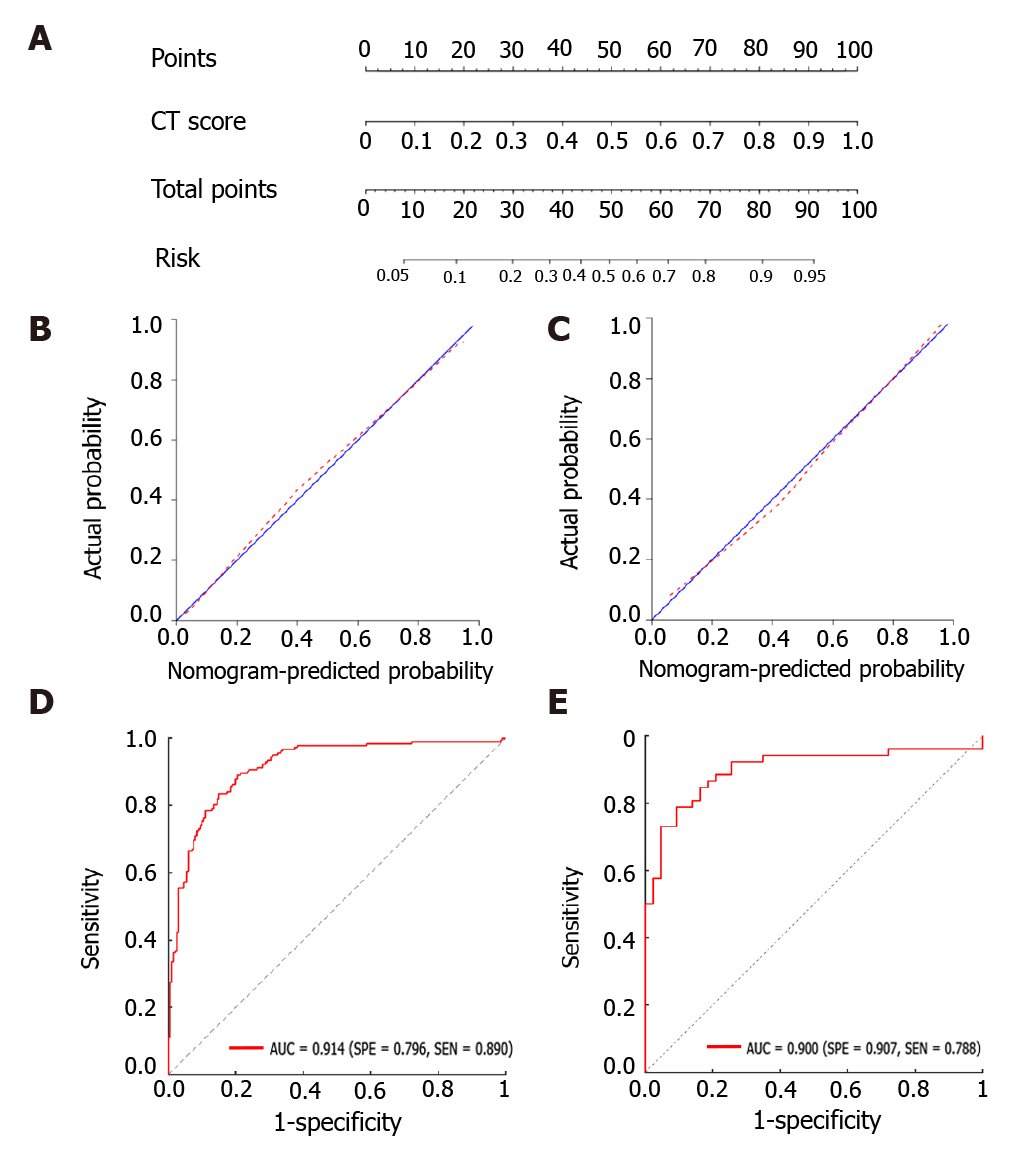
A nomogram model was then constructed (Figure 3A), which includes the radiomics score for differentiating TB and LC in the second row. The favorable calibration of the present radiomics nomogram model was confirmed in the training and validation groups (Figure 3B and C). The calibration curves indicated good agreements between the nomogram-estimated probability and actual outcome. The X and Y axes represented the calculated and actual probabilities, respectively. The diagonal blue line represented the performance of an ideal diagnostic model. Furthermore, the red dotted line represented the performance of the constructed nomogram model. The closer the red dotted line was to the diagonal blue line, the better the discriminative performance achieved by the nomogram model. The nomogram model exhibited a marked discriminative efficacy, with an AUC of 0.914 in the training group and 0.900 in the validation group (Figure 3D and E). Hence, the constructed nomogram model has good discriminative power in differentiating TB from LC.
