Published online Jul 6, 2020. doi: 10.12998/wjcc.v8.i13.2817
Peer-review started: April 7, 2020
First decision: April 28, 2020
Revised: May 5, 2020
Accepted: May 30, 2020
Article in press: May 30, 2020
Published online: July 6, 2020
Processing time: 90 Days and 17.5 Hours
Gut microbiota is an emerging field of research, with related research having breakthrough development in the past 15 years. Bibliometric analysis can be applied to analyze the evolutionary trends and emerging hotspots in this field.
To study the subject trends and knowledge structures of gut microbiota related research fields from 2004 to 2018.
The literature data on gut microbiota were identified and downloaded from the PubMed database. Through biclustering analysis, strategic diagrams, and social network analysis diagrams, the main trend and knowledge structure of research fields concerning gut microbiota were analyzed to obtain and compare the research hotspots in each period.
According to the strategic coordinates and social relationship network map, Clostridium Infections/microbiology, Clostridium Infections/therapy, RNA, Ribosomal, 16S/genetics, Microbiota/genetics, Microbiota/immunology, Dysbiosis/immunology, Infla-mmation/immunology, Fecal Microbiota Transplantation/methods, Fecal Microbiota Transplantation can be used as an emerging research hotspot in the past 5 years (2014-2018).
Some subjects were not yet fully studied according to the strategic coordinates; and the emerging hotspots in the social network map can be considered as directions of future research.
Core tip: In this study, the co-word analysis method was used for the first time to study the subject trends and knowledge structures of gut microbiota related research fields from 2004 to 2018. Our results indicate that some subjects have not been not fully studied yet, and the emerging hotspots in the social network map can be considered as a direction of future research.
- Citation: Yue YY, Fan XY, Zhang Q, Lu YP, Wu S, Wang S, Yu M, Cui CW, Sun ZR. Bibliometric analysis of subject trends and knowledge structures of gut microbiota. World J Clin Cases 2020; 8(13): 2817-2832
- URL: https://www.wjgnet.com/2307-8960/full/v8/i13/2817.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v8.i13.2817
The microbiota is a collection of all microbe groups in a microbial community, such as bacteria, archaea, fungi, and protists. The gut microbiota is a collection of all microbe groups in the intestinal microbial community[1,2]. Gut microbiota is composed of thousands of different types of bacteria, of which more than 98% include four bacterial phyla: Firmicutes, Bacteroides, Proteobacteria, and Actinobacteria (dominantly Firmicutes or Bacteroides)[3-5]. Various important functions are executed by gut microbiota, such as production of vitamins, metabolism of dietary compounds, and prevention of distension and systemic infiltration by gut pathogens[6-8]. The microbiome plays a potential role in human health, including early in life[9,10] and in some special diseases such as cardiac metabolic disorders, inflammatory bowel diseases, nervous/mental diseases, and cancer[11-13]. As shown by numerous recent studies, gut microbiota is very relevant to infection[14-17]. As a new therapy, fecal microbiota transplantation has been applied increasingly to treat Clostridium difficile infection, inflammatory bowel diseases, irritable bowel syndrome, and colon cancer[18-21]. In the past 15 years, there has been great progress in understanding various physiological and pathological functions of gut microbiota[22-24]. With the recent development of bibliometrics, the effects of gut microbiota in obesity and the research hotspots of gastrointestinal microbiota have been further studied[5,25].
In this study, the trends in the research field of gut microbiota were revealed by bibliometric analysis. As a set of special methods, bibliometrics can be used for quantitative analysis of hotspots in the literature. Co-word analysis, as the most common method, can estimate the relationship between two specialized words in relevant literature. In the present study, the specialized words extracted were first classified using biclustering analysis, and then their evolutionary trend and subjects were analyzed by strategic diagram and social network analysis (SNA)[26,27].
Literature data on gut microbiota were identified and downloaded from the PubMed database. As a source of medical words collected by the American National Library of Medicine, Medical Subject Heading (MeSH) was applied in the search index and catalog articles of PubMed. Language was limited to English, literature type was limited to journal article, and human was designated to exclude other species. Relevant articles in PubMed were searched through the following strategies: Gut microbiome OR Gut microflora OR Gut microbiota OR Gut flora OR Intestinal microbiome OR Microbiome microbiota OR Intestinal microflora OR Intestinal flora. In order to analyze the subject trend and knowledge structure of gut microbiota over the past 15 years, literature was divided into three 5-year-periods according to publication date: from 1 January 2004 to 31 December 2008; from 1 January 2009 to 31 December 2013; and from 1 January 2014 to 31 December 2018. The search results were screened by two investigators according to title, abstract, and full text. Finally, data of 1057, 2757, and 7648 articles were obtained from these three periods, respectively.
The data of the three periods were accurately extracted by Bibliographic Item Co-Occurrence Matrix Builder. The following information was extracted: Year of literature, journal, country, author, first author, and major MeSH terms/MeSH subheadings. Finally, the term-source article and co-occurrence matrixes were generated as the data basis for subsequent bibliometric analysis[28,30]. The high-frequency major MeSH terms/MeSH subheadings were firstly counted using the Donohue equation; then the lexical matrix and co-occurrence matrix were constructed through Bibliographic Item Co-Occurrence Matrix Builder according to the high-frequency major MeSH terms/MeSH subheadings[29]. T = [1 + (1 + 8i)1/2]/2. Wherein, i represents major MeSH terms/MeSH subheadings occurring in all extracted data only once.
Biclustering analysis was performed for high-frequency major MeSH terms/MeSH subheadings and PubMed unique identifiers of searched articles on gut microbiota. Through the repeated dichotomization method in gCLUTO software, mountain visualization was clustered, and a visual matrix constructed[30]. In the mountain visualization, one cluster was represented by a peak. The position, volume, height, and color of each peak corresponded to the cluster and its relevant information. The color of each peak indicated the effect of biclustering analysis: A good effect was indicated by red and a poor effect by blue.
The intra-field relations and inter-field interactions in the research field of gut microbiota were analyzed by strategic diagram, and then the research hotspot problems in this field were further analyzed. In strategic diagrams, there are two axes: The X axis indicates the density to represent the capability of this cluster for self-maintenance and self-development; and the Y axis indicates the centrality to represent the interaction degree between the research field of gut microbiota and other fields. In 1991, the calculation methods for density and centrality were given by Callon et al[30]. Four quadrants are formed by the X and Y axes. The biclustering analysis allocates the major MeSH terms/MeSH subheadings into different quadrants. The changes in the research field of gut microbiota were clearly shown by comparing the strategic diagrams in the three periods. The strategic diagrams were generated by GraphPad 5 software (La Jolla, CA, United States).
The knowledge structure data were analyzed by SNA. Centrality measurement is the most important research method for social relationship network analysis. The closeness centrality of a node is associated with the number of its connections with other nodes in this network and to a certain degree represents the importance of this node for the network. The higher the betweenness index is, the stronger is the capability for controlling other nodes. In this study, the betweenness index was selected as an evaluation index. The research field of gut microbiota was clearly shown in the SNA diagram.
The SNA network was constructed by Ucinet 6.0 software (Analytic Technologies Co., Lexington, KY, United States) based on the co-occurrence matrix of high-frequency major MeSH terms/MeSH subheadings and displayed in a 2D map by NetDraw 2.084 software. Major MeSH terms/MeSH subheadings were represented by the nodes, and their co-occurrence frequency was represented by the links.
