Published online Jun 6, 2020. doi: 10.12998/wjcc.v8.i11.2066
Peer-review started: January 21, 2020
First decision: March 15, 2020
Revised: April 10, 2020
Accepted: April 29, 2020
Article in press: April 29, 2020
Published online: June 6, 2020
Processing time: 138 Days and 12.6 Hours
Digestive tract cancer is one of the main diseases that endanger human health. At present, the early diagnosis of digestive tract tumors mainly depends on serology, imaging, endoscopy, and so on. Although tissue specimens are the gold standard for cancer diagnosis, with the rapid development of precision medicine in cancer, the demand for dynamic monitoring of tumor molecular characteristics has increased. Liquid biopsy involves the collection of body fluids via non-invasive approaches, and analyzes biological markers such as circulating tumor cells, circulating tumor DNA, circulating cell-free DNA, microRNAs, and exosomes. In recent years, liquid biopsy has become more and more important in the diagnosis and prognosis of cancer in clinical practice due to its convenience, non-invasiveness, high specificity and it overcomes temporal-spatial heterogeneity. Therefore, this review summarizes the current evidence on liquid biopsies in digestive tract cancers in relation to diagnosis and prognosis.
Core tip: Digestive tract cancers can be divided into several types. In this article, we reviewed the relevant research on liquid biopsies used for the diagnosis and prognosis of each type of cancer.
- Citation: Chen L, Chen Y, Feng YL, Zhu Y, Wang LQ, Hu S, Cheng P. Tumor circulome in the liquid biopsies for digestive tract cancer diagnosis and prognosis. World J Clin Cases 2020; 8(11): 2066-2080
- URL: https://www.wjgnet.com/2307-8960/full/v8/i11/2066.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v8.i11.2066
Digestive tract cancers are those that occur in the esophagus, stomach, intestine, and liver. Approximatively 180000 new esophageal, gastric, and colorectal cancer (CRC) cases and 77000 deaths are predicted in the United States. At present[1], the early diagnosis of digestive tract tumors mainly depends on serology, imaging, endoscopy, etc. Endoscopy is the main diagnostic method. However, due to the medical staff operating technology and personal ability in recognizing different lesions, painful endoscopic examination, and low acceptance by patients, it is necessary to identify new tumor markers to improve the early diagnosis and prognosis of digestive tract tumors[2].
The occurrence of malignant tumors is mainly due to the regulation of cell proliferation and differentiation[3]. Tissue specimens are the gold standard for cancer diagnosis, as well as determining prognosis and monitoring treatment[4]. Tumor molecular expression profiles obtained from tissue samples can only provide a "snapshot" of tumor heterogeneity, and the sub-spectrum often changes as the disease progresses[5]. In the era of precise tumor treatment, limited tissue specimens do not reflect all tumor genome changes, and the opportunity for treatment may be missed. The "liquid biopsy" may make up for these shortcomings[6].
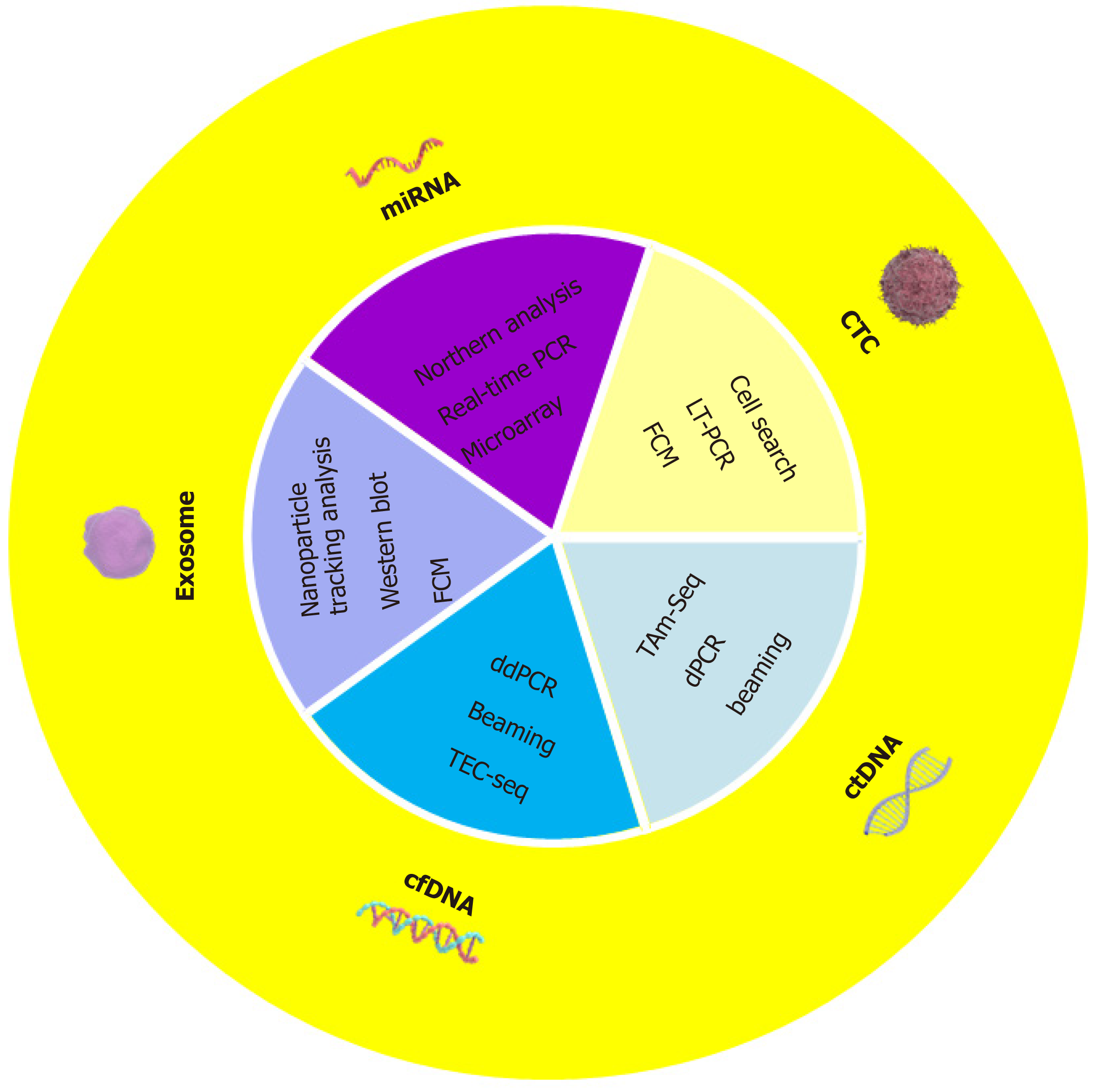
Liquid biopsy technology involves molecular in vitro diagnostics and is the representative diagnostic technology of precision medicine[7]. In recent years, the application of liquid biopsy technology in the early diagnosis, prognosis and management of tumors has increased[8]. In terms of tumor screening, traditional invasive screening methods have not met the necessary requirements[9]. Liquid biopsy is a non-invasive testing method and is used to detect tumors or metastatic foci (Figure 1). Liquid biopsy can be divided into blood-based biomarkers and other humoral biomarkers including circulating free DNA (cfDNA), circulating tumor DNA (ctDNA), circulating tumor cells (CTCs), microRNAs (miRNAs), and exosomes[10]. Other body fluid-derived substances can also be used as biomarkers in tests, such as urine, saliva, cerebrospinal fluid, pleural effusion, ascites, etc. Liquid biopsy technology has become a non-invasive, comprehensive, real-time, accurate and has other advantages in the field of tumor monitoring[9].
In 1948, Mandel[11] first discovered circulating nucleic acids in the blood of healthy humans. In 1977, Leon determined that the average concentration of cfDNA in the plasma of healthy humans was 13 ± 3 g/L and the average concentration was 180 ± 38 g/L[12]. Although the data obtained by different research methods later differed, all data suggested that the cfDNA content in the blood of cancer patients was significantly higher than that of healthy people[13]. At present, the source and production mechanisms of cfDNA are still not clear. Apoptosis, cell necrosis and active secretion are the more popular hypotheses[14]. Suzuki et al[15] found that the length of cfDNA fragments in healthy human blood was mostly 180 bp, which is similar to the length of DNA fragments produced by apoptosis, and the length of cfDNA fragments produced by cell necrosis was greater than 10000 bp. It is speculated that the cfDNA in healthy human blood is mainly from apoptosis. Considering that the integrity of DNA fragments in the circulation of patients with malignant tumors is significantly higher than that in healthy people, some scholars have proposed that necrolysis of tumor cells is the main reason for the significant increase in cfDNA in cancer patients, but this still does not explain the decline in cfDNA levels after radiotherapy[16].
ctDNA is the tumor information code in the blood. Tumor cells in patients' lesions are continuously transferred, and undergo apoptosis and necrosis in the circulatory system[17]. It is released into the blood, and carries biological information such as tumor generation, development, metastasis, and recurrence, which is of great value in the diagnosis, treatment and prognosis evaluation of tumors[18].
ctDNA is mainly released into the blood through a combination of three mechanisms: (1) Apoptosis; (2) Cell necrosis; and (3) Exosomes secreted by tumor cells[19]. In healthy individuals, infiltrating phagocytic cells are responsible for removing apoptotic and necrotic cell debris, including cfDNA[20]. Compared to cfDNA, the content of ctDNA in the blood is quite low, accounting for only 1% or less than 1% of cfDNA, and the length of most ctDNA fragments ranges from 160 to 200 bp, which is shorter than cfDNA without mutations[13]. Some studies have found that ctDNA entering the blood circulation has a half-life of approximately 15 min to 2.5 h, compared with protein markers that may take several weeks[21]. This feature ensures that ctDNA can be used as a "real-time" tumor biomarker, which can accurately reflect tumor burden.
ctDNA detection is a hotspot in the field of liquid biopsy. The detection of ctDNA can not only reveal the comprehensive genetic information of tumors, but also more accurately reflect the heterogeneity of tumor tissues and the dynamic evolution ability of tumor genomes, which has extremely high scientific value and clinical application prospects[22]. However, the content of ctDNA in peripheral blood is very small, and the content and nucleic acid polymorphism in different patients are very different[21]. Therefore, the analysis and detection of ctDNA have great technical challenges. At present, commonly used ctDNA detection methods include those based on digital polymerase chain reaction (PCR) and next-generation sequencing (NGS)[23]. Digital PCR technology has high specificity and sensitivity, usually for known mutant ctDNA, and NGS technology can achieve high-throughput parallel deep sequencing of multiple gene nucleic acid fragments, which can simultaneously detect multiple genes, different forms and unknown mutations[24].
The concept of CTCs was first introduced by the Australian scholar Ashworth[25] in 1969. It refers to tumor cells that enter the peripheral blood circulation from the primary tumor or metastasis of the solid tumor due to spontaneous or diagnostic procedures[26]. Most tumor cells that invade the circulatory system usually die within a short period of time, but a small number of tumor cells with high viability and high metastatic potential survive. They then aggregate with each other to form tiny tumor plugs and form microscopic tumors under certain conditions[27]. Therefore, the detection of CTCs in the peripheral blood circulation indicates that the tumor may have metastasized[28]. CTCs in the blood of patients include not only epithelial tumor cells but also tumor cells and tumor stem cells that undergo epithelial-mesenchymal transition[29]. Although acquiring the phenotype of some mesenchymal cells, malignant tumor cells will lose the phenotype of some epithelial cells, including morphology, surface antigens, and gene expression, etc.
As 107 blood cells contain only 1 CTC, the detection of CTCs in peripheral blood must first involve their enrichment (isolated and purified)[30]. This can be divided into a positive enrichment method and a negative enrichment method based on whether the tumor cells in the blood sample are enriched by techniques such as immunomagnetic beads before detection[31]. Common detection methods include immunocytochemistry, flow cytometry, PCR, and immunomagnetic bead separation[32].
A miRNA is a type of endogenous non-coding small single-stranded RNA consisting of 18 to 24 nucleotides, which is widely found in plants, animals, protozoa, and viruses[33]. miRNAs can bind to gene promoter regions to modify histones and regulate gene expression at the transcriptional level. miRNAs are involved in regulating the physiological processes of a variety of cells, including differentiation, proliferation, and apoptosis, by the above-mentioned mechanisms of action, and are widely considered to be key regulators of gene expression[34]. The change in miRNA expression level is closely related to the occurrence and development of human cancer. It can be used as an oncogene or tumor suppressor to target and control the expression of signal pathway genes, which affects the growth, proliferation, migration, and invasion of various tumor cells[35].
Exosomes are round extracellular vesicles with a lipid bilayer membrane, 30-150 nm in diameter[36]. In 1987, Johnstone et al[37] first discovered that a small membrane vesicle is released during the maturation of reticulocytes. Such vesicles can transfer transferrin receptors between cells and these vesicles were named "exosomes". Unlike microvesicles (100-1000 nm in diameter) that are shed directly from the cell membrane, and are derived from intracellular multivesicular bodies[38]. Early exosomes are also called early endosomes. They are formed by the inward budding of the cytoplasmic membrane, and then the inner body membrane further buds to form various intraluminal vesicles (multivesicular bodies), forming the endosomes of the pupa phase. This process is mainly composed of the endosomal sorting complex (ESCRT) which is initiated and driven, resulting in the production of polysomes[39]; the resulting polysomes can be transported to the lysosome for degradation or can be fused with the plasma membrane of the cell to be released into the extracellular environment. The released intraluminal vesicles are exosomes. The ESCRT contains a variety of proteins, including ESCRT series proteins (ESCRT-0, ESCRT-I, ESCRT-II and ESCRT-III), auxiliary proteins, and interaction protein X, vacuolar protein sorting 4 and vesicle transporter protein[40]; Exosomes secreted by donor cells can be bound and taken up by recipient cells in different ways, including fusion with cell membranes, endocytosis, receptor ligand-mediated recognition and internalization; exosomes are taken up by recipient cells. After that, the substances contained in it can be released into the cytoplasm of the recipient cells and participate in regulating the biological activities of the recipient cells[41].
Hsieh et al[42] found that plasma cfDNA levels in patients with esophageal squamous cell carcinoma were lower than those in normal individuals and that plasma cfDNA levels in patients with esophageal squamous cell carcinoma were not related to tumor location, stage, cell differentiation, lymph node metastasis or the presence of lymph and vascular invasion. However, high levels of plasma cfDNA can lead to shorter disease-free survival (DFS)[43]. Studies have shown that plasma cfDNA levels in patients with esophageal squamous cell carcinoma were not associated with gender, age, family history of tumor, tumor size, tumor stage, or tumor volume[43,44].
CTCs are positive in the blood of patients with esophageal cancer, which may be an indication of early distant metastasis of esophageal cancer. Matsushita's research[45] suggests that the CTC count may be related to the clinical stage of esophageal cancer, but the investigators did not find a statistical correlation between the tumor stage and lymph node metastasis. Therefore, in view of the differences in the above research conclusions, whether the detection of CTCs in the peripheral blood of patients with esophageal cancer can be used as indicators of the clinical stage in esophageal cancer patients still requires further investigation. Some studies found that 2-year progression-free survival was lower than that in patients who were negative for CTCs[46,47].
There are reports in the literature that when early imaging does not find obvious abnormalities, some esophageal cancer cells or fragments of DNA will enter the blood circulation and form ctDNA[48]. These substances can be detected at a very early stage due to the ongoing developments in NGS techniques. For tumors, there are also reports in the literature that the detected ctDNA is used as a clinical-stage for esophageal cancer, and has a significant correlation with the classic tumor staging system (TNM)[49,50].
In recent years, a large number of studies have been conducted on normal esophageal tissues and esophageal cancer tissues, and it was found that the expression of many types of mRNA is significantly different between esophageal cancer tissues and normal tissues[51]. Feber et al[51] demonstrated that the expression of miR-21, miR-93, miR-192, and miR-194 in esophageal squamous cell carcinoma cells was higher than that in normal esophageal epithelial cells using miRNA expression profiling chips. In 2011, Matsushima et al[52] confirmed that miR-205 expression was higher in ESCC cell lines than in normal esophageal squamous epithelium. Li et al[53] found that miR-21 expression was significantly up-regulated in esophageal cancer tissues and cell lines. It was also found that miR-16, miR-25, miR-29c, miR-155, miR-22, and miR-315 are highly expressed in esophageal cancer. In contrast, a large number of down-regulated mRNAs have been found in esophageal cancer, such as miR-133a, miR-138, miR-143, miR195, miR-200b, miR-203, miR-429, miR-375, and miR-625[54].
Lin et al[55] used chip technology to analyze the expression profile of miRNA in esophageal squamous cell carcinoma and found that the high expression of has-miR-103 and has-miR-107 in esophageal squamous cell carcinoma is a potential poor prognostic factor for esophageal squamous cell carcinoma. Nguyen et al[56] analyzed the expression profiles of 10 miRNAs in 10 esophageal cancer cell lines and 158 tissue samples and found that the expression of miR-16-2 and miR-30 was related to a shorter overall survival and DFS rate in esophageal cancer patients. Guo et al[57] analyzed the relationship between the expression of four miRNAs (hsa-miR-31, hsa-miR-1423p, hsa-miR3383p, and hsa-miR-261) and the clinicopathological characteristics and prognosis of 91 patients with esophageal squamous cell carcinoma, and found that the expression of miR-31 and miR-1423p was related to the degree of tissue differentiation of esophageal squamous cell carcinoma (P < 0.05). The prognosis of esophageal squamous cell carcinoma patients with high expression of miR-1423 is poor and is an independent prognostic factor. Hierarchical analysis showed that in esophageal squamous cell carcinoma patients with a good prognosis (smaller tumor size, negative lymph node metastasis, or earlier stage), high expression of miR-1423 also indicated a poor prognosis.
The meta-analysis conducted by Gao et al[58] on mixed-stage gastric cancer (GC) showed that ctDNA detection had obvious advantages in the specificity of GC diagnosis. Studies have found that cfDNA in the circulating blood of tumor patients is usually higher than that in healthy individuals, and the biological characteristics of cfDNA in these patients are the same as those in tumor tissues, which indicate that ctDNA is the main source of cfDNA in tumor patients. Kim et al[59] confirmed that the serum cDNA level in GC patients was higher than that in the healthy control group, and the sensitivity for the diagnosis of GC was 96.67%, and the specificity was 94.11%. These studies suggest that ctDNA can be used as a tumor marker for the diagnosis of GC. Newman et al[60] also confirmed this finding in a study of non-small cell lung cancer. They presented a fundamental new method named CAPP-Seq to detect non-small cell lung cancer ctDNA mutants with a specificity of 96%. It is worth noting that the sensitivity of current ctDNA tests for early cancers is about 30% to 40%[61]. Therefore, most early-stage cancers will not be detected by current ctDNA analysis methods.
Liu et al[62] first identified the expression profile of miRNAs in the serum of GC patients using microarray, and subsequently found that miR-378 expression levels in the serum of GC patients were increased (P < 0.01) compared with 41 healthy controls using quantitative real-time PCR. The area under the curve (AUC) was 0.861, and its sensitivity and specificity were 87.5% and 70.73%, respectively. It is worth noting that miR-378 showed a significant increase in the serum of patients with early GC (P < 0.05), indicating that it has potential value as a new noninvasive biomarker for screening early GC[63]. Song et al[63] also verified the serum miRNA expression profile in GC patients and further confirmed that the combined detection of three serum miRNAs (including miR-221, miR-744, and miR-376c) can diagnose GC with a sensitivity of 82.4% and specificity of 58.8%. A subgroup analysis showed that the combined detection of these three miRNAs could correctly diagnose GC in 22 of 30 early GC patients, and the early diagnosis accuracy rate was 73.3%, suggesting that these three miRNAs can be used as markers for screening early GC. Studies such as those by Cai et al[64] have shown that the AUC of miR-106b, miR-20a, and miR-221 were all greater than 0.75. In addition, these three miRNAs were significantly increased in the four TNM stages of GC (P < 0.05), suggesting that these three miRNAs can be used for screening of early GC. Li et al[65] confirmed that the expression of miR-199a-3p in the plasma of patients with early GC was significantly higher than that in healthy controls (P < 0.01) and patients with precancerous gastric lesions (P < 0.01), and that miR-199a-3 predicted an AUC > 0.8 for early GC, which was significantly higher than CEA, CA72-4, CA19-9, and CA12-5 detection. The diagnostic sensitivity, specificity, and accuracy of miR-199a-3 were greater than 70%, indicating that it is expected to be an ideal diagnostic and screening marker of early GC. Zhu et al[66] detected the expression of miRNAs in the plasma of 40 patients with early gastric non-cardiac adenocarcinoma (Ia/Ib) and 40 healthy controls using a TLDA chip, and screened out 5 miRNAs with elevated expression in the plasma of GC patients. The five miRNAs (miR-16, miR-25, miR-92a, miR-451, and miR-486-5p), had an AUC greater than 0.69 in the diagnosis of early gastric non-cardiac adenocarcinoma; and the combined detection of these five miRNAs was better than a single miRNA, with an AUC of 0.89 detected with the combination in the test phase and the validation phase, which distinguished early gastric non-cardiac adenocarcinoma patients from healthy controls. The above results show that although a specific miRNA in plasma can help distinguish different subpopulations, the combined detection of a set of miRNAs may have higher diagnostic potential.
CTCs may provide the ability to reproducibly and non-invasively detect treatment effect in cancer patients and identify potential lesions not found by imaging, which can aid early diagnosis and individualized treatment, thereby improving the survival rate of GC patients. Shen et al[67] used the receiver operating characteristic curve to determine the optimal threshold level of CTCs for distinguishing GC patients from healthy control groups, and defined the threshold as ≥ 2 CTCs per 7.5 mL of blood, with sensitivity and specificity values of 85.3% and 90.3%, respectively. Some studies[68,69] have found that the positive rate of CTCs in the peripheral blood of patients with GC is significantly higher than that in patients with benign gastric disease. Tang et al[70] conducted a meta-analysis to assess the overall accuracy of the diagnosis of GC using the detection of CTCs. The specificity of the test for GC diagnosis was 99%. Therefore, the outstanding feature of CTCs used for GC diagnosis is that they have better specificity, which may be valuable in the diagnosis and confirmation of GC. These results indicate that CTCs may serve as a biomarker for the early diagnosis of GC.
In studies of GC, the detection of CTCs indicated that patients with GC have shorter progression-free survival and overall survival[71]. Uenosono et al[72] used the cell search system to detect CTCs in 251 GC patients, and the results showed that the relapse-free survival rate and 5-year survival rate of CTCs-positive patients were significantly lower than those of CTCs-negative patients. Wang et al[73] found that GC patients who were positive for CTCs had a higher recurrence rate than GC patients who were negative for CTCs, and their 3-year DFS was shorter. A meta-analysis[74] showed that CTCs are more common in late GC than early GC, and are more common in poorly differentiated GCs than in well-differentiated or moderately differentiated GCs, and are significantly associated with patient progression-free survival and overall survival. These studies suggest that CTCs can be used as an effective indicator in monitoring the recurrence or prognosis of GC patients. If recurrence can be predicted, then information on CTCs could help patients avoid unnecessary adjuvant chemotherapy. Some studies have suggested the predictive value of CTCs detection during GC chemotherapy. Li et al[75] monitored the dynamic changes in CTCs during the treatment of 15 patients with GC. The results showed that patients with persistently low levels of CTCs or patients who transitioned to low levels of CTCs early after treatment had a better prognosis. Patients with persistently higher levels of CTCs or those who transitioned to higher levels of CTCs after treatment had a worse prognosis, which means that the effect of the treatment being administered was limited. A meta-analysis[76] concluded that the status of CTCs has significant prognostic value for patients with GC treated with chemotherapy alone. Therefore, the evaluation of CTCs may be an effective strategy for predicting tumor prognosis and monitoring the curative effect in patients with GC.
In recent years, with the development of exosome extraction technology and the in-depth study of exosomes, research on the relationship between exosomes and GC has been continuously reported. In the early diagnosis of GC, Lin et al[77] found that tumor-derived exosomes IncUEGC1 could be used as a circulating biomarker for early GC. During GC development, Li et al[78] found that the overexpression of miR-217 in GC-derived exosomes promoted GC cell proliferation. Zheng et al[79] found that exosomes-mediated functional ApoE protein promoted metastasis of GC cells. In terms of prognosis, Liu et al[80] found that exosomes miR-451 derived from GC cells can be used as an indicator of poor prognosis.
ctDNA methylation is strongly associated with early CRC tumors. In a large-scale, multi-center, prospective CRC screening test[81] more than 7900 patients who underwent regular colonoscopy were recruited, of which 53 patients were found to have CRC and the SEP9 gene in plasma ctDNA. The sensitivity of abnormal methylation as a diagnostic screening test in asymptomatic CRC patients was 48.2%, and the specificity was 91.5%. Recent studies[82,83] have confirmed that the mutation frequency of ctDNA is correlated with the prognosis of CRC. Lecomte et al[83] analyzed the survival of 37 patients with CRC (average follow-up period of 22 mo). The results showed that the overall annual survival rate of patients with KRAS mutation and CDKN2A promoter hypermethylation was only 48%, while patients without these genetic mutations had an annual overall survival rate which reached 100%.
Reinert et al[84] developed a personalized detection method based on ctDNA and evaluated and quantified the levels of ctDNA and somatic cell variation in the plasma of 6 patients with recurrent CRC and 5 patients with non-recurrent CRC using digital PCR technology. Compared with traditional follow-up methods, ctDNA can detect tumor recurrence 10 months in advance, and its sensitivity and specificity are 100%. Therefore, for non-metastatic patients after radical surgery, ctDNA can capture and monitor small residual lesions before imaging and clinical confirmation of recurrence, and predict tumor recurrence early through genomic changes.
At present, there are more studies on the correlation between CTCs and the prognosis of CRC patients than in other gastrointestinal tumors[85,86]. CTCs can predict the long-term survival of CRC patients. A significant increase in the number of high CTCs or a continuous increase after surgery may indicate a poor prognosis for CRC patients[85]. CTC concentration at 7 d is an independent prognostic factor for local metastasis[86]. In addition, a meta-analysis[86] showed that baseline CTC counts are independent prognostic factors related to progression-free survival and overall survival in patients with CRC.
Hiraiwa et al[87] examined CTCs in 130 patients with gastrointestinal tumors and 41 healthy volunteers and found that the number of CTCs in the peripheral blood of patients with metastatic tumors was significantly higher than the number of CTCs in non-metastatic patients and healthy volunteers. Maestro et al[88] tested peripheral blood CTCs in 438 patients with breast cancer, 195 cases of rectal fever, and 50 patients with previous adenocarcinomas. The rates of tumor metastasis in these three groups of patients were 43.8%, 15.9%, and 48%, respectively. 63.3% of patients with metastasis had more than 2 CTCs, and only 14.0% of patients with tumors who did not metastasize had more than 2 CTCs, and no healthy volunteer exceeded this level. Dalum et al[89] studied 183 newly diagnosed patients with non-diffusive metastatic CRC before surgery. It was found that 44 patients (24%) had CTCs greater than 1 CTC/30 m in 4 preoperative tests. These patients had significantly lower metastasis-free and recurrence-free survival after surgery than those without CTCs. CTCs in the peripheral blood of patients were not related to recurrence-free survival within a few weeks after surgery, but were related to recurrence-free survival 2-3 years after surgery. This evidence shows that CTCs are closely related to the metastasis of CRC.
The expression of different miRNAs is helpful in the prognosis of CRC. A study of CRC patients treated according to the International Union Against Cancer standards found that the expression levels of miR-152 and miR-148 were correlated with the prognosis of CRC patients[90]. Agostini et al[91] compared the miRNA expression profiles of two groups of stage II colon cancer and adjacent normal tissues, and found that there were 37 miRNA expression differences, of which miR-20, miR-21, miR-106, miR-181, and miR-203 may be related to prognosis, and high expression in tumor tissue decreased the survival rate of patients. Another study[92] showed that patients with stage II colon cancer and high levels of miR-320 and miR-498 have a shorter relapse-free survival than those with low expression.
In addition, many studies have shown that abnormal miRNA expression is related to lymph node metastasis[93]. For example, miR-129 and miR-137 are different in colon cancer patients with and without lymph node metastasis[93]. Weissmann-Brenner et al[94] conducted a study of 110 CRC patients with relapse and no recurrence and found that in stage II CRC, the expression of 903 miRNAs was detected in CRC tissues, regardless of recurrence. There was no difference between the relapse group and the non-relapse group; and in the CRC development period, those with down-regulated expression of miRNA-29a in CRC had a better prognosis and longer survival time. The above studies show that the expression of various miRNAs in CRC tissues is related to tumor growth, lymph node and organ metastasis, and patient survival. Thus, miRNAs are expected to be prognostic markers of CRC.
Li et al[95] found that cirRNA was included in the exosomes of patients with colon cancer and that the expression of exosome cirRNA in serum was different to that in the normal population, providing a new marker for the diagnosis of colon cancer. The continuous advancement of exosome analysis and detection technology provides a theoretical and practical basis for the detection, separation and purification of exosomes. The Japanese experimental team used a highly sensitive analytical technique, Exo Screen, to determine the surface protein of extracellular vesicles[96]. CD147 is an antigen on the surface of colon cancer cell lines. Studies have confirmed that the detection of CD147 in extracellular vesicles can be used for the diagnosis of colon cancer. CD147, as a biomarker for colon cancer diagnosis, has better effects than the traditional diagnostic markers CEA and CA19-9 for colon cancer[97].
The level of miR-17-92a in serum exosomes is closely related to the recurrence of CRC[98]. Compared with the normal population, the level of miR-17-92a in the serum of patients with CRC is significantly increased. Matsumura et al[98] divided 209 colon cancer patients into a high-expression group (n = 133) and low-expression group (n = 76) based on serum exosomal miR-17-92a levels. The survival rate of patients in the high-expression group was significantly lower than that in the low-expression group (P = 0.004). In addition, an analysis of 146 patients with colon cancer, which did not include stage IV, found that the DFS of the high-expression group was significantly lower than that of the low-expression group (P = 0.0002). Factor analysis showed that miR-17-92a levels in serum exosomes can be used as independent factors for prognosis. Soldevilla et al[99] found that plasma exosome DNp73 levels in patients with colon cancer were significantly higher than those in the normal population. Moreover, the DNp73 high-level group had a higher stage and shorter DFS. In addition, the serum CEA level of patients in the DNp73 high expression group was significantly increased (P = 0.08). The level of DNp73 in exosomes of patients with colon cancer may be related to the survival of patients and may become an index for judging prognosis.
During the occurrence and development of liver cancer, the content of cfDNA is closely related to the development and prognosis of the tumor. The content of cfDNA is usually higher in patients with hepatocellular carcinoma (HCC) or large liver cancer, but lower in patients with early or small liver cancer[100]. Patients with higher cfDNA content have a worse prognosis and are more likely to develop recurrence and metastasis[101]. Therefore, the content of cfDNA in the blood of liver cancer patients can be used for the diagnosis and prognosis of liver cancer, but this method only calculates the overall cfDNA concentration and does not distinguish the genetic changes of ctDNA[102]. The sensitivity and specificity are not high. By detecting cancer-specific gene mutations or epigenetic changes in the blood of liver cancer patients, this can provide a more accurate marker for the diagnosis of liver cancer. For example, as a classic tumor suppressor gene, the P53 gene has few mutation hot spots and high stability, and the serine mutation at position 249 of this gene often occurs in the liver caused by the chemical carcinogen aflatoxin or hepatitis B virus infection. It can be detected early in the onset of liver cancer[103]. Therefore, the ctDNA of serine 249 in the P53 gene can be used as a marker for early diagnosis of liver cancer[104]. In addition, the detection of epigenetic changes of ctDNA in peripheral blood can be used as another means for early diagnosis of liver cancer. For example, detection of the methylation of RASSE1A, p15, and p16 in ctDNA can be used as an effective biomarker for the diagnosis and prognosis of liver cancer[104]. The diagnostic sensitivity can reach 84% and the specificity can reach 94%, which can diagnose liver cancer earlier than current detection methods[105].
In the early stages of CTC research, the early diagnosis of liver cancer is mainly carried out by assessing the number of CTCs[106]. A significant positive correlation was observed between the number of CTCs and the standard Barcelona (Barcelona Clinic Liver Cancer) stage and serum alpha-fetoprotein (AFP) levels[107]. Not only is the number of CTCs related to staging, but different CTC phenotypes (such as epithelial phenotype, mesenchymal phenotype, hybrid phenotype, etc.) are also correlated with staging[108]. The accuracy of CTCs in the early diagnosis of liver cancer has been further improved. For example, combining CTCs with genomics, researchers used a microfluidic chip to directly filter out red blood cells, platelets, plasma and other components in the blood, while retaining active CTCs[109]. By combining with PCR technology, early liver cancer and CTCs were found in peripheral blood from AFP-negative patients[110]. In addition, the combination of CTC detection and other biomarker detection methods to form a multi-marker model has improved the sensitivity and specificity of diagnosis to more than 90%[111]. Methods using different mathematical models, such as logistic regression and classification and regression tree modeling analysis combined with CTC test results under different etiologies are also highly stable and accurate in the diagnosis of early liver cancer.
Studies have shown that HCC patients with higher peripheral blood CTC counts have a worse prognosis, higher risk of recurrence, and lower DFS and overall survival[112]. In addition, the change in CTC count in peripheral blood after treatment can reflect the treatment effect. The decrease or disappearance of CTC count in peripheral blood after treatment (including surgery, intervention, and targeted drugs) indicates that the treatment effect is positive[111]. Patients with no change in CTC count and an increase after a decline often indicate poor treatment outcomes and tumor recurrence or resistance. Ye et al[113] investigated the relationship between the CTC count and the clinical outcome of patients with hepatitis B virus-related HCC after radical resection and found that patients with lower CTC counts (≤ 2/5 mL) and patients with higher counts (> 2/5 mL) were similar. Both, DFS and OS were significantly prolonged, suggesting that higher CTC counts may be an independent marker of poor prognosis in HCC. Hao et al[114] conducted further research, and the results showed that patients with anterior approach hepatectomy had significantly lower CTC counts in peripheral blood than patients with traditional approach hepatectomy, and the 2-year recurrence rate and 2-year survival rate were also significantly reduced. A significant reduction suggested that anterior approach hepatectomy significantly reduced the postoperative CTC count compared with traditional hepatectomy and significantly improved the patient's prognosis. CTCs have shown great advantages in related studies on HCC prognosis and have benefited some patients[115].
Early HCC is difficult to diagnose, and serum miR-21 and miR-122 can distinguish HCC patients from healthy controls. The AUCs of miR-21 and miR-122 are 0.88 and 0.77, respectively; the sensitivity and specificity of miR-21 are 86.6% and 79.5 %, the sensitivity and specificity of miR-122 are 68.0% and 73.3%[116]. Serum miR-106b level has a certain clinical value for HCC diagnosis and disease assessment[117]. The serum level of miR-106b in patients with advanced HCC (stage III/IV) is higher than that in the early stage[118]. MiR-92-3p, miR-107 and miR-3126-5p are valuable markers for the diagnosis of HCC, especially for patients with early-stage HCC (stage I/II) (AUC = 0.975) and patients with low levels of AFP (AUC = 0.971). The combined application of miR-92-3p, miR-107, and miR-31265p with AFP is more effective in distinguishing patients with early HCC (AUC = 0.988) and patients with low-level AFP (AUC = 0.989)[119].
Exosomal-derived miRNA can be used not only as a marker of early screening and early diagnosis but also as a marker of tumor prognosis[120]. miR-221 is also highly expressed in liver cancer tissues and is related to the size of liver cancer and the prognosis of patients[121]. Other studies have reported that in liver cancer, miR-718 is inhibited in peripheral blood exosomes by regulating the expression of the target gene HOXB8. The tumor cells were differentiated, and the expression of serum exosomes miR-718 after liver transplantation was negatively correlated with tumor recurrence[122].
In summary, liquid biopsy technology has been widely studied and applied in CRC and other tumor types, and can provide more accurate treatment measures for tumor patients and enable patients to obtain the best clinical benefits. Despite this, liquid biopsy still faces many challenges and is still in the clinical trial stage. Large-scale prospective clinical trials are needed for further verification. It is believed that liquid biopsy can bring hope to patients in terms of more accurate diagnosis and prognosis.
Manuscript source: Invited manuscript
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cidon EU S-Editor: Yan JP L-Editor: Webster JR E-Editor: Xing YX
| 1. | Lopez A, Harada K, Mizrak Kaya D, Dong X, Song S, Ajani JA. Liquid biopsies in gastrointestinal malignancies: when is the big day? Expert Rev Anticancer Ther. 2018;18:19-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 2. | De Rubis G, Rajeev Krishnan S, Bebawy M. Liquid Biopsies in Cancer Diagnosis, Monitoring, and Prognosis. Trends Pharmacol Sci. 2019;40:172-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 381] [Article Influence: 63.5] [Reference Citation Analysis (0)] |
| 3. | Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, Ponzetti A, Cremolini C, Amatu A, Lauricella C, Lamba S, Hobor S, Avallone A, Valtorta E, Rospo G, Medico E, Motta V, Antoniotti C, Tatangelo F, Bellosillo B, Veronese S, Budillon A, Montagut C, Racca P, Marsoni S, Falcone A, Corcoran RB, Di Nicolantonio F, Loupakis F, Siena S, Sartore-Bianchi A, Bardelli A. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21:795-801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 572] [Cited by in RCA: 674] [Article Influence: 67.4] [Reference Citation Analysis (0)] |
| 4. | Schøler LV, Reinert T, Ørntoft MW, Kassentoft CG, Árnadóttir SS, Vang S, Nordentoft I, Knudsen M, Lamy P, Andreasen D, Mortensen FV, Knudsen AR, Stribolt K, Sivesgaard K, Mouritzen P, Nielsen HJ, Laurberg S, Ørntoft TF, Andersen CL. Clinical Implications of Monitoring Circulating Tumor DNA in Patients with Colorectal Cancer. Clin Cancer Res. 2017;23:5437-5445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 237] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 5. | Diaz LA, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA, Kinzler KW, Oliner KS, Vogelstein B. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537-540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1259] [Cited by in RCA: 1352] [Article Influence: 104.0] [Reference Citation Analysis (0)] |
| 6. | Krebs MG, Metcalf RL, Carter L, Brady G, Blackhall FH, Dive C. Molecular analysis of circulating tumour cells-biology and biomarkers. Nat Rev Clin Oncol. 2014;11:129-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 483] [Cited by in RCA: 482] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 7. | Wurst T. Liquid Biopsies. Genet Test Mol Biomarkers. 2018;22:207-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 8. | Yamada T, Matsuda A, Koizumi M, Shinji S, Takahashi G, Iwai T, Takeda K, Ueda K, Yokoyama Y, Hara K, Hotta M, Matsumoto S, Yoshida H. Liquid Biopsy for the Management of Patients with Colorectal Cancer. Digestion. 2019;99:39-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 9. | Zhang W, Xia W, Lv Z, Ni C, Xin Y, Yang L. Liquid Biopsy for Cancer: Circulating Tumor Cells, Circulating Free DNA or Exosomes? Cell Physiol Biochem. 2017;41:755-768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 166] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 10. | Mann J, Reeves HL, Feldstein AE. Liquid biopsy for liver diseases. Gut. 2018;67:2204-2212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 78] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 11. | MANDEL P, METAIS P. Les acides nucléiques du plasma sanguin chez l'homme. C R Seances Soc Biol Fil. 1948;142:241-243. [PubMed] |
| 12. | Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646-650. [PubMed] |
| 13. | Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1954] [Cited by in RCA: 2143] [Article Influence: 153.1] [Reference Citation Analysis (0)] |
| 14. | Corcoran RB, Chabner BA. Application of Cell-free DNA Analysis to Cancer Treatment. N Engl J Med. 2018;379:1754-1765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 481] [Cited by in RCA: 670] [Article Influence: 95.7] [Reference Citation Analysis (0)] |
| 15. | Suzuki N, Kamataki A, Yamaki J, Homma Y. Characterization of circulating DNA in healthy human plasma. Clin Chim Acta. 2008;387:55-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 135] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 16. | Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, Azad NS, Bardelli A, Brem H, Cameron JL, Lee CC, Fecher LA, Gallia GL, Gibbs P, Le D, Giuntoli RL, Goggins M, Hogarty MD, Holdhoff M, Hong SM, Jiao Y, Juhl HH, Kim JJ, Siravegna G, Laheru DA, Lauricella C, Lim M, Lipson EJ, Marie SK, Netto GJ, Oliner KS, Olivi A, Olsson L, Riggins GJ, Sartore-Bianchi A, Schmidt K, Shih lM, Oba-Shinjo SM, Siena S, Theodorescu D, Tie J, Harkins TT, Veronese S, Wang TL, Weingart JD, Wolfgang CL, Wood LD, Xing D, Hruban RH, Wu J, Allen PJ, Schmidt CM, Choti MA, Velculescu VE, Kinzler KW, Vogelstein B, Papadopoulos N, Diaz LA Jr. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2770] [Cited by in RCA: 3545] [Article Influence: 322.3] [Reference Citation Analysis (0)] |
| 17. | Yao Y, Liu J, Li L, Yuan Y, Nan K, Wu X, Zhang Z, Wu Y, Li X, Zhu J, Meng X, Wei L, Chen J, Jiang Z. Detection of circulating tumor DNA in patients with advanced non-small cell lung cancer. Oncotarget. 2017;8:2130-2140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 18. | Gormally E, Caboux E, Vineis P, Hainaut P. Circulating free DNA in plasma or serum as biomarker of carcinogenesis: practical aspects and biological significance. Mutat Res. 2007;635:105-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 315] [Cited by in RCA: 333] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 19. | Underhill HR, Kitzman JO, Hellwig S, Welker NC, Daza R, Baker DN, Gligorich KM, Rostomily RC, Bronner MP, Shendure J. Fragment Length of Circulating Tumor DNA. PLoS Genet. 2016;12:e1006162. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 374] [Cited by in RCA: 498] [Article Influence: 55.3] [Reference Citation Analysis (0)] |
| 20. | Batth IS, Mitra A, Manier S, Ghobrial IM, Menter D, Kopetz S, Li S. Circulating tumor markers: harmonizing the yin and yang of CTCs and ctDNA for precision medicine. Ann Oncol. 2017;28:468-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 21. | Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong AS, Marass F, Humphray S, Hadfield J, Bentley D, Chin TM, Brenton JD, Caldas C, Rosenfeld N. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1288] [Cited by in RCA: 1311] [Article Influence: 109.3] [Reference Citation Analysis (0)] |
| 22. | Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20:71-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 969] [Cited by in RCA: 925] [Article Influence: 154.2] [Reference Citation Analysis (0)] |
| 23. | Akbariqomi M, Heidari R, Kooshki H, Akbari Nakhjavani S, Sadat Lavasani P, Absalan M, Shafie S, Samadi N, Mortazavy M, Motaei J. Clinical Application of Circulating Tumor DNA in Diagnosis of Breast Cancer. Multidisciplinary Cancer Invest. 2017;Suppl 1. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 24. | Rothwell DG, Ayub M, Cook N, Thistlethwaite F, Carter L, Dean E, Smith N, Villa S, Dransfield J, Clipson A, White D, Nessa K, Ferdous S, Howell M, Gupta A, Kilerci B, Mohan S, Frese K, Gulati S, Miller C, Jordan A, Eaton H, Hickson N, O'Brien C, Graham D, Kelly C, Aruketty S, Metcalf R, Chiramel J, Tinsley N, Vickers AJ, Kurup R, Frost H, Stevenson J, Southam S, Landers D, Wallace A, Marais R, Hughes AM, Brady G, Dive C, Krebs MG. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat Med. 2019;25:738-743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 210] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 25. | Ashworth T. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust Med J. 1869;14:146-147. |
| 26. | Dittmar T, Heyder C, Gloria-Maercker E, Hatzmann W, Zänker KS. Adhesion molecules and chemokines: the navigation system for circulating tumor (stem) cells to metastasize in an organ-specific manner. Clin Exp Metastasis. 2008;25:11-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 56] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 27. | Chi KR. The tumour trail left in blood. Nature. 2016;532:269-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 72] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 28. | Torino F, Bonmassar E, Bonmassar L, De Vecchis L, Barnabei A, Zuppi C, Capoluongo E, Aquino A. Circulating tumor cells in colorectal cancer patients. Cancer Treat Rev. 2013;39:759-772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 29. | Hardingham JE, Grover P, Winter M, Hewett PJ, Price TJ, Thierry B. Detection and Clinical Significance of Circulating Tumor Cells in Colorectal Cancer--20 Years of Progress. Mol Med. 2015;21 Suppl 1:S25-S31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 97] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 30. | Tan SJ, Yobas L, Lee GY, Ong CN, Lim CT. Microdevice for the isolation and enumeration of cancer cells from blood. Biomed Microdevices. 2009;11:883-892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 257] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 31. | Alvarez Cubero MJ, Lorente JA, Robles-Fernandez I, Rodriguez-Martinez A, Puche JL, Serrano MJ. Circulating Tumor Cells: Markers and Methodologies for Enrichment and Detection. Methods Mol Biol. 2017;1634:283-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 32. | Masuda T, Hayashi N, Iguchi T, Ito S, Eguchi H, Mimori K. Clinical and biological significance of circulating tumor cells in cancer. Mol Oncol. 2016;10:408-417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 114] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 33. | Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172-3183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 517] [Cited by in RCA: 491] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 34. | Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143-1149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 741] [Cited by in RCA: 789] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 35. | Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2105] [Cited by in RCA: 2356] [Article Influence: 130.9] [Reference Citation Analysis (0)] |
| 36. | Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17-24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1443] [Cited by in RCA: 1556] [Article Influence: 155.6] [Reference Citation Analysis (0)] |
| 37. | Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. 1987;262:9412-9420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 38. | Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529-539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 503] [Cited by in RCA: 504] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 39. | van Niel G, Raposo G, Candalh C, Boussac M, Hershberg R, Cerf-Bensussan N, Heyman M. Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology. 2001;121:337-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 512] [Cited by in RCA: 554] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 40. | Chaput N, Taïeb J, Schartz NE, André F, Angevin E, Zitvogel L. Exosome-based immunotherapy. Cancer Immunol Immunother. 2004;53:234-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 99] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 41. | Yu X, Odenthal M, Fries JW. Exosomes as miRNA Carriers: Formation-Function-Future. Int J Mol Sci. 2016;17:2028. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 299] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 42. | Hsieh CC, Hsu HS, Chang SC, Chen YJ. Circulating Cell-Free DNA Levels Could Predict Oncological Outcomes of Patients Undergoing Esophagectomy for Esophageal Squamous Cell Carcinoma. Int J Mol Sci. 2016;17:2131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 43. | Nygaard AD, Holdgaard PC, Spindler KL, Pallisgaard N, Jakobsen A. The correlation between cell-free DNA and tumour burden was estimated by PET/CT in patients with advanced NSCLC. Br J Cancer. 2014;110:363-368. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 96] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 44. | Banki F, Mason RJ, Oh D, Hagen JA, DeMeester SR, Lipham JC, Tanaka K, Danenberg KD, Yacoub WN, Danenberg PV, DeMeester TR. Plasma DNA as a molecular marker for completeness of resection and recurrent disease in patients with esophageal cancer. Arch Surg. 2007;142:533-8; discussion 538-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 45. | Matsushita D, Uenosono Y, Arigami T, Yanagita S, Nishizono Y, Hagihara T, Hirata M, Haraguchi N, Arima H, Kijima Y, Kurahara H, Maemura K, Okumura H, Ishigami S, Natsugoe S. Clinical Significance of Circulating Tumor Cells in Peripheral Blood of Patients with Esophageal Squamous Cell Carcinoma. Ann Surg Oncol. 2015;22:3674-3680. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 46. | Riethdorf S, Fritsche H, Müller V, Rau T, Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Jänicke F, Jackson S, Gornet T, Cristofanilli M, Pantel K. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin Cancer Res. 2007;13:920-928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 996] [Cited by in RCA: 1029] [Article Influence: 57.2] [Reference Citation Analysis (0)] |
| 47. | Liu Z, Jiang M, Zhao J, Ju H. Circulating tumor cells in perioperative esophageal cancer patients: quantitative assay system and potential clinical utility. Clin Cancer Res. 2007;13:2992-2997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 52] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 48. | Creemers A, Krausz S, Strijker M, van der Wel MJ, Soer EC, Reinten RJ, Besselink MG, Wilmink JW, van de Vijver MJ, van Noesel CJM, Verheij J, Meijer SL, Dijk F, Bijlsma MF, van Oijen MGH, van Laarhoven HWM. Clinical value of ctDNA in upper-GI cancers: A systematic review and meta-analysis. Biochim Biophys Acta Rev Cancer. 2017;1868:394-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 49. | Azad TD, Chaudhuri A, Newman AM, Stehr H, Schroers-Martin J, Chabon JJ, Fang P, Qiao Y, Liao ZX, Komaki R. Circulating tumor DNA analysis for outcome prediction in localized esophageal cancer. J Clin Oncol. 2017;35:4055-4055. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 50. | Jia R, Li P, Zhao C, Wang Y, Liu R, Chang L, Guan Y, Yi X, Xu J-M. Postradiation ctDNA status as a prognostic factor in locally advanced esophageal squamous cell carcinoma. J Clin Oncol. 2018;36:e16053. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 51. | Feber A, Xi L, Luketich JD, Pennathur A, Landreneau RJ, Wu M, Swanson SJ, Godfrey TE, Litle VR. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135:255-260; discussion 260. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 298] [Cited by in RCA: 304] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 52. | Matsushima K, Isomoto H, Yamaguchi N, Inoue N, Machida H, Nakayama T, Hayashi T, Kunizaki M, Hidaka S, Nagayasu T, Nakashima M, Ujifuku K, Mitsutake N, Ohtsuru A, Yamashita S, Korpal M, Kang Y, Gregory PA, Goodall GJ, Kohno S, Nakao K. MiRNA-205 modulates cellular invasion and migration via regulating zinc finger E-box binding homeobox 2 expression in esophageal squamous cell carcinoma cells. J Transl Med. 2011;9:30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 117] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 53. | Li H, Zheng D, Zhang B, Liu L, Ou J, Chen W, Xiong S, Gu Y, Yang J. Mir-208 promotes cell proliferation by repressing SOX6 expression in human esophageal squamous cell carcinoma. J Transl Med. 2014;12:196. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 54. | Feber A, Xi L, Pennathur A, Gooding WE, Bandla S, Wu M, Luketich JD, Godfrey TE, Litle VR. MicroRNA prognostic signature for nodal metastases and survival in esophageal adenocarcinoma. Ann Thorac Surg. 2011;91:1523-1530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 70] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 55. | Lin RJ, Xiao DW, Liao LD, Chen T, Xie ZF, Huang WZ, Wang WS, Jiang TF, Wu BL, Li EM, Xu LY. MiR-142-3p as a potential prognostic biomarker for esophageal squamous cell carcinoma. J Surg Oncol. 2012;105:175-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 93] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 56. | Nguyen GH, Schetter AJ, Chou DB, Bowman ED, Zhao R, Hawkes JE, Mathé EA, Kumamoto K, Zhao Y, Budhu A, Hagiwara N, Wang XW, Miyashita M, Casson AG, Harris CC. Inflammatory and microRNA gene expression as prognostic classifier of Barrett's-associated esophageal adenocarcinoma. Clin Cancer Res. 2010;16:5824-5834. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 56] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 57. | Guo Y, Chen Z, Zhang L, Zhou F, Shi S, Feng X, Li B, Meng X, Ma X, Luo M, Shao K, Li N, Qiu B, Mitchelson K, Cheng J, He J. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res. 2008;68:26-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 268] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 58. | Gao Y, Zhang K, Xi H, Cai A, Wu X, Cui J, Li J, Qiao Z, Wei B, Chen L. Diagnostic and prognostic value of circulating tumor DNA in gastric cancer: a meta-analysis. Oncotarget. 2017;8:6330-6340. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 59. | Kim K, Shin DG, Park MK, Baik SH, Kim TH, Kim S, Lee S. Circulating cell-free DNA as a promising biomarker in patients with gastric cancer: diagnostic validity and significant reduction of cfDNA after surgical resection. Ann Surg Treat Res. 2014;86:136-142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 114] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 60. | Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE, Shrager JB, Loo BW, Alizadeh AA, Diehn M. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548-554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1341] [Cited by in RCA: 1642] [Article Influence: 149.3] [Reference Citation Analysis (0)] |
| 61. | Webb S. The cancer bloodhounds. Nat Biotechnol. 2016;34:1090-1094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 62. | Liu H, Zhu L, Liu B, Yang L, Meng X, Zhang W, Ma Y, Xiao H. Genome-wide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer. Cancer Lett. 2012;316:196-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 208] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 63. | Song MY, Pan KF, Su HJ, Zhang L, Ma JL, Li JY, Yuasa Y, Kang D, Kim YS, You WC. Identification of serum microRNAs as novel non-invasive biomarkers for early detection of gastric cancer. PLoS One. 2012;7:e33608. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 159] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 64. | Cai H, Yuan Y, Hao YF, Guo TK, Wei X, Zhang YM. Plasma microRNAs serve as novel potential biomarkers for early detection of gastric cancer. Med Oncol. 2013;30:452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 79] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 65. | Li C, Li JF, Cai Q, Qiu QQ, Yan M, Liu BY, Zhu ZG. MiRNA-199a-3p: A potential circulating diagnostic biomarker for early gastric cancer. J Surg Oncol. 2013;108:89-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 70] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 66. | Zhu C, Ren C, Han J, Ding Y, Du J, Dai N, Dai J, Ma H, Hu Z, Shen H, Xu Y, Jin G. A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br J Cancer. 2014;110:2291-2299. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 187] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 67. | Shen W, Song Y, Burklund A, Le B, Zhang R, Wang L, Xi Y, Qian K, Shen T, Zhang JXJ. Combined immunomagnetic capture coupled with ultrasensitive plasmonic detection of circulating tumor cells in blood. Biomed Microdevices. 2018;20:99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 68. | Kang HM, Kim GH, Jeon HK, Kim DH, Jeon TY, Park DY, Jeong H, Chun WJ, Kim MH, Park J, Lim M, Kim TH, Cho YK. Circulating tumor cells detected by lab-on-a-disc: Role in early diagnosis of gastric cancer. PLoS One. 2017;12:e0180251. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 69. | Wan QS, Zhang KH. Noninvasive detection of gastric cancer. Tumour Biol. 2016;37:11633-11643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 70. | Tang L, Zhao S, Liu W, Parchim NF, Huang J, Tang Y, Gan P, Zhong M. Diagnostic accuracy of circulating tumor cells detection in gastric cancer: systematic review and meta-analysis. BMC Cancer. 2013;13:314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 71. | Matsusaka S, Chìn K, Ogura M, Suenaga M, Shinozaki E, Mishima Y, Terui Y, Mizunuma N, Hatake K. Circulating tumor cells as a surrogate marker for determining response to chemotherapy in patients with advanced gastric cancer. Cancer Sci. 2010;101:1067-1071. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 93] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 72. | Uenosono Y, Arigami T, Kozono T, Yanagita S, Hagihara T, Haraguchi N, Matsushita D, Hirata M, Arima H, Funasako Y, Kijima Y, Nakajo A, Okumura H, Ishigami S, Hokita S, Ueno S, Natsugoe S. Clinical significance of circulating tumor cells in peripheral blood from patients with gastric cancer. Cancer. 2013;119:3984-3991. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 71] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 73. | Wang S, Zheng G, Cheng B, Chen F, Wang Z, Chen Y, Wang Y, Xiong B. Circulating tumor cells (CTCs) detected by RT-PCR and its prognostic role in gastric cancer: a meta-analysis of published literature. PLoS One. 2014;9:e99259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 74. | Wang HY, Wei J, Zou ZY, Qian XP, Liu BR. Circulating tumour cells predict survival in gastric cancer patients: a meta-analysis. Contemp Oncol (Pozn). 2015;19:451-457. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 75. | Li Y, Gong J, Zhang Q, Lu Z, Gao J, Li Y, Cao Y, Shen L. Dynamic monitoring of circulating tumour cells to evaluate therapeutic efficacy in advanced gastric cancer. Br J Cancer. 2016;114:138-145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 78] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 76. | Zou K, Yang S, Zheng L, Wang S, Xiong B. Prognostic Role of the Circulating Tumor Cells Detected by Cytological Methods in Gastric Cancer: A Meta-Analysis. Biomed Res Int. 2016;2016:2765464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 77. | Lin LY, Yang L, Zeng Q, Wang L, Chen ML, Zhao ZH, Ye GD, Luo QC, Lv PY, Guo QW, Li BA, Cai JC, Cai WY. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol Cancer. 2018;17:84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 181] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 78. | Li W, Gao YQ. MiR-217 is involved in the carcinogenesis of gastric cancer by down-regulating CDH1 expression. Kaohsiung J Med Sci. 2018;34:377-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 79. | Zheng P, Luo Q, Wang W, Li J, Wang T, Wang P, Chen L, Zhang P, Chen H, Liu Y, Dong P, Xie G, Ma Y, Jiang L, Yuan X, Shen L. Tumor-associated macrophages-derived exosomes promote the migration of gastric cancer cells by transfer of functional Apolipoprotein E. Cell Death Dis. 2018;9:434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 283] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 80. | Liu F, Bu Z, Zhao F, Xiao D. Increased T-helper 17 cell differentiation mediated by exosome-mediated microRNA-451 redistribution in gastric cancer infiltrated T cells. Cancer Sci. 2018;109:65-73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 71] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 81. | Church TR, Wandell M, Lofton-Day C, Mongin SJ, Burger M, Payne SR, Castaños-Vélez E, Blumenstein BA, Rösch T, Osborn N, Snover D, Day RW, Ransohoff DF; PRESEPT Clinical Study Steering Committee, Investigators and Study Team. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63:317-325. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 486] [Cited by in RCA: 589] [Article Influence: 53.5] [Reference Citation Analysis (0)] |
| 82. | Arena S, Siravegna G, Mussolin B, Kearns JD, Wolf BB, Misale S, Lazzari L, Bertotti A, Trusolino L, Adjei AA, Montagut C, Di Nicolantonio F, Nering R, Bardelli A. MM-151 overcomes acquired resistance to cetuximab and panitumumab in colorectal cancers harboring EGFR extracellular domain mutations. Sci Transl Med. 2016;8:324ra14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 84] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 83. | Lecomte T, Berger A, Zinzindohoué F, Micard S, Landi B, Blons H, Beaune P, Cugnenc PH, Laurent-Puig P. Detection of free-circulating tumor-associated DNA in plasma of colorectal cancer patients and its association with prognosis. Int J Cancer. 2002;100:542-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 230] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 84. | Reinert T, Schøler LV, Thomsen R, Tobiasen H, Vang S, Nordentoft I, Lamy P, Kannerup AS, Mortensen FV, Stribolt K, Hamilton-Dutoit S, Nielsen HJ, Laurberg S, Pallisgaard N, Pedersen JS, Ørntoft TF, Andersen CL. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut. 2016;65:625-634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 341] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 85. | Galizia G, Gemei M, Orditura M, Romano C, Zamboli A, Castellano P, Mabilia A, Auricchio A, De Vita F, Del Vecchio L, Lieto E. Postoperative detection of circulating tumor cells predicts tumor recurrence in colorectal cancer patients. J Gastrointest Surg. 2013;17:1809-1818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 86. | Nesteruk D, Rutkowski A, Fabisiewicz S, Pawlak J, Siedlecki JA, Fabisiewicz A. Evaluation of prognostic significance of circulating tumor cells detection in rectal cancer patients treated with preoperative radiotherapy: prospectively collected material data. Biomed Res Int. 2014;2014:712827. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 87. | Hiraiwa K, Takeuchi H, Hasegawa H, Saikawa Y, Suda K, Ando T, Kumagai K, Irino T, Yoshikawa T, Matsuda S, Kitajima M, Kitagawa Y. Clinical significance of circulating tumor cells in blood from patients with gastrointestinal cancers. Ann Surg Oncol. 2008;15:3092-3100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 150] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 88. | Maestro LM, Sastre J, Rafael SB, Veganzones SB, Vidaurreta M, Martín M, Olivier C, DE La Orden VB, Garcia-Saenz JA, Alfonso R, Arroyo M, Diaz-Rubio E. Circulating tumor cells in solid tumor in metastatic and localized stages. Anticancer Res. 2009;29:4839-4843. [PubMed] |
| 89. | van Dalum G, Stam GJ, Scholten LF, Mastboom WJ, Vermes I, Tibbe AG, De Groot MR, Terstappen LW. Importance of circulating tumor cells in newly diagnosed colorectal cancer. Int J Oncol. 2015;46:1361-1368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 90] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 90. | Chen Y, Song Y, Wang Z, Yue Z, Xu H, Xing C, Liu Z. Altered expression of MiR-148a and MiR-152 in gastrointestinal cancers and its clinical significance. J Gastrointest Surg. 2010;14:1170-1179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 174] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 91. | Agostini M, Pucciarelli S, Calore F, Bedin C, Enzo M, Nitti D. miRNAs in colon and rectal cancer: A consensus for their true clinical value. Clin Chim Acta. 2010;411:1181-1186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 92. | Schepeler T, Reinert JT, Ostenfeld MS, Christensen LL, Silahtaroglu AN, Dyrskjøt L, Wiuf C, Sørensen FJ, Kruhøffer M, Laurberg S, Kauppinen S, Ørntoft TF, Andersen CL. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68:6416-6424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 388] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 93. | Huang ZM, Yang J, Shen XY, Zhang XY, Meng FS, Xu JT, Zhang BF, Gao HJ. MicroRNA expression profile in non-cancerous colonic tissue associated with lymph node metastasis of colon cancer. J Dig Dis. 2009;10:188-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 34] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 94. | Weissmann-Brenner A, Kushnir M, Lithwick Yanai G, Aharonov R, Gibori H, Purim O, Kundel Y, Morgenstern S, Halperin M, Niv Y, Brenner B. Tumor microRNA-29a expression and the risk of recurrence in stage II colon cancer. Int J Oncol. 2012;40:2097-2103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 95. | Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981-984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1303] [Cited by in RCA: 1749] [Article Influence: 174.9] [Reference Citation Analysis (0)] |
| 96. | Yoshioka Y, Kosaka N, Konishi Y, Ohta H, Okamoto H, Sonoda H, Nonaka R, Yamamoto H, Ishii H, Mori M, Furuta K, Nakajima T, Hayashi H, Sugisaki H, Higashimoto H, Kato T, Takeshita F, Ochiya T. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat Commun. 2014;5:3591. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 347] [Cited by in RCA: 443] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 97. | Weidle UH, Scheuer W, Eggle D, Klostermann S, Stockinger H. Cancer-related issues of CD147. Cancer Genomics Proteomics. 2010;7:157-169. [PubMed] |
| 98. | Matsumura T, Sugimachi K, Iinuma H, Takahashi Y, Kurashige J, Sawada G, Ueda M, Uchi R, Ueo H, Takano Y, Shinden Y, Eguchi H, Yamamoto H, Doki Y, Mori M, Ochiya T, Mimori K. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer. 2015;113:275-281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 332] [Cited by in RCA: 408] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 99. | Soldevilla B, Rodríguez M, San Millán C, García V, Fernández-Periañez R, Gil-Calderón B, Martín P, García-Grande A, Silva J, Bonilla F, Domínguez G. Tumor-derived exosomes are enriched in ΔNp73, which promotes oncogenic potential in acceptor cells and correlates with patient survival. Hum Mol Genet. 2014;23:467-478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 90] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 100. | Liao W, Mao Y, Ge P, Yang H, Xu H, Lu X, Sang X, Zhong S. Value of quantitative and qualitative analyses of circulating cell-free DNA as diagnostic tools for hepatocellular carcinoma: a meta-analysis. Medicine (Baltimore). 2015;94:e722. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 55] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 101. | Openshaw MR, Harvey RA, Sebire NJ, Kaur B, Sarwar N, Seckl MJ, Fisher RA. Circulating Cell Free DNA in the Diagnosis of Trophoblastic Tumors. EBioMedicine. 2016;4:146-152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 102. | Labgaa I, Villanueva A. Liquid biopsy in liver cancer. Discov Med. 2015;19:263-273. [PubMed] |
| 103. | Kirk GD, Camus-Randon AM, Mendy M, Goedert JJ, Merle P, Trépo C, Bréchot C, Hainaut P, Montesano R. Ser-249 p53 mutations in plasma DNA of patients with hepatocellular carcinoma from The Gambia. J Natl Cancer Inst. 2000;92:148-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 115] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 104. | Kirk GD, Lesi OA, Mendy M, Szymañska K, Whittle H, Goedert JJ, Hainaut P, Montesano R. 249(ser) TP53 mutation in plasma DNA, hepatitis B viral infection, and risk of hepatocellular carcinoma. Oncogene. 2005;24:5858-5867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 120] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 105. | Chan KC, Lai PB, Mok TS, Chan HL, Ding C, Yeung SW, Lo YM. Quantitative analysis of circulating methylated DNA as a biomarker for hepatocellular carcinoma. Clin Chem. 2008;54:1528-1536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 124] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 106. | Paterlini-Brechot P, Benali NL. Circulating tumor cells (CTC) detection: clinical impact and future directions. Cancer Lett. 2007;253:180-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 696] [Cited by in RCA: 701] [Article Influence: 38.9] [Reference Citation Analysis (0)] |
| 107. | Schulze K, Gasch C, Staufer K, Nashan B, Lohse AW, Pantel K, Riethdorf S, Wege H. Presence of EpCAM-positive circulating tumor cells as biomarker for systemic disease strongly correlates to survival in patients with hepatocellular carcinoma. Int J Cancer. 2013;133:2165-2171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 192] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 108. | Vona G, Estepa L, Béroud C, Damotte D, Capron F, Nalpas B, Mineur A, Franco D, Lacour B, Pol S, Bréchot C, Paterlini-Bréchot P. Impact of cytomorphological detection of circulating tumor cells in patients with liver cancer. Hepatology. 2004;39:792-797. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 163] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 109. | Lv P, Tang Z, Liang X, Guo M, Han RP. Spatially gradated segregation and recovery of circulating tumor cells from peripheral blood of cancer patients. Biomicrofluidics. 2013;7:34109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 110. | Sauzay C, Petit A, Bourgeois AM, Barbare JC, Chauffert B, Galmiche A, Houessinon A. Alpha-foetoprotein (AFP): A multi-purpose marker in hepatocellular carcinoma. Clin Chim Acta. 2016;463:39-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 187] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 111. | Suo Y, Xie C, Zhu X, Fan Z, Yang Z, He H, Wei X. Proportion of circulating tumor cell clusters increases during cancer metastasis. Cytometry A. 2017;91:250-253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 44] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 112. | Lu S, Luo Y. Research on the diagnostic value of serum tumor markers and CTC in the diagnosis of liver cancer. Biomed Res. 2018;Special issue:S223-S226. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 113. | Ye X, Li G, Han C, Han Q, Shang L, Su H, Han B, Gong Y, Lu G, Peng T. Circulating tumor cells as a potential biomarker for postoperative clinical outcome in HBV-related hepatocellular carcinoma. Cancer Manag Res. 2018;10:5639-5647. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 114. | Hao HC, Yao DJ. Detection of cancer cells on a chip. Curr Top Med Chem. 2015;15:1543-1550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 115. | Hao SJ, Wan Y, Xia YQ, Zou X, Zheng SY. Size-based separation methods of circulating tumor cells. Adv Drug Deliv Rev. 2018;125:3-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 149] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 116. | Huang JT, Liu SM, Ma H, Yang Y, Zhang X, Sun H, Zhang X, Xu J, Wang J. Systematic Review and Meta-Analysis: Circulating miRNAs for Diagnosis of Hepatocellular Carcinoma. J Cell Physiol. 2016;231:328-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 64] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 117. | Shi BM, Lu W, Ji K, Wang YF, Xiao S, Wang XY. Study on the value of serum miR-106b for the early diagnosis of hepatocellular carcinoma. World J Gastroenterol. 2017;23:3713-3720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 16] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 118. | Li Y, Tan W, Neo TW, Aung MO, Wasser S, Lim SG, Tan TM. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci. 2009;100:1234-1242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 218] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 119. | Zhang Y, Li T, Qiu Y, Zhang T, Guo P, Ma X, Wei Q, Han L. Serum microRNA panel for early diagnosis of the onset of hepatocellular carcinoma. Medicine (Baltimore). 2017;96:e5642. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 76] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 120. | Cazzoli R, Buttitta F, Di Nicola M, Malatesta S, Marchetti A, Rom WN, Pass HI. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol. 2013;8:1156-1162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 365] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 121. | Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:264-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 542] [Cited by in RCA: 613] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 122. | Sugimachi K, Matsumura T, Hirata H, Uchi R, Ueda M, Ueo H, Shinden Y, Iguchi T, Eguchi H, Shirabe K, Ochiya T, Maehara Y, Mimori K. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br J Cancer. 2015;112:532-538. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 212] [Cited by in RCA: 243] [Article Influence: 24.3] [Reference Citation Analysis (0)] |