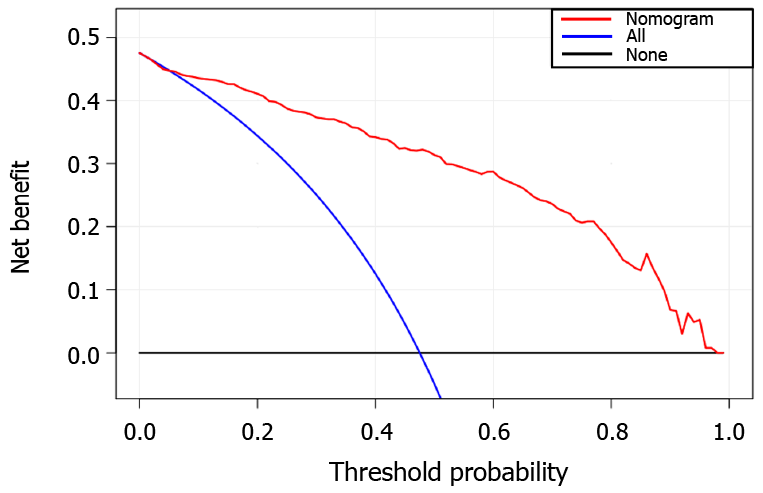
The decision curve analysis showed that our nomogram model for distinguishing TB and LC patients was advantageous, which indicates the good performance of the nomogram in terms of clinical application (Figure 4).
Prompt diagnosis, such as the discrimination of TB from other chronic lung disorders, including LC, is important in providing appropriate and timely treatment[24]. Reports have shown that the delay in the diagnosis and treatment of LC frequently leads to poor outcome and survival[25]. However, LC often exhibits similarities to TB, requiring invasive biopsy for distinguishing these two diseases[4,26]. In clinical practice, even radiologists with decades of experience may still misdiagnose TB and LC using CT imaging data, or even miss the diagnosis altogether. There is little understanding on the differentiation of the two diseases in CT images using computer-aided methods, with no reported attempts. Therefore, these radiomic methods were proven to improve the differentiation between TB and LC.
The selected radiomics features from CT images included two local binary patterns, two Laplacian of Gaussian, one square root and three wavelet-filtered features. The original images were first filtered with corresponding filters, then used to extract handcrafted features. The first-order features describe the distribution of voxel intensities in images. The GLCM features quantify the second-order joint probabilities of images. The GLDM and GLRLM features describe gray-level dependencies and gray-level runs in an image, respectively. Our findings might partially explain the fact that radiologists find it hard to distinguish between TB and LC, since the discriminative CT markers all belonged to high-dimensional space that can hardly be understood by naked eye examination. However, the selected features were all closely related with gray intensities, which indicate that changes in gray levels in lung lesion CT images can potentially assist in the differential diagnosis of TB and LC. The discriminative power of peritumoral tissues was also evaluated. We extracted imaging features from several radial dilation distances (up to a radial distance of 10 mm outside the lung lesion) from the original ROI. Our results revealed that the peritumoral area exhibited more discriminative power than the intratumoral area. The radiomics model with lowest overfitting and best AUCs were obtained at 4 mm outside the lesion for the training and validation cohorts. The results were consistent with previous studies and showed that CT-based peritumoral radiomics are important in the diagnosis of lung lesions[27,28].