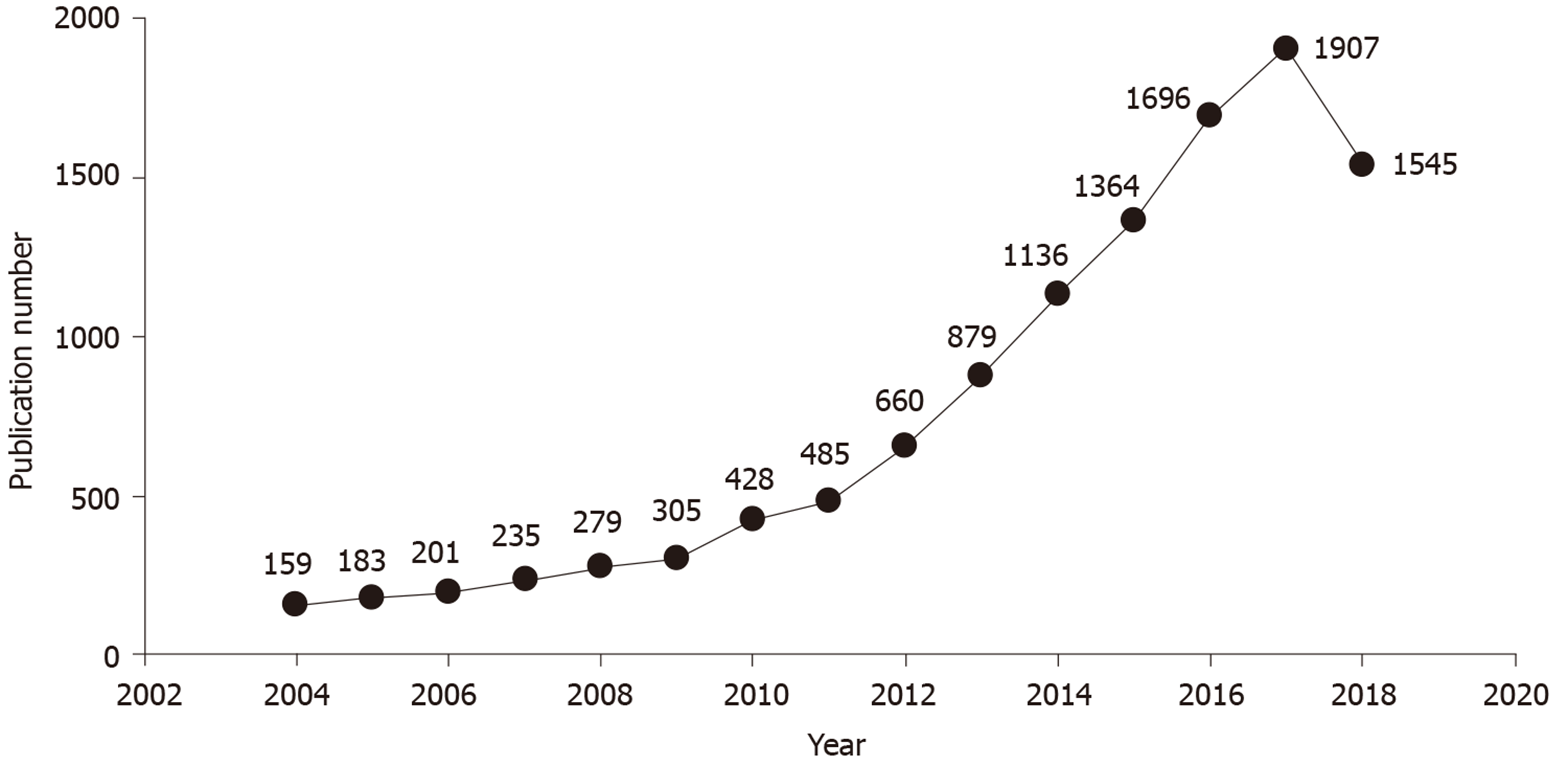
From the three periods (1 January 2004 to 31 December 2008; 1 January 2009 to 31 December 2013; and 1 January 2014 to 31 December 2018), 1057, 2757, and 7648 articles were obtained, respectively, and a comparison analysis was performed. The annual number of articles on gut microbiota increased by nearly 10 fold, from 159 in 2004 to 1545 in 2018 (Figure 1). It was noteworthy that the number of articles on gut microbiota in 2018 decreased compared with 2017. The top 10 publication countries, journals, and authors of articles in each period (Table 1) showed the core change of literature in the research field of gut microbiota in the past 15 years. The first and second highest publication rates in these three periods were in the United States and the United Kingdom; the proportion of publications decreased gradually in the Russian Federation; and increasing numbers of contributions were made in Australia and the United Arab Emirates.
| Period | Rank | Country | Publications, n (%) | Top journal | Publications, n (%) | Author, number of papers |
| 2004-2008 | 1 | United States | 406 (37.3) | The Journal of Nutrition | 35 (3.2) | Isolauri E (27) |
| 2 | England | 307 (28.2) | The British Journal of Nutrition | 30 (2.7) | Salminen S (25) | |
| 3 | Netherlands | 73 (6.7) | Journal of Clinical Gastroenterology | 25 (2.2) | Gibson GR (20) | |
| 4 | Russia (Federation) | 45 (4.1) | Applied and Environmental Microbiology | 23 (2.1) | Doré J (17) | |
| 5 | Germany | 40 (3.6) | Zhurnal Mikrobiologii, Epidemiologii, I immunobiologii | 20 (1.8) | Knol J (14) | |
| 6 | Switzerland | 31 (2.8) | Journal of Agricultural and Food Chemistry | 19 (1.7) | Blaut M (13) | |
| 7 | France | 27 (2.4) | Journal of Pediatric Gastroenterology and Nutrition | 19 (1.7) | Gordon JI (13) | |
| 8 | Japan | 26 (2.3) | Alimentary Pharmacology and Therapeutics | 15 (1.3) | Boehm G (11) | |
| 9 | China | 18 (1.6) | The American Journal of Gastroenterology | 12 (1.0) | Shanahan F (11) | |
| 10 | Italy | 12 (1.1) | Clinical and Experimental Allergy: Journal of the British Society for Allergy and Clinical Immunology | 12 (1.0) | Kim DH (10) | |
| Total | 985 (90.5) | 210 (19.2) | ||||
| 2009-2013 | 1 | United States | 1174 (40.4) | PLoS One | 145 (4.9) | de Vos WM (44) |
| 2 | England | 884 (30.4) | Gut Microbes | 57 (1.9) | O′Toole PW (34) | |
| 3 | Netherlands | 177 (6.1) | The British Journal of Nutrition | 53 (1.8) | Doré J (32) | |
| 4 | Switzerland | 125 (4.3) | Journal of Agricultural and Food Chemistry | 48 (1.6) | Knight R (28) | |
| 5 | Germany | 99 (3.4) | Anaerobe | 38 (1.3) | Gordon JI (25) | |
| 6 | Russia (Federation) | 49 (1.6) | Proceedings of the National Academy of Sciences of the United States of America | 36 (1.2) | Gibson GR (25) | |
| 7 | France | 46 (1.5) | Inflammatory Bowel Diseases | 33 (1.1) | Shanahan F (24) | |
| 8 | China | 32 (1.1) | Digestive Diseases (Basel, Switzerland) | 31 (1.0) | Raoult D (24) | |
| 9 | United Arab Emirates | 26 (0.8) | Current Opinion in Gastroenterology | 31 (1.0) | Salminen S (24) | |
| 10 | Spain | 26 (0.8) | Nature Reviews. Gastroenterology and Hepatology | 27 (0.9) | Isolauri E (23) | |
| Total | 2638 (90.9) | 499 (17.1) | ||||
| 2014-2018 | 1 | United States | 3013 (39.4) | PLoS One | 247 (3.2) | Wang J (87) |
| 2 | England | 2394 (31.3) | Scientific Reports | 149 (1.9) | Li J (80) | |
| 3 | Netherlands | 467 (6.1) | Nutrients | 148 (1.9) | Wang X (68) | |
| 4 | Switzerland | 446 (5.8) | Gut Microbes | 138 (1.8) | de Vos WM (60) | |
| 5 | Germany | 312 (4.0) | World Journal of Gastroenterology | 132 (1.7) | Wang Y (59) | |
| 6 | China | 119 (1.5) | Microbiome | 100 (1.3) | Li Y (58) | |
| 7 | Japan | 105 (1.3) | Food and Function | 93 (1.2) | Zhang X (50) | |
| 8 | France | 97 (1.2) | Gut | 80 (1.0) | Gasbarrini A (49) | |
| 9 | United Arab Emirates | 76 (0.9) | International Journal of Molecular Sciences | 67 (0.8) | Li L (49) | |
| 10 | Australia | 62 (0.8) | Journal of Agricultural and Food Chemistry | 67 (0.8) | Cryan JF (48) | |
| Total | 7091 (92.8) | 1221 (15.9) |
During 2004-2008, the top three journals were Journal of Nutrition, British Journal of Nutrition, and Journal of Clinical Gastroenterology, accounting for 8.1% of all publications. During 2009-2013, the first and third journals were replaced by PLoS One and Gut Microbes. During 2014-2018, the top three journals were PLoS One, Scientific Reports, and Nutrients. Overall, most articles in this field were published by PLoS One during the 15 years. In terms of authors, the greatest contributors were Isolauri E during 2004-2008, de Vos WM during 2009-2013, and Wang J during 2014-2018.
In this study, 50, 44, and 66 high-frequency major MeSH terms/MeSH subheadings were extracted from relevant literature in the three periods, with occurrence frequencies of 33.6886%, 33.8091% and 33.8794%, respectively. These major MeSH terms/MeSH subheadings were regarded as research hotspots of gut microbiota in the recent 15 years.
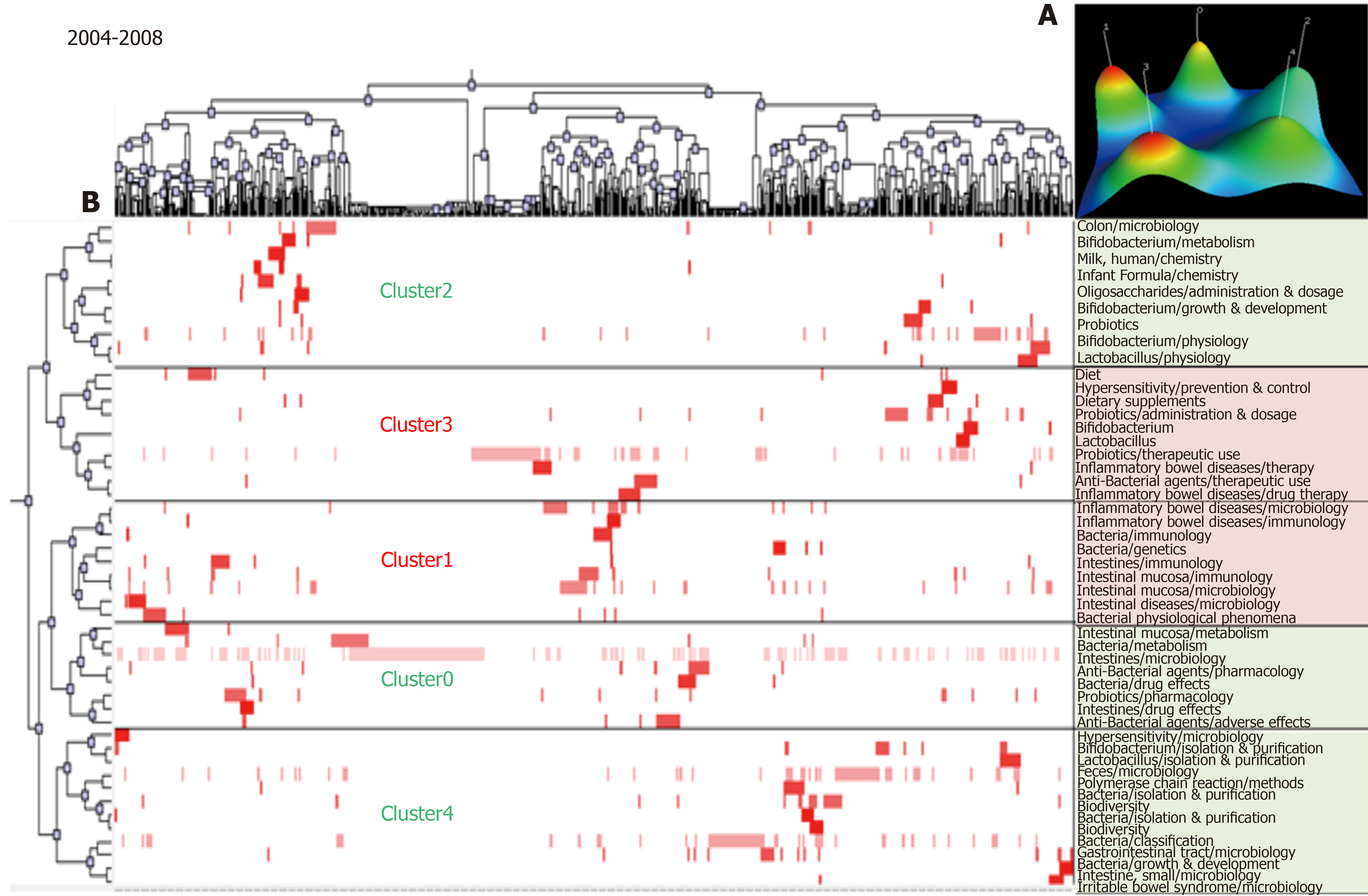
The MeSH terms in the articles during 2004–2008 were classified into five clusters by biclustering analysis. The mountain and matrix visualization of these major MeSH terms/MeSH subheadings are shown in Figure 2. According to the matrix visualization, 50 high-frequency major MeSH terms/MeSH subheadings were allocated into five groups. The relationship of these major MeSH terms/MeSH subheadings with articles is shown by the hierarchical tree at the left and top. The representative articles are clearly indicated by the identified subjects in these clusters. Fifty high-frequency major MeSH terms/MeSH subheadings are shown in the right part, representing the MeSH terms involved in each cluster; the mountain visualization is shown at the right upper corner. Clusters 1 and 3 with red peaks indicate the most significant results.
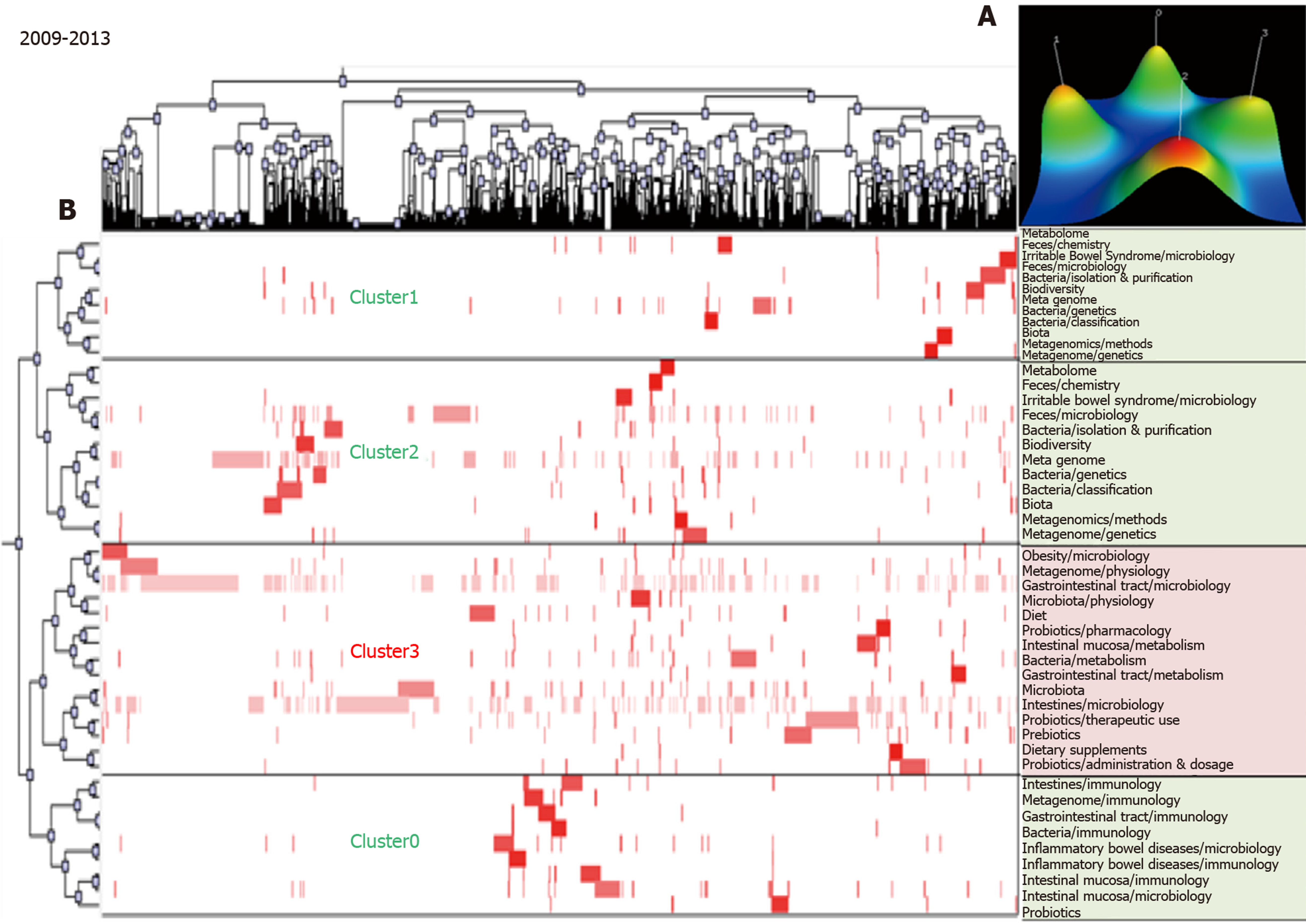
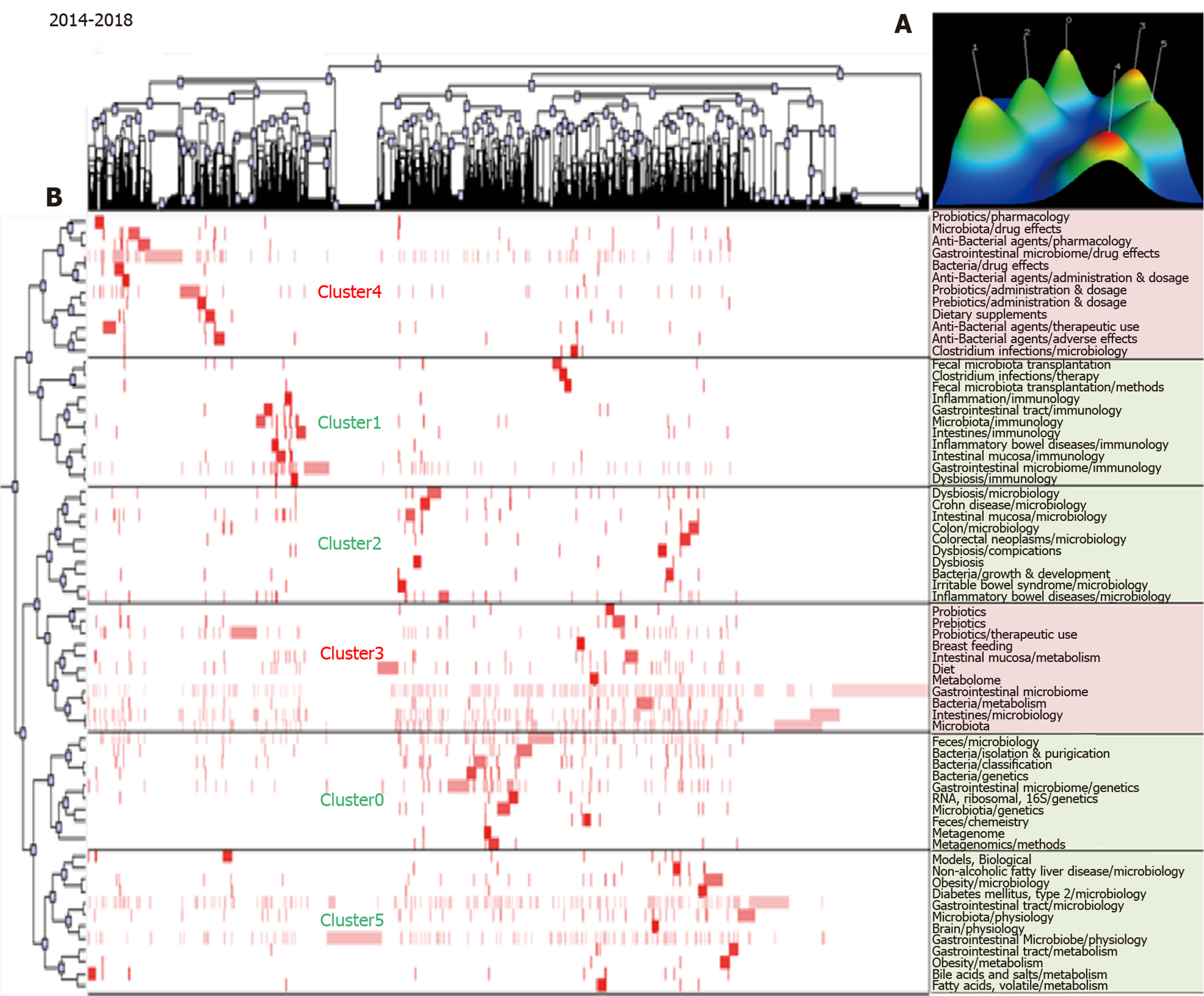
The biclustering analyses of high-frequency major MeSH terms/MeSH subheadings during the periods 2009–2013 and 2014–2018 are shown in Figure 3 and Figure 4, respectively. In these two periods, 44 and 66 high-frequency major MeSH terms/MeSH subheadings were classified into four and six clusters, respectively. The biclustering analysis results on high-frequency major MeSH terms/MeSH subheadings in the research field of gut microbiota in three periods are shown in Table 2.
| Period | Cluster | Number of MeSH terms1 | Cluster analysis |
| 2004-2008 | 0 | 1, 8, 12, 15, 19, 23, 31, 46 | (1) Intestinal mucosa and intestines metabolism; and (2) Probiotics pharmacology. |
| 1 | 6, 7, 13, 17, 22, 28, 34, 36, 44 | (1) Inflammatory bowel diseases microbiology and immunology; (2) Gut microbiota immunology; and (3) Intestinal mucosa immunology and microbiology. | |
| 2 | 5, 9, 20, 25, 26, 29, 35, 39, 40, 41, 48 | (1) Bifidobacterium metabolism, physiology and drug effects; and (2) Bifidobacterium growth and development. | |
| 3 | 2, 10, 16, 21, 27, 32, 33, 38, 42, 49 | (1) Inflammatory bowel disease drug therapy; and (2) Probiotics therapeutic use. | |
| 4 | 3, 4, 11, 14, 18, 24, 30, 37, 43, 45, 47, 50 | (1) Feces microbiology and gastrointestinal tract microbiology; (2) Gut microbiota isolation, purification classification, growth and development; and (3) Irritable bowel syndrome microbiology. | |
| 2009-2013 | 0 | 8, 17, 21, 25, 26, 29, 31, 34, 35 | (1) Gastrointestinal tract immunology; (2) Inflammatory bowel diseases microbiology and immunology; and (3) Intestinal mucosa microbiology and immunology. |
| 1 | 10, 20, 23, 30, 33, 39, 42, 44 | (1) Anti-bacterial agents pharmacology, therapeutic use and adverse effects; and (2) Gut microbiota growth and development. | |
| 2 | 3, 4, 14, 15, 18, 19, 22, 28, 32, 38, 40, 43 | (1) Irritable bowel syndrome microbiology; (2) Gut microbiota classification, isolation and purification; and (3) Feces microbiology. | |
| 3 | 1, 2, 5, 6, 7, 9, 11, 12, 13, 16, 24, 27, 36, 37, 41 | (1) Intestines and intestinal mucosa metabolism; (2) Gastrointestinal tract microbiology; (3) Obesity microbiology; and (4) Probiotics pharmacology and therapeutic use. | |
| 2014-2018 | 0 | 7, 9, 15, 17, 27, 28, 31, 46, 55, 65 | (1) Gut microbiota genetics; (2) Metagenomics methods; and (3) Feces microbiology. |
| 1 | 8, 23, 30, 32, 38, 40, 57, 58, 59, 60, 66 | (1) Inflammatory bowel diseases immunology; (2) Fecal microbiota transplantation methods; (3) Gastrointestinal tract and intestinal mucosa immunology; and (4) Intestinal mucosa immunology. | |
| 2 | 18, 20, 21, 29, 34, 42, 44, 53, 54, 56 | (1) Irritable bowel syndrome microbiology; (2) Dysbiosis microbiology; (3) Gut microbiota growth and development; and (4) Intestinal mucosa microbiology. | |
| 3 | 1, 4, 5, 10, 11, 14, 19, 25, 45, 51, 61 | (1) Probiotics therapeutic use; and (2) Intestines and intestinal mucosa metabolism. | |
| 4 | 6, 13, 22, 24, 26, 35, 37, 41, 47, 48, 62, 63 | (1) Anti-bacterial agents pharmacology, therapeutic use and adverse effects; and (2) Prebiotics therapeutic use and pharmacology. | |
| 5 | 2, 3, 12, 16, 33, 36, 39, 43, 49, 50, 52, 64 | (1) Gastrointestinal tract microbiology; and (2) Non-alcoholic fatty liver disease, obesity and diabetes mellitus (type2) microbiology. |
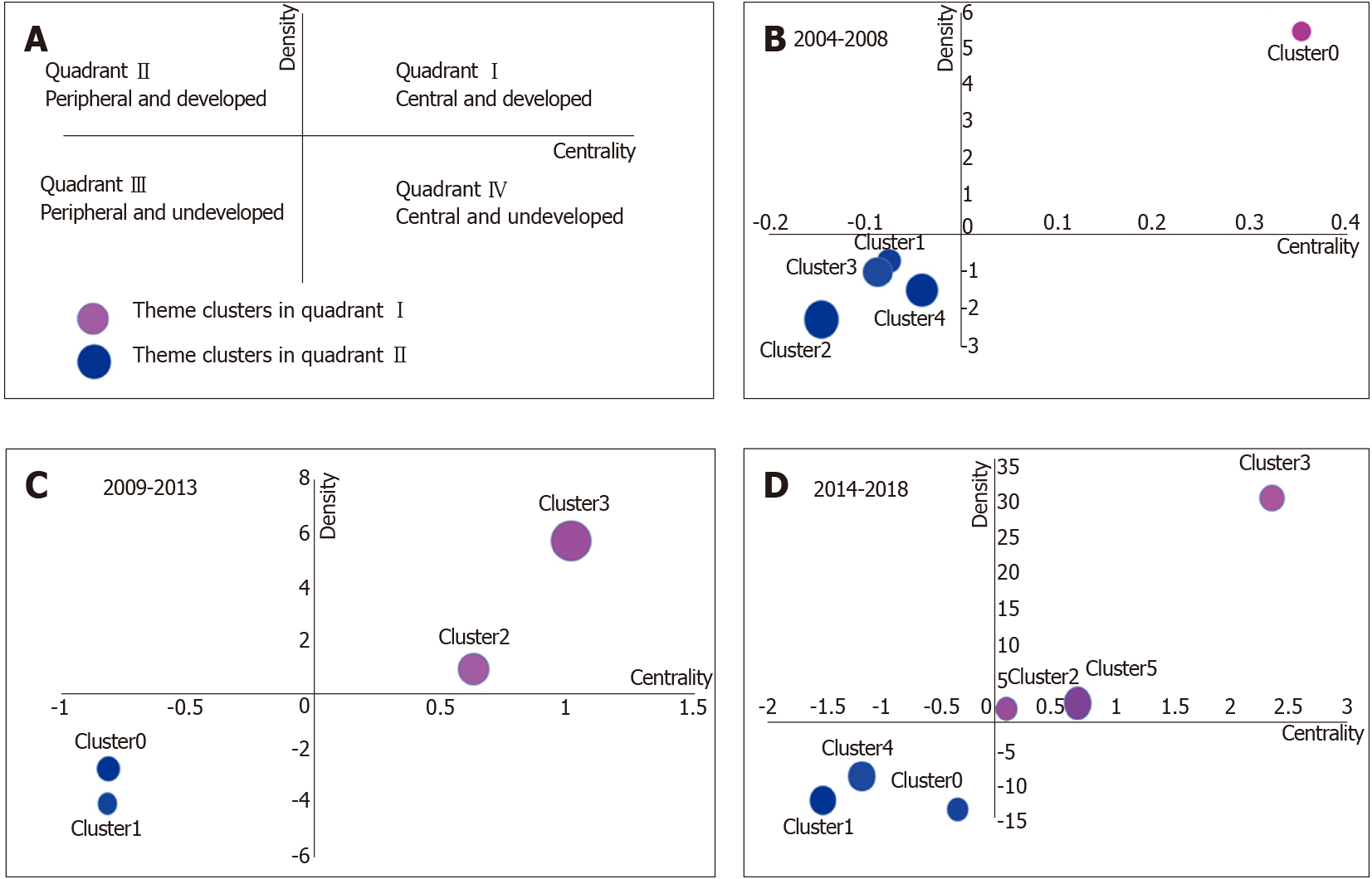
Strategic diagrams were defined by Callon et al[30]. As shown in Figure 5, the motor-themes lie in quadrant I (at the right upper corner), which has strong centrality and high density. As well as these themes generally have high development and strong internal relationship with each other. The specialized themes lie in quadrant II (at the left upper corner), which has insufficient external interaction and high density. These themes are generally considered as close to perfection. The subjects in quadrant III (at the left lower corner) have a low density and insufficient centrality, and are generally considered as emerging or disappearing subjects. The subjects in quadrant IV (at the right lower corner) have very strong centrality but low internal density, and the internal clusters are mature to a certain degree.
In the strategic diagram, the subjects are identified in different quadrants by spheres according to the density and centrality that separately represent the internal and external cohesiveness. The development trends of gut microbiota in the different periods are shown in three strategic diagrams. The clusters in the strategic diagrams show the results of biclustering analysis from Table 2. The areas of spheres represent the number of high-frequency major MeSH terms/MeSH subheadings in each cluster.
In 2004-2008, cluster 0 was in quadrant I with strong centrality and high density, indicating that intestinal mucosa and intestines metabolism and probiotics pharmacology were at the core of all aspects of research during this period. Clusters 1-4 in quadrant III showed that studies in inflammatory bowel diseases microbiology and immunology, gut microbiota immunology, intestinal mucosa immunology and microbiology, Bifidobacterium metabolism, physiology and drug effects, Bifidobacterium growth and development, feces microbiology and gastrointestinal tract microbiology, inflammatory bowel disease drug therapy, probiotics therapeutic use, irritable bowel syndrome microbiology, gut microbiota isolation, purification classification, growth and development, and Bifidobacterium isolation and purification were not yet mature.
During 2009-2013, intestinal mucosa and intestine metabolism were still research emphases, and there were also some new hotspot subjects, such as gut microbiota physiology, gastrointestinal tract microbiology and metabolism, obesity microbiology, and probiotics therapeutic use. Some immature subjects in the previous period had developed continuously, and so were considered as developing subjects in this period, such as irritable bowel syndrome microbiology, feces microbiology and gastro-intestinal tract microbiology, probiotics therapeutic use, and gut microbiota classification, isolation, and purification. Compared with 2009–2013, some subjects remained as key research points during 2014–2018, such as intestinal mucosa and intestine metabolism, probiotics therapeutic use, gastrointestinal tract microbiology, obesity microbiology, and irritable bowel syndrome microbiology. However, some hotspot subjects also emerged in quadrant I, such as dysbiosis microbiology, non-alcoholic fatty liver disease, and diabetes mellitus (type 2) microbiology. Likewise, some immature subjects in the previous period became mature, such as intestinal mucosa microbiology and gut microbiota growth and development. It was noteworthy that some new immature subjects occurred in quadrant III in this period, such as gut microbiota genetics, metagenomics methods, fecal microbiota transplantation methods, and prebiotics therapeutic use and pharmacology.
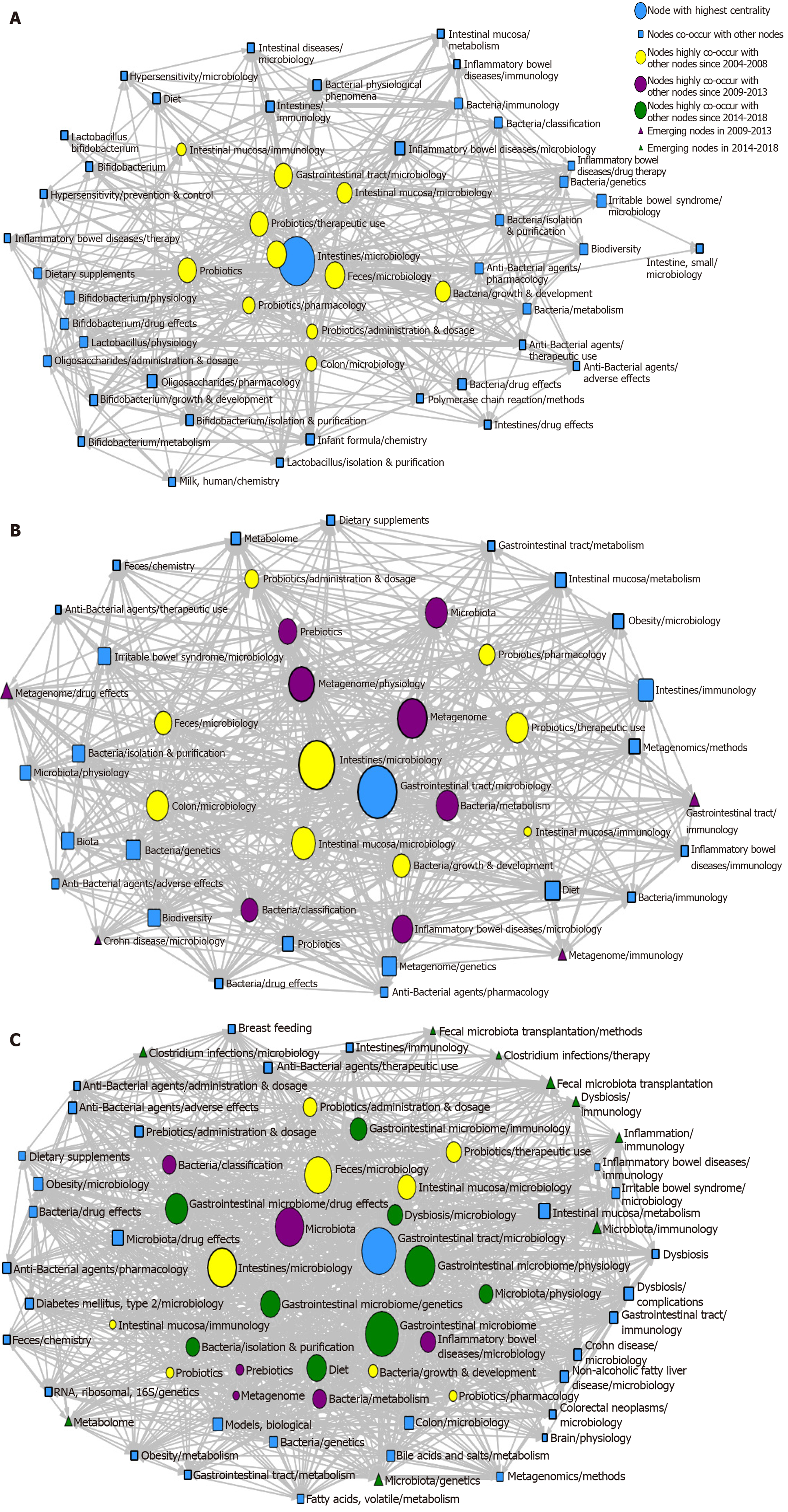
Three SNA diagrams are shown in Figure 6. In order to understand better the results, the knowledge structure of the SNA network was analyzed with three indices (i.e. degree, betweenness, and closeness); the dimension of nodes was directly proportional to the centrality of major MeSH terms/MeSH subheadings; and the size of lines represented collinear frequency.
In the SNA network for 2004–2008, 11 major MeSH terms/MeSH subheadings (i.e. blue and yellow spheres in Figure 6A) had a high centrality (> 16.4), of which Intestines/microbiology was the highest centrality (315, Supplementary Table 1). The first two betweenness indices were 160.08 and 73.69 (Supplementary Table 1), representing Intestines/microbiology and Feces/microbiology, respectively. These two major MeSH terms/MeSH subheadings played a strong mediation effect in the network, with closeness values of 47.50 and 42.50, respectively, indicating that they were closely connected with other nodes (Supplementary Table 1).
In addition to the above two major MeSH terms/MeSH subheadings, other major MeSH terms/MeSH subheadings had a strong mediation effect, such as Gastrointestinal tract/microbiology, Probiotics/therapeutic use, Probiotics, Intestinal mucosa/microbiology, Bacteria/growth and development, Probiotics/pharmacology, Probiotics/administration and dosage, Colon/microbiology, and Intestinal mucosa/immunology. Therefore, these terms played an important mediation role in the network. The mean betweenness value was 16.46 ± 1.40 (Supplementary Table 2).
Compared with 2004-2008, seven new major MeSH terms/MeSH subheadings were added (purple circles in Figure 6B) in the SNA diagram during 2009–2013, including Metagenome/physiology, Metagenome, Bacteria/classification, Inflammatory bowel diseases/microbiology, Microbiota, and Bacteria/metabolism and prebiotics. Gastrointestinal tract/microbiology had the highest betweenness value (Supplementary Table 3).
A total of four new nodes (purple triangles in Figure 6B) emerged at the edge of the network, including Metagenome/drug effects, Crohn’s disease/microbiology, Metagenome/immunology, and Gastrointestinal tract/immunology. These four terms were considered key research points of gut microbiota during 2009–2013.
In the SNA of 2014-2018, nine new major MeSH terms/MeSH subheadings (green circles in Figure 6C) were present: Gastrointestinal microbiome/drug effects, Gastrointestinal microbiome/genetics, Diet, Bacteria/isolation and purification, Microbiota/Physiology, Gastrointestinal Microbiome, Gastrointestinal Microbiome/immunology, Dysbiosis/microbiology, and Gastrointestinal Microbiome/physiology.
There were nine new nodes in the edge of the network (green triangles in Figure 6C): Clostridium, Infections/microbiology, Clostridium Infections/therapy, RNA Ribosomal 16S/genetics, Microbiota/genetics, Microbiota/immunology, Dysbiosis/immunology, Inflammation/immunology, Fecal Microbiota Transplantation/methods, and Fecal microbiota transplantation. Thus, these nine terms were emerging hotspots for gut microbiota research in 2014-2018.
As the potential clinical applications of gut microbiota are understood more deeply, the number of studies on gut microbiota has gradually increased, and this has become an emerging research field. In this study, the evolution of subject trend and knowledge structure in the past 15 years was analyzed in detail using co-word analysis, biclustering analysis, strategic diagrams, and SNA diagrams. Our assessment of global research on gut microbiota showed that relevant publications have increased rapidly in the past 15 years.
In 1977, the gut microbiota was discovered by Breznak et al[31] in wood-eating termites. Now, the gut microbiota is recognized as an overlooked system, and it plays an indispensable role in development of human biology. In the past 15 years, due to the rapid development of new-generation sequencing technology and metabolomics, understanding of the composition and functions of human gut microbiota is increasing exponentially[32]. In this period, there have been numerous gut microbiota relevant publications.
In this study, the subject trends in three periods were analyzed by strategic diagrams. In quadrant I during 2004–2008, there was only one cluster, indicating that this cluster was developed very well in this period; cluster 0 represented probiotics pharmacology and intestinal mucosa and intestines metabolism, which are considered as the research focus of gut microbiota in this period. The gut microbiota plays an important role in human health and occurrence of diseases; and the adjustment of antibiotics and probiotics may become a new therapeutic method[33-35]. Clusters 1–4 were in quadrant III and included numerous studies focused on these themes: gut microbiota immunology; gut microbiota isolation, purification, classification, growth, and development; probiotics therapeutic use (e.g., Bifidobacterium); inflammatory bowel diseases microbiology, immunology, and drug therapy; intestinal mucosa immunology and microbiology; feces microbiology; gastrointestinal tract microbiology; and irritable bowel syndrome microbiology. These subjects are immature and should be further studied. According to Noverr et al[34], gut microbiota can regulate immune responses outside the gut. High-throughput diversity and functionality of gut microbiota have been increasingly analyzed, showing that Bifidobacterium plays an important role in the intestinal tract. Notably, gut microbiota has become more important in studies on inflammatory bowel diseases and irritable bowel syndrome[36-40].
Some subjects in clusters 2 and 3 during 2009-2013 were still key points of research, such as probiotics pharmacology and intestines and intestinal mucosa metabolism. The interaction mechanism between gut microbiota and intestinal mucosa is still in strong dispute; and probiotics have become more popular and are used for prevention and treatment of various diseases[41,42]. However, the immature subjects during 2004-2008 had developed smoothly, such as probiotics therapeutic use, fecal microbiology, gastrointestinal tract microbiology, irritable bowel syndrome microbiology, and gut microbiota classification, isolation, and purification. The subjects from quadrant I became mature[43-46]. As reported[47], fecal microbiota differ between patients with irritable bowel syndrome and healthy people; and microorganisms might be the basis for intestinal symptoms of irritable bowel syndrome. In addition, gastrointestinal tract immunology; inflammatory bowel diseases microbiology and immunology; intestinal mucosa microbiology and immunology; anti-bacterial agents pharmacology, therapeutic use, adverse effects; and gut microbiota growth and development are still not mature subjects. Gut microbiota were shown to be closely related to inflammatory bowel diseases[48]. In the study of Pérez-Cobas et al[49], the effects and action mode of antibiotics were found, which played an important role in regulating the composition and functions of gut microbiota. As shown in mounting evidence[50], gut microbiota can regulate intestinal immune function, but the potential molecular mechanism has not been clarified.
The strategic diagram for 2014-2018 represented the knowledge structure for the past 5 years, and offered vast information on new hotspot problems. As a new subject in cluster 1, fecal microbiota transplantation methods were immature. In fecal microbiota transplantation methods, donor bacteria are transplanted into intestines of patients[51]; but both the controlling force and results of such transplantation remain unknown. In cluster 4, newly emerged prebiotics therapeutic use and pharmacology were immature. Human understanding of interactions among diets, microorganisms, and hosts is increased gradually and prebiotics have become an important direction of research[52]. In cluster 2, irritable bowel syndrome microbiology was a mature subject. In the study of Halkjær et al[53], the change of gut microbiota did not result in clinical improvement of irritable bowel syndrome. In cluster 5, non-alcoholic fatty liver disease and diabetes mellitus (type 2) were two mature subjects. Boursier et al[54] showed that the severity of non-alcoholic fatty liver disease was related to ecological disorder of intestines and change in the metabolic function of gut microbiota. As shown by a study using a mouse model[55-57], gut microbiota could be a driving factor for development of diabetes mellitus (type 2). In cluster 3, probiotics therapeutic use was a mature subject. As shown in multiple studies[58-60], rheumatoid arthritis, Helicobacter pylori infection, and hepatic encephalopathy could be better treated by the intervention of gut microbiota and probiotics. In cluster 0, gut microbiota genetics was a mature subject. In the study of Lim et al[61], the development of MeSH could be promoted by changing the composition of gut microbiota through the mediation of specific host genes.
The SNA diagram showed that 11, 16, and 19 major MeSH terms/MeSH subheadings had a higher centrality in the three consecutive periods, respectively. In the three periods, Intestines/microbiology and Gastrointestinal tract/microbiology were directly connected with the largest number of other nodes, indicating that they played the greatest potential role in regulating the co-occurrence of other nodes.
In addition, four MeSH terms were newly emerged and lay at the edge of the network during 2009–2013 and were immature subjects. Crohn’s disease research developed rapidly and became a mature subject during 2014–2018. Based on this phenomenon, the nine MeSH terminologies, Clostridium infections/microbiology, Clostridium infections/therapy, RNA ribosomal 16S/genetics, Microbiota/genetics, Microbiota/immunology, Dysbiosis/immunology, Inflammation/immunology, Fecal microbiota transplantation/methods, and Fecal microbiota transplantation were emerging hotspots in the third period.
This study is the first comprehensive bibliometric analysis concerning gut microbiota. Study of the gut microbiota is still in the development stage and will be studied more deeply in the future. The abovementioned emerging hotspots offer a basis and guidance for scientific researchers, clinicians, and medical educators to initiate new projects.
However, our study has certain limitations. Firstly, the inclusion and exclusion criteria only retained journals, and reviews and other types of literature were excluded; thus, some research hotspots were omitted. Secondly, since high-frequency MeSH terms were the basis for co-word analysis, the number of these terms might somewhat influence the cluster analysis results; and new subjects of low concern might not be included. Therefore, a range of databases should be used for analysis in future studies.
Aimed at high-frequency MeSH terminology, we performed co-word analysis on gut microbiota by combining bicluster analysis, strategic maps, and SNA. This study showed that gut microbiota genetics, metagenomics methods, inflammatory bowel disease immunology, fecal microbiota transplantation methods, gastrointestinal tract and intestinal mucosa immunology, intestinal mucosa immunology, anti-bacterial agents pharmacology, therapeutic use and adverse effects, and prebiotics therapeutic use and pharmacology were the topics of most interest in the past 5 years. Probiotics therapeutic use, feces microbiology and gastrointestinal tract microbiology, irritable bowel syndrome microbiology, inflammatory bowel disease microbiology and immunology, intestinal mucosa microbiology and immunology, and gut microbiota isolation, purification, classification, growth, and development were core themes that evolved during 2004–2018. Clostridium infections/microbiology, Clostridium infections/therapy, RNA ribosomal 16S/genetics, Microbiota/genetics, Microbiota/immunology, Dysbiosis/immunology, Inflammation/immunology, Fecal microbiota transplantation/methods, and Fecal microbiota transplantation were emerging hotspots for gut microbiota research in the past 5 years.
Gut microbiota is an emerging field of research, and related research has shown breakthrough development in the past 15 years. Bibliometric analysis can be applied to analyze the evolutionary trends and emerging hotspots in this field.
As the potential clinical application of gut microbiota is understood more and more deeply, the number of studies on gut microbiota has increased rapidly. The study of gut microbiota is an emerging research field, and our findings will offer guidance to scholars in this field.
To study the subject trends and knowledge structures of gut microbiota related research fields from 2004 to 2018.
Through the biclustering analysis, strategic diagram, and social network analysis diagram, main trends and knowledge structure of research fields concerning gut microbiota were analyzed to obtain and compare the research hotspots in each period.
According to the strategic coordinates and social relationship network map, Clostridium Infections/microbiology, Clostridium Infections/therapy, RNA, Ribosomal, 16S/genetics, Microbiota/genetics, Microbiota/immunology, Dysbiosis/immunology, Inflammation/immunology, Fecal Microbiota Transplantation/methods, and Fecal Microbiota Transplantation were emerging research hotspot in the past 5 years (2014-2018).
Using strategic coordinates, our results identified which subjects have not been not fully studied yet, and the emerging hotspots in the social network map provide a direction for future research.
This study is the first comprehensive bibliometric analysis of gut microbiota. Research on microbiota is still at the developing stage, and it will continue to be studied more deeply in the future. In our view, the abovementioned emerging hotspot problems can offer the basis for future research and can guide scientific researchers, clinicians, and medical educators to initiate new projects.
Manuscript source: Unsolicited manuscript
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewers: Antaki N, Lion M, Pavlovic M S-Editor: Zhang H L-Editor: Filipodia E-Editor: Liu JH
| 1. | Knight R, Callewaert C, Marotz C, Hyde ER, Debelius JW, McDonald D, Sogin ML. The Microbiome and Human Biology. Annu Rev Genomics Hum Genet. 2017;18:65-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 216] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 2. | Cani PD. Human gut microbiome: hopes, threats and promises. Gut. 2018;67:1716-1725. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 821] [Cited by in RCA: 930] [Article Influence: 132.9] [Reference Citation Analysis (0)] |
| 3. | Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780-13785. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3731] [Cited by in RCA: 3430] [Article Influence: 190.6] [Reference Citation Analysis (1)] |
| 4. | Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635-1638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5700] [Cited by in RCA: 5587] [Article Influence: 279.4] [Reference Citation Analysis (2)] |
| 5. | Huang X, Fan X, Ying J, Chen S. Emerging trends and research foci in gastrointestinal microbiome. J Transl Med. 2019;17:67. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 129] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 6. | Magnúsdóttir S, Ravcheev D, de Crécy-Lagard V, Thiele I. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front Genet. 2015;6:148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 453] [Cited by in RCA: 516] [Article Influence: 51.6] [Reference Citation Analysis (0)] |
| 7. | Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci USA. 2008;105:20858-20863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 829] [Cited by in RCA: 764] [Article Influence: 44.9] [Reference Citation Analysis (0)] |
| 8. | Belkaid Y, Naik S. Compartmentalized and systemic control of tissue immunity by commensals. Nat Immunol. 2013;14:646-653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 283] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 9. | Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes S, Mancabelli L, Lugli GA, Rodriguez JM, Bode L, de Vos W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol Mol Biol Rev. 2017;81:e00036-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1057] [Cited by in RCA: 1141] [Article Influence: 142.6] [Reference Citation Analysis (0)] |
| 10. | Sprockett D, Fukami T, Relman DA. Role of priority effects in the early-life assembly of the gut microbiota. Nat Rev Gastroenterol Hepatol. 2018;15:197-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 257] [Article Influence: 36.7] [Reference Citation Analysis (0)] |
| 11. | Routy B, Gopalakrishnan V, Daillère R, Zitvogel L, Wargo JA, Kroemer G. The gut microbiota influences anticancer immunosurveillance and general health. Nat Rev Clin Oncol. 2018;15:382-396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 390] [Article Influence: 65.0] [Reference Citation Analysis (0)] |
| 12. | Kong F, Kong X, Zhu J, Sun T, Du Y, Wang K, Jin Z, Li Z, Wang D. A prospective comparison of conventional cytology and digital image analysis for the identification of pancreatic malignancy in patients undergoing EUS-FNA. Endosc Ultrasound. 2019;8:269-276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 13. | Tilg H, Adolph TE, Gerner RR, Moschen AR. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell. 2018;33:954-964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 548] [Article Influence: 78.3] [Reference Citation Analysis (0)] |
| 14. | Seekatz AM, Rao K, Santhosh K, Young VB. Dynamics of the fecal microbiome in patients with recurrent and nonrecurrent Clostridium difficile infection. Genome Med. 2016;8:47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 93] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 15. | Zuo T, Wong SH, Cheung CP, Lam K, Lui R, Cheung K, Zhang F, Tang W, Ching JYL, Wu JCY, Chan PKS, Sung JJY, Yu J, Chan FKL, Ng SC. Gut fungal dysbiosis correlates with reduced efficacy of fecal microbiota transplantation in Clostridium difficile infection. Nat Commun. 2018;9:3663. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 203] [Cited by in RCA: 183] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 16. | Yang F, Wang H, Liu X, Ge N, Guo J, Wang S, Song X, Cao L, Sun S. EUS-guided fine-needle technique-derived cancer organoids: A tailored "Shennong deity" for every patient with cancer. Endosc Ultrasound. 2019;8:73-75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 17. | Schuijt TJ, Lankelma JM, Scicluna BP, de Sousa e Melo F, Roelofs JJ, de Boer JD, Hoogendijk AJ, de Beer R, de Vos A, Belzer C, de Vos WM, van der Poll T, Wiersinga WJ. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut. 2016;65:575-583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 443] [Cited by in RCA: 589] [Article Influence: 65.4] [Reference Citation Analysis (0)] |
| 18. | D'Haens GR, Jobin C. Fecal Microbial Transplantation for Diseases Beyond Recurrent Clostridium Difficile Infection. Gastroenterology. 2019;157:624-636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 71] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 19. | Benech N, Kapel N, Sokol H. Fecal Microbiota Transplantation for Ulcerative Colitis. JAMA. 2019;321:2240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 20. | Borody TJ, Eslick GD, Clancy RL. Fecal microbiota transplantation as a new therapy: from Clostridioides difficile infection to inflammatory bowel disease, irritable bowel syndrome, and colon cancer. Curr Opin Pharmacol. 2019;49:43-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 21. | Burrello C, Giuffrè MR, Macandog AD, Diaz-Basabe A, Cribiù FM, Lopez G, Borgo F, Nezi L, Caprioli F, Vecchi M, Facciotti F. Fecal Microbiota Transplantation Controls Murine Chronic Intestinal Inflammation by Modulating Immune Cell Functions and Gut Microbiota Composition. Cells. 2019;8:517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 22. | Sommer F, Bäckhed F. The gut microbiota--masters of host development and physiology. Nat Rev Microbiol. 2013;11:227-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2011] [Cited by in RCA: 2402] [Article Influence: 200.2] [Reference Citation Analysis (0)] |
| 23. | Wang W, Lin L, Du Y, Song Y, Peng X, Chen X, Yang CJ. Assessing the viability of transplanted gut microbiota by sequential tagging with D-amino acid-based metabolic probes. Nat Commun. 2019;10:1317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 69] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 24. | Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2488] [Cited by in RCA: 3424] [Article Influence: 311.3] [Reference Citation Analysis (1)] |
| 25. | Yao H, Wan JY, Wang CZ, Li L, Wang J, Li Y, Huang WH, Zeng J, Wang Q, Yuan CS. Bibliometric analysis of research on the role of intestinal microbiota in obesity. PeerJ. 2018;6:e5091. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 26. | Shi B, Wei W, Qin X, Zhao F, Duan Y, Sun W, Li D, Cao Y. Mapping theme trends and knowledge structure on adipose-derived stem cells: a bibliometric analysis from 2003 to 2017. Regen Med. 2019;14:33-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 27. | Zhang Q, Yue Y, Shi B, Yuan Z. A Bibliometric Analysis of Cleft Lip and Palate-Related Publication Trends From 2000 to 2017. Cleft Palate Craniofac J. 2019;56:658-669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 28. | Li F, Li M, Guan P, Ma S, Cui L. Mapping publication trends and identifying hot spots of research on Internet health information seeking behavior: a quantitative and co-word biclustering analysis. J Med Internet Res. 2015;17:e81. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 115] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 29. | Donohue JC. Understanding scientific literature: a bibliometric approach. In: Infromation Storage and Retrieval: Farradane J, editor. Manchester (UK): The Massachusetts Institute of Technology, 1974: 101. |
| 30. | Callon M, Courtial JP, Laville F. Co-word analysis as a tool for describing the network of interactions between basic and technological research: The case of polymer chemsitry. Scientometrics. 1991;22:155-205. [RCA] [DOI] [Full Text] [Cited by in Crossref: 733] [Cited by in RCA: 462] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 31. | Breznak JA, Pankratz HS. In situ morphology of the gut microbiota of wood-eating termites [Reticulitermes flavipes (Kollar) and Coptotermes formosanus Shiraki]. Appl Environ Microbiol. 1977;33:406-426. [PubMed] |
| 32. | Agace WW. T-cell recruitment to the intestinal mucosa. Trends Immunol. 2008;29:514-522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 130] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 33. | Iapichino G, Callegari ML, Marzorati S, Cigada M, Corbella D, Ferrari S, Morelli L. Impact of antibiotics on the gut microbiota of critically ill patients. J Med Microbiol. 2008;57:1007-1014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 63] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 34. | Noverr MC, Huffnagle GB. Does the microbiota regulate immune responses outside the gut? Trends Microbiol. 2004;12:562-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 331] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 35. | Jia W, Li H, Zhao L, Nicholson JK. Gut microbiota: a potential new territory for drug targeting. Nat Rev Drug Discov. 2008;7:123-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 353] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 36. | Servin AL. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol Rev. 2004;28:405-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 749] [Cited by in RCA: 770] [Article Influence: 36.7] [Reference Citation Analysis (0)] |
| 37. | Parkes GC, Brostoff J, Whelan K, Sanderson JD. Gastrointestinal microbiota in irritable bowel syndrome: their role in its pathogenesis and treatment. Am J Gastroenterol. 2008;103:1557-1567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 113] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 38. | Scanlan PD, Marchesi JR. Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J. 2008;2:1183-1193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 229] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 39. | Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1339] [Cited by in RCA: 1376] [Article Influence: 80.9] [Reference Citation Analysis (1)] |
| 40. | Zoetendal EG, Rajilic-Stojanovic M, de Vos WM. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut. 2008;57:1605-1615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 454] [Cited by in RCA: 456] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 41. | Gareau MG, Sherman PM, Walker WA. Probiotics and the gut microbiota in intestinal health and disease. Nat Rev Gastroenterol Hepatol. 2010;7:503-514. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 707] [Cited by in RCA: 623] [Article Influence: 41.5] [Reference Citation Analysis (0)] |
| 42. | Sánchez de Medina F, Ortega-González M, González-Pérez R, Capitán-Cañadas F, Martínez-Augustin O. Host-microbe interactions: the difficult yet peaceful coexistence of the microbiota and the intestinal mucosa. Br J Nutr. 2013;109 Suppl 2:S12-S20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 43. | Sanders ME, Guarner F, Guerrant R, Holt PR, Quigley EM, Sartor RB, Sherman PM, Mayer EA. An update on the use and investigation of probiotics in health and disease. Gut. 2013;62:787-796. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 345] [Cited by in RCA: 338] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 44. | Dominguez-Bello MG, Blaser MJ, Ley RE, Knight R. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology. 2011;140:1713-1719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 273] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 45. | Greer RL, Morgun A, Shulzhenko N. Bridging immunity and lipid metabolism by gut microbiota. J Allergy Clin Immunol. 2013;132:253-262; quiz 263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 46. | Simrén M, Barbara G, Flint HJ, Spiegel BM, Spiller RC, Vanner S, Verdu EF, Whorwell PJ, Zoetendal EG; Rome Foundation Committee. Intestinal microbiota in functional bowel disorders: a Rome foundation report. Gut. 2013;62:159-176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 655] [Cited by in RCA: 649] [Article Influence: 54.1] [Reference Citation Analysis (0)] |
| 47. | Jalanka-Tuovinen J, Salojärvi J, Salonen A, Immonen O, Garsed K, Kelly FM, Zaitoun A, Palva A, Spiller RC, de Vos WM. Faecal microbiota composition and host-microbe cross-talk following gastroenteritis and in postinfectious irritable bowel syndrome. Gut. 2014;63:1737-1745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 248] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 48. | Nagalingam NA, Lynch SV. Role of the microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2012;18:968-984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 215] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 49. | Pérez-Cobas AE, Artacho A, Knecht H, Ferrús ML, Friedrichs A, Ott SJ, Moya A, Latorre A, Gosalbes MJ. Differential effects of antibiotic therapy on the structure and function of human gut microbiota. PLoS One. 2013;8:e80201. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 174] [Cited by in RCA: 149] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 50. | Kanauchi O, Andoh A, Mitsuyama K. Effects of the modulation of microbiota on the gastrointestinal immune system and bowel function. J Agric Food Chem. 2013;61:9977-9983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 51. | Smillie CS, Sauk J, Gevers D, Friedman J, Sung J, Youngster I, Hohmann EL, Staley C, Khoruts A, Sadowsky MJ, Allegretti JR, Smith MB, Xavier RJ, Alm EJ. Strain Tracking Reveals the Determinants of Bacterial Engraftment in the Human Gut Following Fecal Microbiota Transplantation. Cell Host Microbe. 2018;23:229-240.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 287] [Cited by in RCA: 264] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 52. | Bindels LB, Delzenne NM, Cani PD, Walter J. Towards a more comprehensive concept for prebiotics. Nat Rev Gastroenterol Hepatol. 2015;12:303-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 531] [Cited by in RCA: 591] [Article Influence: 59.1] [Reference Citation Analysis (0)] |
| 53. | Halkjær SI, Christensen AH, Lo BZS, Browne PD, Günther S, Hansen LH, Petersen AM. Faecal microbiota transplantation alters gut microbiota in patients with irritable bowel syndrome: results from a randomised, double-blind placebo-controlled study. Gut. 2018;67:2107-2115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 234] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 54. | Boursier J, Mueller O, Barret M, Machado M, Fizanne L, Araujo-Perez F, Guy CD, Seed PC, Rawls JF, David LA, Hunault G, Oberti F, Calès P, Diehl AM. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016;63:764-775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 763] [Cited by in RCA: 1049] [Article Influence: 116.6] [Reference Citation Analysis (0)] |
| 55. | Meijnikman AS, Gerdes VE, Nieuwdorp M, Herrema H. Evaluating Causality of Gut Microbiota in Obesity and Diabetes in Humans. Endocr Rev. 2018;39:133-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 193] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 56. | Abad-Belando R, Varas-Lorenzo MJ, Pons-Vilardell C, Puig-Torrus X, Pla-Alcaraz M, Monleón-Getino A, Sánchez-Vizcaíno-Mengual E. Canalization technique to obtain deep tissue biopsy of gastrointestinal subepithelial tumors as an alternative to conventional known techniques. Endosc Ultrasound. 2018;7:184-190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 57. | Oh B, Kim BS, Kim JW, Kim JS, Koh SJ, Kim BG, Lee KL, Chun J. The Effect of Probiotics on Gut Microbiota during the Helicobacter pylori Eradication: Randomized Controlled Trial. Helicobacter. 2016;21:165-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 123] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 58. | Mohammed AT, Khattab M, Ahmed AM, Turk T, Sakr N, M Khalil A, Abdelhalim M, Sawaf B, Hirayama K, Huy NT. The therapeutic effect of probiotics on rheumatoid arthritis: a systematic review and meta-analysis of randomized control trials. Clin Rheumatol. 2017;36:2697-2707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 70] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 59. | Pesenti C, Bories E, Caillol F, Ratone JP, Godat S, Monges G, Poizat F, Raoul JL, Ries P, Giovannini M. Characterization of subepithelial lesions of the stomach and esophagus by contrast-enhanced EUS: A retrospective study. Endosc Ultrasound. 2019;8:43-49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 60. | Djiambou-Nganjeu H. Hepatic Encephalopathy in Patients in Lviv (Ukraine). J Transl Int Med. 2018;6:146-151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 61. | Lim MY, You HJ, Yoon HS, Kwon B, Lee JY, Lee S, Song YM, Lee K, Sung J, Ko G. The effect of heritability and host genetics on the gut microbiota and metabolic syndrome. Gut. 2017;66:1031-1038. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 250] [Article Influence: 31.3] [Reference Citation Analysis (0)] |