To the best of our knowledge, there is no previous report on differentiating TB and LC using CT radiomics. Our findings indicated the diagnostic value of peritumoral regions that have a dilation distance of approximately 4 mm outside the lesions. From the peritumoral area, 1967 imaging features were extracted. A radiomics signature was obtained using the LASSO algorithm by reducing high-dimensional and overfitting data[19]. The constructed nomogram model exhibited favorable discrimination of TB and LC, with AUCs of 0.914 and 0.900 in the training and validation groups, respectively. In addition, good sensitivity and specificity were also obtained, which revealed the low misdiagnosis rate and missed diagnosis rates of our model. The calibration of the present nomogram model was confirmed by calibration-curve-based analysis, which revealed excellent agreement with the actual outcome. To further evaluate whether our nomogram-assisted diagnosis method improved patient outcomes, the clinical usefulness of the model was assessed by DCA, which quantified net benefits for a range of threshold probabilities in the training and validation cohorts. If radiologists use the proposed radiomics nomogram model for differentiating TB and LC, they need first to manually segment the lesions in the CT thoracic images for each patient, and calculate the probabilities of TB or LC based on the nomogram model. Then, the radiologists could consider the clinical information, calculate the probabilities for these patients, and accordingly make a comprehensive decision on medical treatment.
There are a few limitations in the present study. First, all CT data were obtained from a single hospital, which may be inherently biased. Second, only CT images were used to perform the radiomics analyses. Clinical parameters should be incorporated in future studies[29]. Third, the ROIs in each image were manually segmented, which is time-consuming. We found a recent study that developed a 3D U-net algorithm for lesion segmentation in CT thoracic images[21]. This enlightened us to explore automatic segmentation and classification methods in future studies.
The radiomic methods for differentiating LC and TB using CT thoracic images are presented in this study. The established nomogram model exhibited favorable classification performance, indicating its potential as an assisting tool in future clinical applications.
Pulmonary tuberculosis (TB) and lung cancer (LC) are common pulmonary diseases with high incidence and similar symptoms, which may be misdiagnosed by radiologists, thus delaying the best treatment opportunity for patients.
Due to the radiological similarities of TB and LC, even highly trained radiologists relying on computed tomography (CT) data are often prone to misdiagnosis, or missed diagnosis. Therefore, the determination of TB or LC is based on histopathological analysis, such as invasive biopsy, with the associated inherent risk of these invasive procedures. Thus, noninvasive and computer-aided alternatives are required to improve the discrimination of TB and LC.
This study aimed to develop and validate radiomic methods for distinguishing pulmonary TB from LC based on CT images.
Radiomics features were extracted and selected from the CT images to establish a logistic regression model. A radiomics nomogram model was constructed, with the receiver operating characteristic, decision and calibration curves plotted to evaluate the discriminative performance.
This study found that radiomics features extracted from the lesion with 4 mm radial dilation distances outside the lesion showed the best discriminative performance. The radiomics nomogram model exhibited good discrimination performance, and decision curve analysis revealed that the constructed nomogram had clinical usefulness.
The proposed radiomic methods can be used as noninvasive tools for differentiating TB and LC based on preoperative CT data.
This study confirms the predictive performance of our proposed radiomics model. In the future, multimodal data combined with deep learning characteristics are desirable.
Manuscript source: Unsolicited manuscript
Specialty type: Engineering, biomedical
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Charco R, Snowdon VK, Voigt M S-Editor: Wang JL L-Editor: Webster JR P-Editor: Wu YXJ
| 1. | Dheda K, Booth H, Huggett JF, Johnson MA, Zumla A, Rook GA. Lung remodeling in pulmonary tuberculosis. J Infect Dis. 2005;192:1201-1209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 173] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 2. | Yu YH, Liao CC, Hsu WH, Chen HJ, Liao WC, Muo CH, Sung FC, Chen CY. Increased lung cancer risk among patients with pulmonary tuberculosis: a population cohort study. J Thorac Oncol. 2011;6:32-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 151] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 3. | Littman AJ, Thornquist MD, White E, Jackson LA, Goodman GE, Vaughan TL. Prior lung disease and risk of lung cancer in a large prospective study. Cancer Causes Control. 2004;15:819-827. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 77] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 4. | Bloom CI, Graham CM, Berry MP, Rozakeas F, Redford PS, Wang Y, Xu Z, Wilkinson KA, Wilkinson RJ, Kendrick Y, Devouassoux G, Ferry T, Miyara M, Bouvry D, Valeyre D, Gorochov G, Blankenship D, Saadatian M, Vanhems P, Beynon H, Vancheeswaran R, Wickremasinghe M, Chaussabel D, Banchereau J, Pascual V, Ho LP, Lipman M, O'Garra A. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLoS One. 2013;8:e70630. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 208] [Cited by in RCA: 228] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 5. | Wu CY, Hu HY, Pu CY, Huang N, Shen HC, Li CP, Chou YJ. Pulmonary tuberculosis increases the risk of lung cancer: a population-based cohort study. Cancer. 2011;117:618-624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 128] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 6. | Zhang Y, Xie F, Mao X, Zheng X, Li Y, Zhu L, Sun J. Determining factors of endobronchial ultrasound-guided transbronchial needle aspiration specimens for lung cancer subtyping and molecular testing. Endosc Ultrasound. 2019;8:404-411. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 7. | Adler DG, Gabr M, Taylor LJ, Witt B, Pleskow D. Initial report of transesophageal EUS-guided intraparenchymal lung mass core biopsy: Findings and outcomes in two cases. Endosc Ultrasound. 2018;7:413-417. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 8. | Liran L, Rottem K, Gregorio FZ, Avi A, Neville B. A novel, stepwise approach combining conventional and endobronchial ultrasound needle aspiration for mediastinal lymph node sampling. Endosc Ultrasound. 2019;8:31-35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 9. | Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F, Rietbergen MM, Leemans CR, Dekker A, Quackenbush J, Gillies RJ, Lambin P. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2262] [Cited by in RCA: 3245] [Article Influence: 295.0] [Reference Citation Analysis (0)] |
| 10. | Zhou H, Dong D, Chen B, Fang M, Cheng Y, Gan Y, Zhang R, Zhang L, Zang Y, Liu Z, Zheng H, Li W, Tian J. Diagnosis of Distant Metastasis of Lung Cancer: Based on Clinical and Radiomic Features. Transl Oncol. 2018;11:31-36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 11. | Song JH, Venkatesh SS, Conant EA, Arger PH, Sehgal CM. Comparative analysis of logistic regression and artificial neural network for computer-aided diagnosis of breast masses. Acad Radiol. 2005;12:487-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 34] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 12. | Lohmann P, Bousabarah K, Hoevels M, Treuer H. Radiomics in radiation oncology-basics, methods, and limitations. Strahlenther Onkol. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 68] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 13. | Brem RF. Clinical versus research approach to breast cancer detection with CAD: where are we now? AJR Am J Roentgenol. 2007;188:234-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 14. | Aviram G, Shmueli H, Adam SZ, Bendet A, Ziv-Baran T, Steinvil A, Berliner AS, Nesher N, Ben-Gal Y, Topilsky Y. Pulmonary Hypertension: A Nomogram Based on CT Pulmonary Angiographic Data for Prediction in Patients without Pulmonary Embolism. Radiology. 2015;277:236-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 15. | Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278:563-577. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4541] [Cited by in RCA: 5542] [Article Influence: 615.8] [Reference Citation Analysis (3)] |
| 16. | Coroller TP, Grossmann P, Hou Y, Rios Velazquez E, Leijenaar RT, Hermann G, Lambin P, Haibe-Kains B, Mak RH, Aerts HJ. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol. 2015;114:345-350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 446] [Cited by in RCA: 520] [Article Influence: 52.0] [Reference Citation Analysis (2)] |
| 17. | Wu W, Parmar C, Grossmann P, Quackenbush J, Lambin P, Bussink J, Mak R, Aerts HJ. Exploratory Study to Identify Radiomics Classifiers for Lung Cancer Histology. Front Oncol. 2016;6:71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 168] [Cited by in RCA: 257] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 18. | van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RGH, Fillion-Robin JC, Pieper S, Aerts HJWL. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017;77:e104-e107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1922] [Cited by in RCA: 3897] [Article Influence: 487.1] [Reference Citation Analysis (0)] |
| 19. | Sauerbrei W, Royston P, Binder H. Selection of important variables and determination of functional form for continuous predictors in multivariable model building. Stat Med. 2007;26:5512-5528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 870] [Cited by in RCA: 823] [Article Influence: 45.7] [Reference Citation Analysis (0)] |
| 20. | Yang X, Pan X, Liu H, Gao D, He J, Liang W, Guan Y. A new approach to predict lymph node metastasis in solid lung adenocarcinoma: a radiomics nomogram. J Thorac Dis. 2018;10:S807-S819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 21. | Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, Forster K, Aerts HJ, Dekker A, Fenstermacher D, Goldgof DB, Hall LO, Lambin P, Balagurunathan Y, Gatenby RA, Gillies RJ. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30:1234-1248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1505] [Cited by in RCA: 1553] [Article Influence: 119.5] [Reference Citation Analysis (0)] |
| 22. | Ruopp MD, Perkins NJ, Whitcomb BW, Schisterman EF. Youden Index and optimal cut-point estimated from observations affected by a lower limit of detection. Biom J. 2008;50:419-430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 925] [Cited by in RCA: 866] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 23. | Zhou Y, Xu J, Liu Q, Li C, Liu Z, Wang M, Zheng H, Wang S. A Radiomics Approach With CNN for Shear-Wave Elastography Breast Tumor Classification. IEEE Trans Biomed Eng. 2018;65:1935-1942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 85] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 24. | Patil S, Jadhav A. Short Course of High-dose Steroids for Anaphylaxis Caused Flare Up of Tuberculosis: A Case Report. J Transl Int Med. 2019;7:39-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 25. | Bhatt M, Kant S, Bhaskar R. Pulmonary tuberculosis as differential diagnosis of lung cancer. South Asian J Cancer. 2012;1:36-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 26. | Ho JC, Leung CC. Management of co-existent tuberculosis and lung cancer. Lung Cancer. 2018;122:83-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 27. | Wang X, Zhao X, Li Q, Xia W, Peng Z, Zhang R, Li Q, Jian J, Wang W, Tang Y, Liu S, Gao X. Can peritumoral radiomics increase the efficiency of the prediction for lymph node metastasis in clinical stage T1 lung adenocarcinoma on CT? Eur Radiol. 2019;29:6049-6058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 124] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 28. | Akinci D'Antonoli T, Farchione A, Lenkowicz J, Chiappetta M, Cicchetti G, Martino A, Ottavianelli A, Manfredi R, Margaritora S, Bonomo L, Valentini V, Larici AR. CT Radiomics Signature of Tumor and Peritumoral Lung Parenchyma to Predict Nonsmall Cell Lung Cancer Postsurgical Recurrence Risk. Acad Radiol. 2020;27:497-507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 29. | Sak K. A Hypothetical Approach on Gender Differences in Cancer Diagnosis. J Transl Int Med. 2019;7:90-92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |