Published online Sep 26, 2019. doi: 10.12998/wjcc.v7.i18.2734
Peer-review started: April 4, 2019
First decision: June 19, 2019
Revised: July 17, 2019
Accepted: July 27, 2019
Article in press: July 27, 2019
Published online: September 26, 2019
Processing time: 179 Days and 16.7 Hours
Systemic-onset juvenile idiopathic arthritis (SoJIA) is one of most serious subtypes of juvenile idiopathic arthritis. Although the pathogenesis of SoJIA remains unclear, several studies have suggested a correlation between gut dysbiosis and JIA. Further understanding of the intestinal microbiome may help to establish alternative ways to treat, or even prevent, the disease.
To explore alterations in fecal microbiota profiles in SoJIA patients and to evaluate the correlations between microbiota and clinical parameters.
We conducted an observational single-center study at the Pediatric Department of Peking Union Medical College Hospital. Children who were diagnosed with SoJIA at our institution and followed for a minimum period of six months after diagnosis were recruited for the study. Healthy children were recruited as a control group (HS group) during the same period. Clinical data and stool samples were collected from SoJIA patients when they visited the hospital.
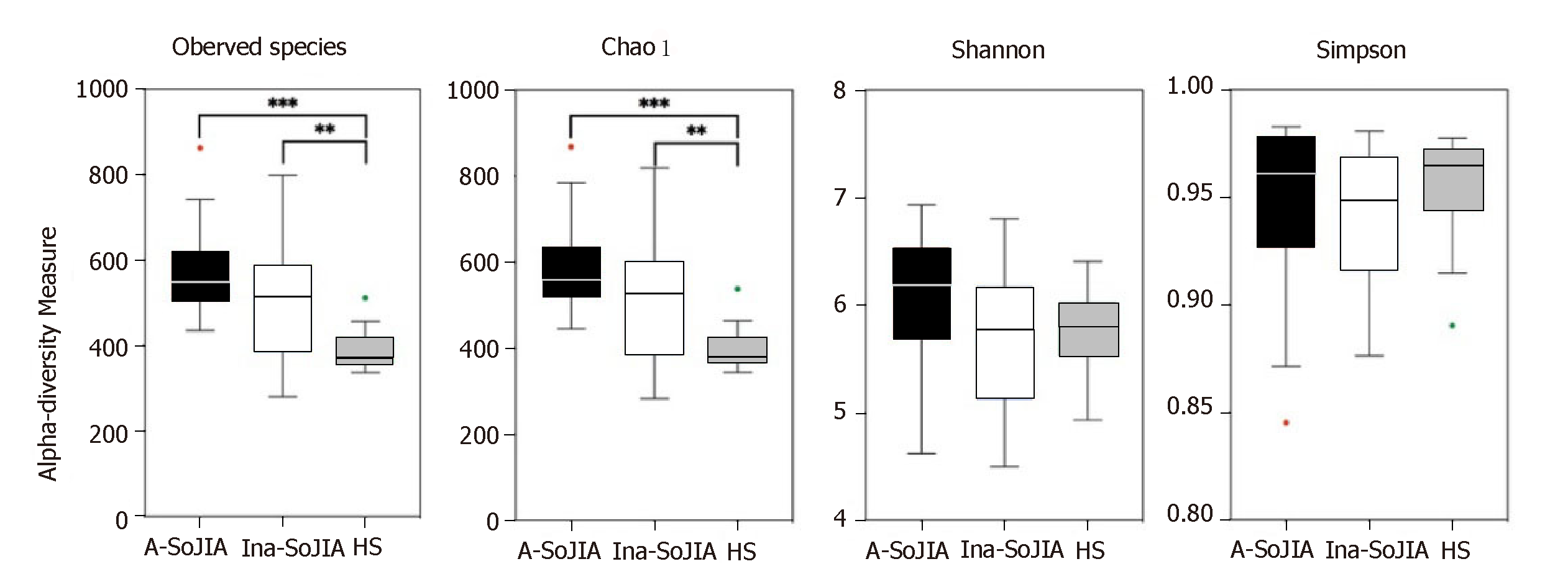
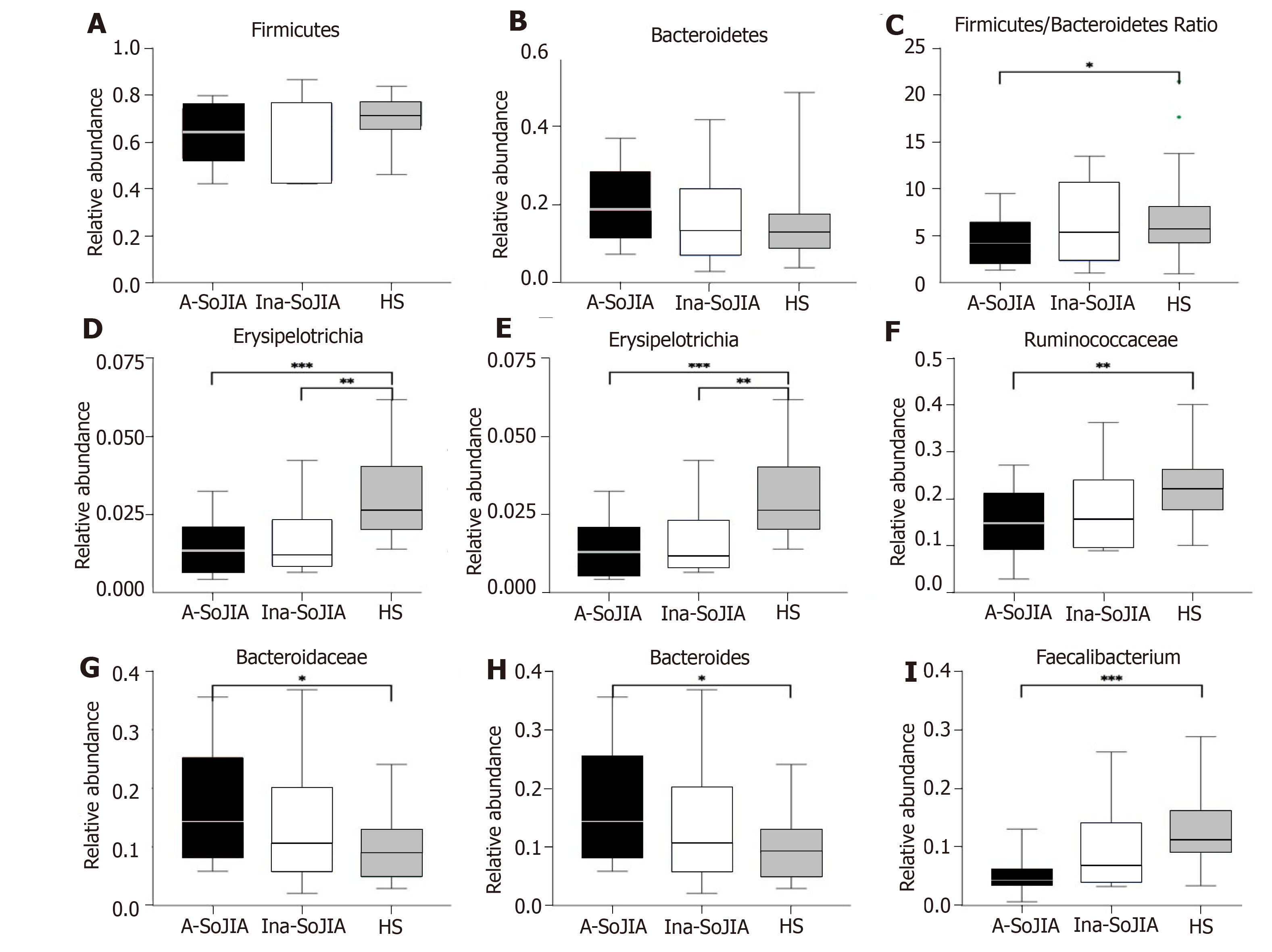
The SoJIA group included 17 active and 15 inactive consecutively recruited children; the control group consisted of 32 children. Firmicutes and Bacteroidetes were the two most abundant phyla among the total sample of SoJIA children and controls. There was a significant difference among the three groups in observed species, which was the highest in the Active-SoJIA group, followed by the Inactive-SoJIA group and then HS group (Active-SoJIA vs HS: P = 0.000; and Inactive-SoJIA vs HS: P = 0.005). We observed a lower Firmicutes/Bacteroidetes ratio in SoJIA patients (3.28 ± 4.47 in Active-SoJIA, 5.36 ± 8.39 in Inactive-SoJIA, and 5.67 ± 3.92 in HS). We also observed decreased abundances of Ruminococcaceae (14.9% in Active-SoJIA, 17.3% in Inactive-SoJIA, and 22.8% in HS; Active-SoJIA vs HS: P = 0.005) and Faecalibacterium (5.1% in Active-SoJIA, 9.9% in Inactive-SoJIA, and 13.0% in HS; Active-SoJIA vs HS: P = 0.000) in SoJIA compared with HS. By contrast, the abundance of Bacteroidaceae was the highest in the Active-SoJIA group, followed by the Inactive-SoJIA and HS groups (16.5% in Active-SoJIA, 12.8% in Inactive-SoJIA, and 9.7% in HS; Active-SoJIA vs HS: P = 0.03). The Spearman correlation analysis revealed a negative correlation between Proteobacteria or Enterobacteriaceae and juvenile arthritis disease activity score on 27 joints (JADAS-27).
The composition of the intestinal microbiota is different in SoJIA patients compared with healthy children. The dysbiosis presents partial restoration in inactive status patients.
Core tip: In recent decades, the potential role of gut microbiome in modulating host homeostasis has gained considerable attention. This is the first report of microbiota composition in systemic-onset juvenile idiopathic arthritis (SoJIA) children. Our results demonstrate that the composition of the intestinal microbiota is different in SoJIA patients compared with healthy children. The perturbed microbiota present partial restoration in inactive status patients. Characterizing intestinal microbiomes may help to understand the pathogenesis of SoJIA. Modifications of the microbiota may be a new way of preventing and managing SoJIA.
- Citation: Dong YQ, Wang W, Li J, Ma MS, Zhong LQ, Wei QJ, Song HM. Characterization of microbiota in systemic-onset juvenile idiopathic arthritis with different disease severities. World J Clin Cases 2019; 7(18): 2734-2745
- URL: https://www.wjgnet.com/2307-8960/full/v7/i18/2734.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v7.i18.2734
Juvenile idiopathic arthritis (JIA) is the most common chronic inflammatory joint disease in children[1]. According to the International League of Associations for Rheumatology (ILAR) classification criteria, there are seven different JIA categories: Systemic-onset juvenile idiopathic arthritis (SoJIA), oligoarticular arthritis (affecting less than four joints at onset), polyarticular arthritis (affecting more than four joints at onset), rheumatoid factor negative and positive arthritis, enthesitis-related arthritis (ERA), psoriatic arthritis, and undifferentiated arthritis. SoJIA is the most severe subtype among the seven subtypes of JIA, distinguished from other subtypes by its predominant systemic inflammation and extraarticular features, including spiking fevers, macular rash, lymphadenopathy, hepatosplenomegaly, and serositis[3]. Although the pathogenesis of SoJIA remains unclear, it is widely believed that complex interactions between host genetics, immunological dysregulation, and environmental influences (lifestyle, diet, antibiotics, and infections) are involved[4].
In recent decades, the potential role of the gut microbiome in modulating host homeostasis has gained considerable attention[5]. There is an increasing number of studies revealing the presence of gut dysbiosis in various diseases, such as inflammatory bowel disease, arthritis, multiple sclerosis, systemic lupus erythematosus, and type 2 diabetes[6-10]. Understanding of the intestinal microbiome may help to establish alternative ways to treat, or even prevent, these diseases. However, data regarding the role of the intestinal microbiota in JIA patients are not conclusive and changes in gut microbes over the JIA disease course remain uncertain. To date, there are no studies on gut microbiota in SoJIA children. Therefore, we used fecal samples to characterize the intestinal microbiomes of SoJIA patients and healthy children. In order to understand the changes in the gut microbiome in different disease states, we also compared the microbiota composition in active-SoJIA patients versus inactive-SoJIA patients. Correlations between the gut microbiota composition and clinical parameters of disease activity were also evaluated.
The protocol for this study was approved by the local ethics committee of the Peking Union Medical College Hospital (Protocol number: JS-1659). Written informed consent was obtained from one of the parents of each participant on the day of sample collection. All procedures were performed in accordance with the Declaration of Helsinki.
Children who were diagnosed with SoJIA according to the ILAR classification criteria and followed for a minimum period of six months were recruited from the Peking Union Medical College Hospital[2]. During the same period, healthy children were recruited as a control group. Inactive disease (ID) status was defined according to the Wallace criteria: No active arthritis; no fever, rash, serositis, splenomegaly, or generalized lymphadenopathy attributable to JIA; no active uveitis; no abnormal erythrocyte sedimentation rate (ESR) or C-reactive protein (CRP); best possible physician’s global assessment (PGA) disease activity score[11]. Patients suffering from cancer, heart failure, renal failure, chronic infectious disease, macrophage activation syndrome (MAS), and overlapping other rheumatic immune diseases were all excluded. Individuals were also excluded if they had received antibiotics or had a symptomatic gastrointestinal infection within one month prior to participating in this study.
Initially, from June 1, 2017 to December 31, 2018, a total of 33 patients with SoJIA agreed to participate in this study; however, one patient was excluded from the study because of comorbid systemic lupus erythematosus (SLE) during follow-up. Therefore, a total of 32 patients with SoJIA were investigated. The control group comprised 32 children aged from 5 to 16 years. Patients were divided into two groups according to activity status at the time of sample collection: 17 were classified as active SoJIA and 15 were inactive. Clinical data and stool samples were collected from the SoJIA patients when they visited the hospital. Each stool specimen was collected in a sterile vial; it was then transported immediately to the laboratory and frozen at -80 °C until analysis. Collected clinical data included age of onset, disease duration, active joint count (AJC), ESR, CRP, PGA, parent/patient visual analogue scale (VAS) of well-being, and juvenile arthritis disease activity score on 27 joints (JADAS-27)[12].
Bacterial DNA was extracted at Beijing Novogene Bioinformatics Technology Co. Ltd using the SDS method. DNA concentration and purity were monitored on 1% agarose gels. Based on the initial concentration, DNA was diluted to 1 ng/μL using sterile water. The specific primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) were used to amplify the V4 region of 16S rRNA genes. All PCR reactions were carried out in 30 μL reactions with 15 μL of Phusion® High-Fidelity PCR Master Mix (New England Biolabs, Ipswich, MA, United States), 0.2 μmol/L of forward and reverse primers, and about 10 ng of template DNA. Thermal cycling consisted of initial denaturation at 98 °C for 1 min, followed by 30 cycles of denaturation at 98 °C for 10 s, annealing at 50 °C for 30 s, and elongation at 72 °C for 30 s, with a final cycle of 72 °C for 5 min. The same volume of 1 × loading buffer (containing SYB green) was mixed with PCR products and electrophoresis was performed on 2% agarose gels for detection. Samples with a bright main band between 400-450 bp were chosen for further experiments. PCR products were mixed in equidensity ratios. Then, mixture PCR products were purified using a Gene JET Gel Extraction Kit (Thermo Scientific).
Sequencing libraries were generated using a TruSeq® DNA PCR-Free Sample Preparation Kit, following the manufacturer’s recommendations, and index codes were added. Library quality was assessed on a Qubit@2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. Finally, the library was sequenced on an Illumina HiSeq 2500 and 250 bp paired-end reads were generated. Paired-end reads from the original DNA fragments were merged using FLASH and were then assigned to each sample according to the unique barcodes. Sequences were analyzed using the QIIME software package (Quantitative Insights Into Microbial Ecology), and in-house Perl scripts were used to analyze alpha- (within samples) and beta- (among samples) diversity[14]. Sequences with a ≥ 97% similarity were assigned to the same operational taxonomic units (OTUs). For each representative sequence, the SILVA132 database was used to annotate taxonomic information.
Alpha diversity analysis, including observed species, Chao1 index, Shannon index, Simpson index, and beta diversity, including both unweighted and weighted Unifrac distances, were performed with QIIME (version 1.9.1). Principal Coordinate Analysis (PCoA) was performed and displayed with the WGCNA package, stats package, and ggplot2 package in R software (version 2.15.3). The significance of between-group differences in UniFrac distances was assessed with PERMANOVA using the adonis function of the R package vegan. Correlations between gut microbiota composition and clinical parameters of disease activity were also evaluated.
Quantitative data are presented as the mean (SD) or median (IQR) in the case of skewed data. Qualitative data are presented as frequencies and proportions. When comparing continuous data between two groups, the Welch's test or Mann-Whitney U test was applied to calculate intergroup differences. Pearson chi-square test was employed for categorical variables. ANOVA was used for comparisons of parametric data among three groups. Differences in abundance of bacterial communities between Active-SoJIA, Inactive-SoJIA, and healthy subjects (HS) were tested by the Kruskal-Wallis test, and P-values with Bonferroni correction for multiple testing were evaluated. A two-sided P-value less than 0.05 was considered statistically significant. Analyses were performed using SPSS version 25.0 (IBM Corp., Armonk, NY, United States) and Graphpad Prism version 8 software (La Jolla, CA, United States).
The total cohort comprised 64 children. The SoJIA group included 17 active and 15 inactive children, aged from 6 to 16 years, who were consecutively recruited into the study. The control group comprised 32 children aged from 5 to 16 years. No significant differences in age or gender were observed among the three groups. SoJIA patients received treatment with steroids/disease-modifying anti-rheumatic drugs (DMARDs)/biologics (Tocilizumab or Infliximab) in different combinations. Demographic and clinical characteristics at the time of sample collection are presented in Table 1 (more details in Supplementary Table S1).
A total of 5270328 effective tags were obtained by high-throughput sequencing from 64 specimens obtained from SoJIA patients and control subjects. The average number of effective tags per sample was 82349 (SoJIA: 82875; HS: 81823). Each effective tag had Q20 > 98% and Q30 > 97%. Among all samples, tags with a similarly level of 97% were clustered into 2216 OTUs. Through annotation of the sequence of the OTUs with the Silva132 database, a total of 1097 (49.5%) OTU notes to the genus level were performed.
To characterize bacterial richness, rarefaction analysis was performed (Supplementary Figure S1). The curve in each group was near saturation, indicating sufficient sampling of the microbial communities. In order to evaluate differences in microbial biodiversity (alpha-diversity) among the three groups, we calculated the observed species, Chao1, Shannon, and Simpson indices. The results indicated that there were significant differences among the three groups in terms of observed species; the number of observed species was highest in the Active-SoJIA group, followed by the Inactive-SoJIA group, and then the HS group (Active-SoJIA vs HS: P = 0.000; Inactive-SoJIA vs HS: P = 0.005; Figure 1). The Chao1 index in both the Active-SoJIA and Inactive-SoJIA groups was significantly increased compared to the HS group (Active-SoJIA vs HS: P = 0.000; Inactive-SoJIA vs HS: P = 0.006; Supplementary Table S2 and Figure 1). This indicates that there was an increased richness in the microbiota of SoJIA patients. Assessment of the Shannon and Simpson indices showed no remarkable differences among the three groups (Figure 1). The significance of the alpha-diversity differences was calculated using the Kruskal-Wallis test with Bonferroni correction (P-value).
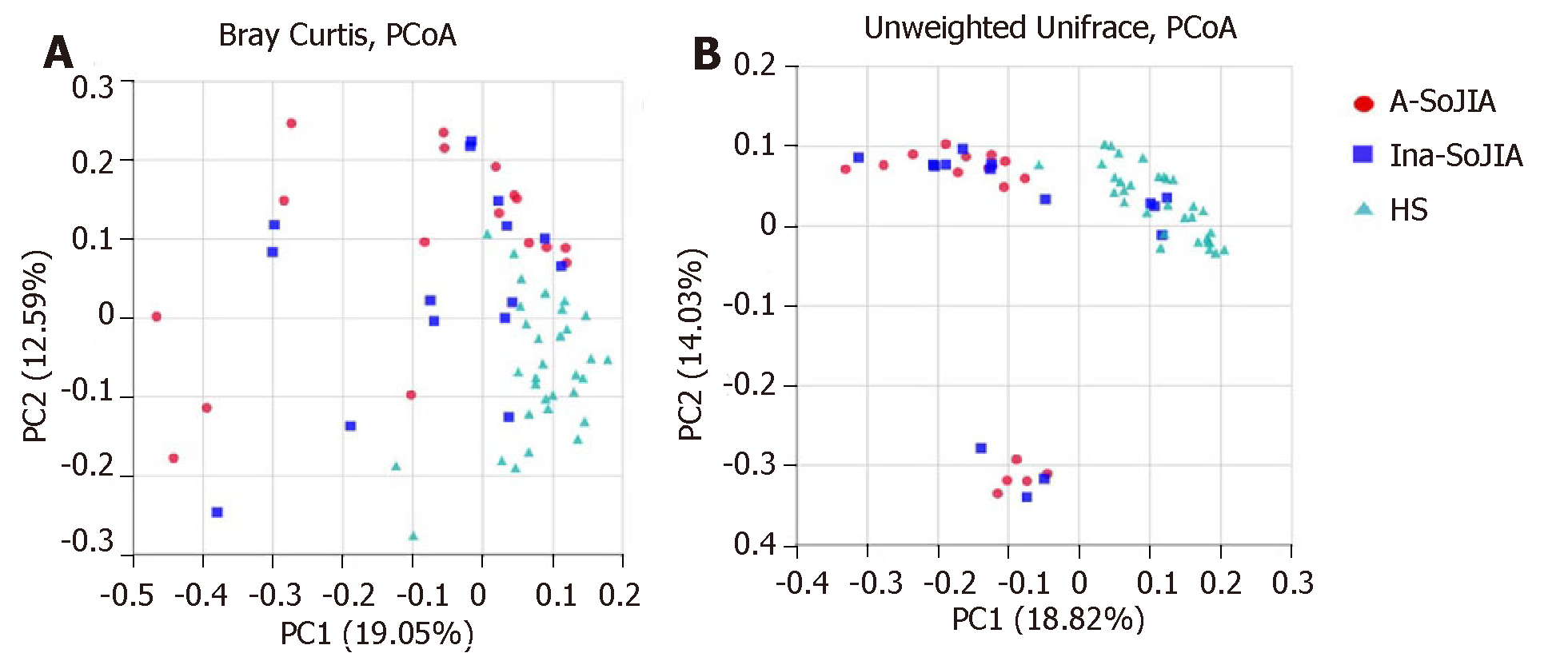
To estimate the between-sample (beta-diversity) variability in microbial communities, we calculated the Bray-Curtis and unweighted UniFrac dissimilarities. PCoA on unweighted UniFrac and Bray-Curtis dissimilarities showed that both SoJIA samples (active and inactive) were more similar to each other than to the HS sample. PERMANOVA confirmed the differences among samples: Active-SoJIA vs HS (P = 0.001) and Inactive-SoJIA vs HS (P = 0.001); there was no significant difference between Active-SoJIA and Inactive-SoJIA (Figure 2A and 2B).
At the phylum level, Firmicutes and Bacteroidetes were the two most abundant phyla among the whole sample of both SoJIA children and controls (Supplementary Figure S2). The abundance of Firmicutes was lowest in Active-SoJIA, followed by Inactive-SoJIA, and was highest in HS (62.6% in Active-SoJIA, 64.7% in Inactive-SoJIA, and 71.0% in HS; Figure 3A and B). In contrast, the abundance of Bacteroidetes was highest in Active-SoJIA and lowest in HS (20.4% in Active-SoJIA, 16.3% in Inactive-SoJIA, and 14.8% in HS; Figure 3A and 3B). We then computed the Firmicutes/Bacteroidetes ratio for the three groups. A Firmicutes/Bacteroidetes ratio gradient from lowest (Active-SoJIA) to higher (Inactive-SoJIA) to highest (HS) was observed, though the only significant difference was noted between Active-SoJIA and HS (3.28 ± 4.47 in Active-SoJIA, 5.36 ± 8.39 in Inactive-SoJIA, and 5.67 ± 3.92 in HS; Active-SoJIA vs HS: P = 0.048; Figure 3C). The decreased Firmicutes/Bacteroidetes ratio in SoJIA is similar to previous findings[16]. The abundance of Erysipelotrichia and Erysipelotrichales in both active and inactive SoJIA populations was significantly decreased compared to HS (Active-SoJIA vs HS: P = 0.000; Inactive-SoJIA vs HS: P = 0.004; Figure 3). Family Ruminococcaceae was less abundant in Active-SoJIA compared with HS (14.9% in Active-SoJIA, 17.3% in Inactive-SoJIA, and 22.8% in HS; Active-SoJIA vs HS: P = 0.005; Figure 3). The abundance of Bacteroidaceae and Bacteroides was as follows: Active-SoJIA > Inactive-SoJIA > HS; there was a statistically significant difference between Active-SoJIA vs HS (16.5% in Active-SoJIA, 12.8% in Inactive-SoJIA, and 9.7% in HS; Active-SoJIA vs HS: P = 0.03; Figure 3). We observed a statistically significant predominance of Faecalibacterium in HS compared with Active-SoJIA (5.1% in Active-SoJIA, 9.9% in Inactive-SoJIA, and 13.0% in HS; Active-SoJIA vs HS P = 0.000; Figure 3). It is worth mentioning that most bacteria described above in the Inactive-SoJIA group showed a moderate abundance in the Active-SoJIA group and the HS group. Differential abundances of bacterial communities between Active-SoJIA, Inactive-SoJIA, and HS were tested by the Kruskal Wallis test; P-values were submitted to Bonferroni correction for multiple testing.
In order to determine whether microbiota composition is associated with clinical parameters in SoJIA patients, we performed Spearman rank correlations between clinical parameters and microorganisms at the phylum, family, and genus levels.
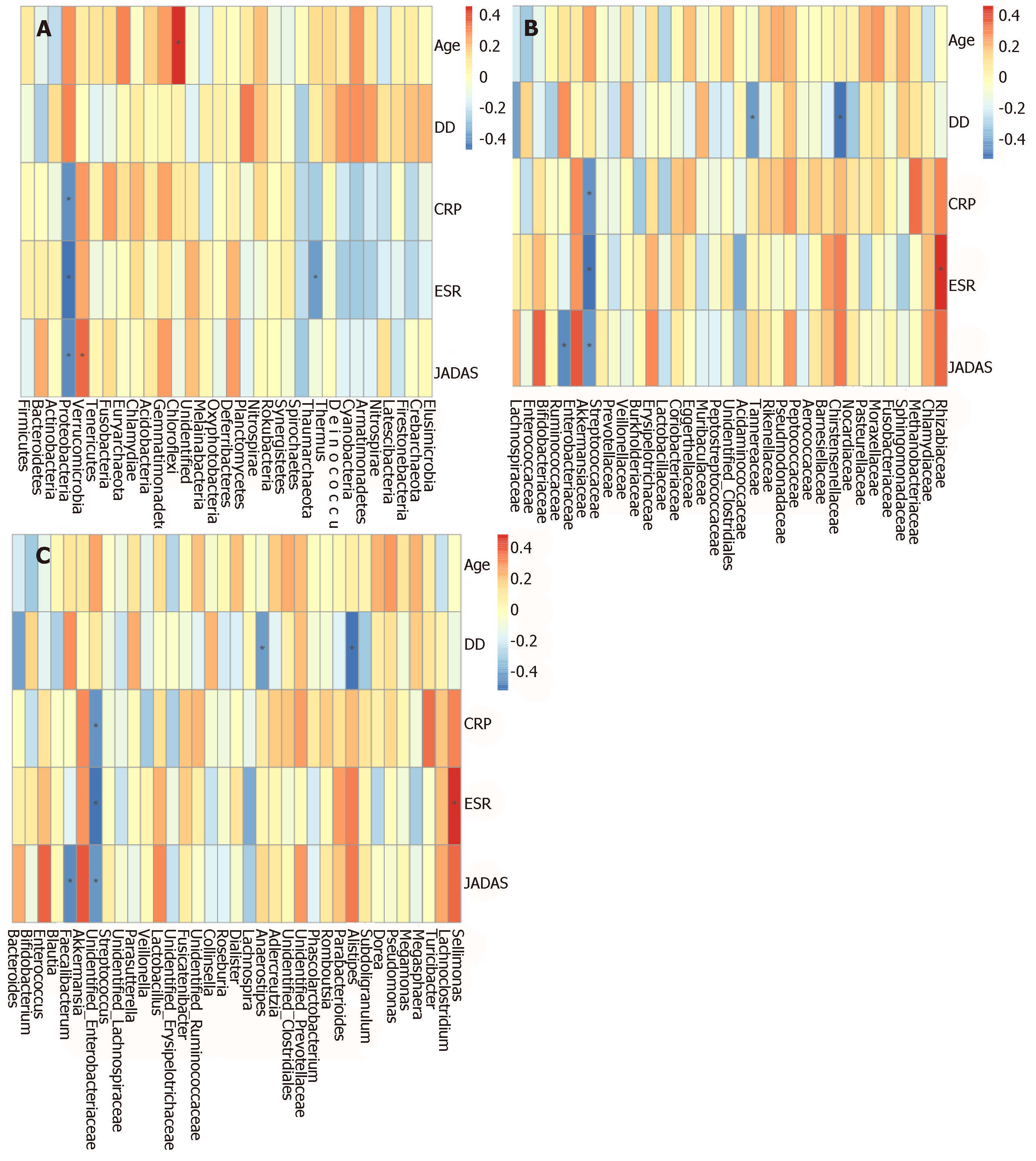
The Spearman correlation analyses revealed a direct association between patient age and phylum Chloroflexi. Disease duration was positively associated with family Burkholderiaceae, and negatively associated with genus Alistipes. CRP showed a positive association with Rhizobiaceae, and negative associations with Proteobacteria and Enterobacteriaceae. ESR was positively associated with Sellimonas and negatively correlated to Proteobacteria and Enterobacteriaceae. JADAS was positively correlated with Eggerthellaceae and Muribaculaceae, while Proteobacteria, Enterobacteriaceae, and Faecalibacterium were negatively correlated with JADAS (Supplementary Figure S3 and Figure 4).
This is the first report on the microbiota composition in SoJIA children. In our study, we applied 16S rDNA sequence analysis to characterize the intestinal microbiota of patients with SoJIA, taking into account the activity state of SoJIA disease. Our results confirmed that the composition of the intestinal microbiota is different in SoJIA patients compared to healthy children. In contrast to previous studies which have shown decreased microbiota richness or did not find any differences between JIA patients and control children[17,18], our results showed that observed species and Chao1 index were highest in Active-SoJIA patients, followed by Inactive-SoJIA patients, and were lowest in healthy children. These findings indicate that there was an increase in microbiota richness in SoJIA patients, and the richness appeared to be decreased in patients who had achieved inactivity status. No significant differences in Shannon and Simpson indices were observed among the three groups. Measures of beta-diversity showed that both SoJIA samples (active and inactive) were more similar to each other than to the control group.
At the phylum level, compared to control children, the Active-SoJIA patients showed a lower abundance of Firmicutes and a higher abundance of Bacteroidetes. The low ratio of Firmicutes compared to the Bacteroidetes phylum in SoJIA is in accordance with another recent study in new-onset JIA patients (mainly oligoarticular or polyarticular) who were not treated with corticosteroids and DMARDs[19]. In the United States and Asia, researchers have also observed an abundance in family Bacteroidaceae and genus Bacteroides in ERA patients[17,19]. In addition, we computed the Firmicutes/Bacteroidetes ratio in the three groups, and a gradient from lowest (Active-SoJIA) to higher (Inactive-SoJIA) to highest (HS) was observed. Studies of patients with systemic lupus erythematosus (SLE) and type 1 diabetes have also shown a low Firmicutes/Bacteroidetes ratio[9,20,21].
Bacteroides are primarily commensal organisms in the gut. Multiple species, including B. fragilis in the Bacteroides genus, can degrade mucin, an important component of the primary mucosal defense, leading to increased gut permeability[22]. Agglutinins and histolytic enzymes of B. fragilis help bacteria adhere to the host mucosa, increasing the chance of bacteria entering the gut immune system, and thereby promoting the inflammatory process. In animal models of arthritis in germ-free conditions, articular inflammation can be activated by the introduction of Bacteroides spp., especially B. vulgatus and B. fragilis[24]. Increased intestinal permeability has been identified both in children with JIA and in adults with ankylosing spondylitis[25,26].
The decreased abundance of Faecalibacterium found in our analysis is in agreement with previous studies on polyarticular JIA and ERA[17,27]. As butyrate-producing bacteria, Faecalibacterium helps in maintaining the integrity and health of the gut epithelial barrier, and is considered to be an anti-inflammatory microorganism[28-30]. Thus, a reduction in Faecalibacterium may induce an inflammatory status. When compared to healthy children, we found a statistically significant reduction in Ruminococcaceae in both SoJIA groups; this is similar to previous findings in oligoarticular and polyarticular JIA[18]. In contrast to our results, two previous studies of JIA have shown depletion of Ruminococcaceae[27,31]. Several factors can explain the partial inconsistency between our findings and other studies, such as different cohort size, JIA categories, and disease status, as well as different geographical, environmental, or dietary habits.
Another important finding of our study is that the Firmicutes/Bacteroidetes ratio and other bacterial alterations in the Inactive-SoJIA group were at a moderate level in the Active-SoJIA group and healthy children. This implies that the perturbed microbiota present in the Active-SoJIA group demonstrated partial restoration towards normal in inactive status patients. This result is consistent with a previous study. In 2016, Berntson et al[16] described a child with polyarticular JIA refractory to multiple medicines, yet the girl showed remarkable clinical improvement accompanied by an elevation in Firmicutes/Bacteroidetes ratio during exclusive enteral nutrition treatment. Thus, we suspect that a low Firmicutes/Bacteroidetes ratio may be related to SoJIA activity status, and differences in microbial composition associated with disease activity may be useful as markers for disease monitoring.
In order to determine whether the microbiota composition is associated with clinical parameters in SoJIA patients, we performed correlation analyses between microorganisms and clinical parameters. Among the most remarkable results, Proteobacteria and Enterobacteriaceae displayed negative correlations with disease activity (JADAS-27), ESR, and CRP. The function of these microorganisms in humans requires further exploration.
Two registry-based case-control studies have suggested a significant association between early life antibiotic use and subsequent JIA; the relationship was dose-dependent and the more antibiotic categories that were used, the higher the risk. This suggests that the greater the overall perturbation in the microbiota, the greater the risk of JIA[28,32,33]. A study of children at risk for type 1 diabetes showed that changes in gut microbiota composition and diversity preceded the development of the clinical disease[34]. Another study of hypertension showed that compared with healthy people, the microbiome characteristics of individuals with pre-hypertension were quite similar to those of individuals with hypertension, indicating that gut dysbiosis had already occurred in the pre-hypertension stage[35]. Zhang et al[7] reported that dysbiosis had been partially restored to normality in rheumatoid arthritis patients showing clinical improvement after prescription of disease modifying antirheumatic drugs. Our data showed that an improvement in clinical symptoms after treatment was associated with partial reversal of the disease-related dysbiosis. Taken together, the above findings suggest that the intestinal microbiota plays a crucial role in disease promotion and clinical course, and that maintaining the homeostasis of intestinal flora is essential for the host’s health.
Diet is one of the main factors contributing to the modulation of gut microbiota composition. Previous studies have reported that the balance of Bacteroidetes and Firmicutes can be modified by dietary patterns[36,37]. Therefore, we speculate that diet, which affects the microbiota composition, can in turn influence these disorders. Modifications to the microbiota through dietary interventions could be a new approach to preventing and managing SoJIA.
There were several strengths of our study. First, we studied SoJIA patients with differing disease activity states and compared these to healthy controls. We also conducted regular follow-up of patients, as the clinical manifestations of SoJIA may be similar to the early manifestations of other diseases, such as tumors. The shortest follow-up period in this study was 10 months, and the patient was in a good state following treatment.
A limitation of our study was that our patients were not new-onset SoJIA patients and had received treatment for arthritis or systemic symptoms with DMARDs/steroids/biologics in different combinations prior to sample collection. This could have had an effect on gut microbiota. However, it is difficult to obviate this limitation in this disease. Due to the wide variation in signs and symptoms, the diagnosis of SoJIA at disease onset is challenging and diagnosis is usually made through exclusion. Infectious agents are considered to be one of potential environmental triggers of SoJIA pathogenesis; thus, antibiotics are often used at disease onset of SoJIA. Numerous studies have shown that antibiotics have an effect on the contents of the microbiota. Thus, in order to reduce the interference associated with the effects of antibiotic use on intestinal microbiota, we excluded new-onset patients who had used antibiotics within one month before enrollment. The small sample was another limitation, and is a result of the rarity of SoJIA. Therefore, further studies are required to draw comprehensive conclusions in the future.
In summary, our results demonstrated that the composition of the intestinal microbiota is different in SoJIA patients compared to healthy children. The perturbed microbiota demonstrated partial restoration in inactive status patients after treatment.
Systemic-onset juvenile idiopathic arthritis (SoJIA) is a serious chronic rheumatic disease of childhood. The pathogenesis of SoJIA remains unclear, and several studies suggest that perturbation of gut microbiota, dysbiosis, could contribute to development of JIA. Understanding the intestinal microbial characteristics may contribute to the prevention and treatment of SoJIA.
We aimed to characterize the gut microbiota in SoJIA and to analyze the changing trend of intestinal flora as the disease improved.
Our main purpose was to characterize the gut microbiota in SoJIA and investigate the correlation between the abundance of intestinal microorganisms and clinical indicators as well as the pathogenesis of SoJIA from the perspective of microorganisms.
We carried out an observational single-center study. A total of 32 patients and 32 healthy children were enrolled. The patients were divided into two groups: Active-SoJIA and Inactive-SoJIA. Clinical data and stool samples were collected from SoJIA patients when they visited the hospital.
Alpha-diversity analysis indicated that there was an increased richness in the microbiota of SoJIA patients. Measures of beta-diversity showed that characteristics of gut microbiota of both SoJIA samples (active and inactive) were more similar to each other than to the control group. The Firmicutes/Bacteroidetes ratio and the abundance of Erysipelotrichales, Ruminococcaceae, and Faecalibacterium were decreased in SoJIA. By contrast, the abundance of Bacteroides and Bacteroidaceae was increased in SoJIA. The Firmicutes/Bacteroidetes ratio and other bacterial abundance in the Inactive-SoJIA group were at a moderate level between the Active-SoJIA group and healthy children. Proteobacteria and Enterobacteriaceae were negatively correlated with disease activity.
The composition of the intestinal microbiota is different in SoJIA patients compared to healthy children. The perturbed microbiota demonstrated partial restoration in inactive status patients after treatment.
There are many factors affecting gut microbiota composition. Future studies should prospectively collect multicenter data on new-onset SoJIA patients, and analyze the changing trend of intestinal flora of each patient as the disease improves to eliminate the interference of geographical, environmental, or dietary habits on intestinal flora.
The authors are grateful to Chao-Yang Gu for technical support.
Manuscript source: Unsolicited Manuscript
Specialty type: Medicine, research and experimental
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gheita TA, Wang T S-Editor: Cui LJ L-Editor: Wang TQ E-Editor: Zhou BX
| 1. | Ravelli A, Martini A. Juvenile idiopathic arthritis. Lancet. 2007;369:767-778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 983] [Cited by in RCA: 1051] [Article Influence: 58.4] [Reference Citation Analysis (0)] |
| 2. | Petty RE, Southwood TR, Manners P, Baum J, Glass DN, Goldenberg J, He X, Maldonado-Cocco J, Orozco-Alcala J, Prieur AM, Suarez-Almazor ME, Woo P; International League of Associations for Rheumatology. International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J Rheumatol. 2004;31:390-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 19] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 3. | De Benedetti F, Brunner HI, Ruperto N, Kenwright A, Wright S, Calvo I, Cuttica R, Ravelli A, Schneider R, Woo P, Wouters C, Xavier R, Zemel L, Baildam E, Burgos-Vargas R, Dolezalova P, Garay SM, Merino R, Joos R, Grom A, Wulffraat N, Zuber Z, Zulian F, Lovell D, Martini A; PRINTO; PRCSG. Randomized trial of tocilizumab in systemic juvenile idiopathic arthritis. N Engl J Med. 2012;367:2385-2395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 608] [Cited by in RCA: 612] [Article Influence: 47.1] [Reference Citation Analysis (0)] |
| 4. | Scher JU. Intestinal dysbiosis and potential consequences of microbiome-altering antibiotic use in the pathogenesis of human rheumatic disease. J Rheumatol. 2015;42:355-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 5. | Van de Wiele T, Van Praet JT, Marzorati M, Drennan MB, Elewaut D. How the microbiota shapes rheumatic diseases. Nat Rev Rheumatol. 2016;12:398-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 109] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 6. | Huang H, Vangay P, McKinlay CE, Knights D. Multi-omics analysis of inflammatory bowel disease. Immunol Lett. 2014;162:62-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 7. | Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y, Lan Z, Chen B, Li Y, Zhong H, Xie H, Jie Z, Chen W, Tang S, Xu X, Wang X, Cai X, Liu S, Xia Y, Li J, Qiao X, Al-Aama JY, Chen H, Wang L, Wu QJ, Zhang F, Zheng W, Li Y, Zhang M, Luo G, Xue W, Xiao L, Li J, Chen W, Xu X, Yin Y, Yang H, Wang J, Kristiansen K, Liu L, Li T, Huang Q, Li Y, Wang J. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21:895-905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 903] [Cited by in RCA: 1182] [Article Influence: 118.2] [Reference Citation Analysis (0)] |
| 8. | Jangi S, Gandhi R, Cox LM, Li N, von Glehn F, Yan R, Patel B, Mazzola MA, Liu S, Glanz BL, Cook S, Tankou S, Stuart F, Melo K, Nejad P, Smith K, Topçuolu BD, Holden J, Kivisäkk P, Chitnis T, De Jager PL, Quintana FJ, Gerber GK, Bry L, Weiner HL. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun. 2016;7:12015. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 703] [Cited by in RCA: 929] [Article Influence: 103.2] [Reference Citation Analysis (0)] |
| 9. | Hevia A, Milani C, López P, Cuervo A, Arboleya S, Duranti S, Turroni F, González S, Suárez A, Gueimonde M, Ventura M, Sánchez B, Margolles A. Intestinal dysbiosis associated with systemic lupus erythematosus. MBio. 2014;5:e01548-e01514. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 341] [Cited by in RCA: 444] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 10. | Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto JM, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K, Wang J. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3971] [Cited by in RCA: 4812] [Article Influence: 370.2] [Reference Citation Analysis (1)] |
| 11. | Wallace CA, Giannini EH, Huang B, Itert L, Ruperto N; Childhood Arthritis Rheumatology Research Alliance; Pediatric Rheumatology Collaborative Study Group; Paediatric Rheumatology International Trials Organisation. American College of Rheumatology provisional criteria for defining clinical inactive disease in select categories of juvenile idiopathic arthritis. Arthritis Care Res (Hoboken. ). 2011;63:929-936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 376] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 12. | Consolaro A, Ruperto N, Bazso A, Pistorio A, Magni-Manzoni S, Filocamo G, Malattia C, Viola S, Martini A, Ravelli A; Paediatric Rheumatology International Trials Organisation. Development and validation of a composite disease activity score for juvenile idiopathic arthritis. Arthritis Rheum. 2009;61:658-666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 441] [Cited by in RCA: 538] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 13. | Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957-2963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7679] [Cited by in RCA: 9171] [Article Influence: 655.1] [Reference Citation Analysis (0)] |
| 14. | Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335-336. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29318] [Cited by in RCA: 23675] [Article Influence: 1578.3] [Reference Citation Analysis (0)] |
| 15. | Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996-998. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8951] [Cited by in RCA: 9922] [Article Influence: 826.8] [Reference Citation Analysis (0)] |
| 16. | Berntson L, Agback P, Dicksved J. Changes in fecal microbiota and metabolomics in a child with juvenile idiopathic arthritis (JIA) responding to two treatment periods with exclusive enteral nutrition (EEN). Clin Rheumatol. 2016;35:1501-1506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 17. | Stoll ML, Kumar R, Morrow CD, Lefkowitz EJ, Cui X, Genin A, Cron RQ, Elson CO. Altered microbiota associated with abnormal humoral immune responses to commensal organisms in enthesitis-related arthritis. Arthritis Res Ther. 2014;16:486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 142] [Cited by in RCA: 151] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 18. | Tejesvi MV, Arvonen M, Kangas SM, Keskitalo PL, Pirttilä AM, Karttunen TJ, Vähäsalo P. Faecal microbiome in new-onset juvenile idiopathic arthritis. Eur J Clin Microbiol Infect Dis. 2016;35:363-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 74] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 19. | Aggarwal A, Sarangi AN, Gaur P, Shukla A, Aggarwal R. Gut microbiome in children with enthesitis-related arthritis in a developing country and the effect of probiotic administration. Clin Exp Immunol. 2017;187:480-489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 20. | He Z, Shao T, Li H, Xie Z, Wen C. Alterations of the gut microbiome in Chinese patients with systemic lupus erythematosus. Gut Pathog. 2016;8:64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 181] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 21. | Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, Casella G, Drew JC, Ilonen J, Knip M, Hyöty H, Veijola R, Simell T, Simell O, Neu J, Wasserfall CH, Schatz D, Atkinson MA, Triplett EW. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5:82-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 555] [Cited by in RCA: 603] [Article Influence: 40.2] [Reference Citation Analysis (0)] |
| 22. | Martínez-González O, Cantero-Hinojosa J, Paule-Sastre P, Gómez-Magán JC, Salvatierra-Ríos D. Intestinal permeability in patients with ankylosing spondylitis and their healthy relatives. Br J Rheumatol. 1994;33:644-647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 23. | Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54:1469-1476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1124] [Cited by in RCA: 1455] [Article Influence: 72.8] [Reference Citation Analysis (0)] |
| 24. | Sinkorová Z, Capková J, Niederlová J, Stepánková R, Sinkora J. Commensal intestinal bacterial strains trigger ankylosing enthesopathy of the ankle in inbred B10.BR (H-2(k)) male mice. Hum Immunol. 2008;69:845-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 55] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 25. | Picco P, Gattorno M, Marchese N, Vignola S, Sormani MP, Barabino A, Buoncompagni A. Increased gut permeability in juvenile chronic arthritides. A multivariate analysis of the diagnostic parameters. Clin Exp Rheumatol. 2000;18:773-778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 26. | Lionetti P, Pupi A, Veltroni M, Fonda C, Cavicchi MC, Azzari C, Falcini F. Evidence of subclinical intestinal inflammation by 99m technetium leukocyte scintigraphy in patients with HLA-B27 positive juvenile onset active spondyloarthropathy. J Rheumatol. 2000;27:1538-1541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 27. | Di Paola M, Cavalieri D, Albanese D, Sordo M, Pindo M, Donati C, Pagnini I, Giani T, Simonini G, Paladini A, Lionetti P, De Filippo C, Cimaz R. Alteration of Fecal Microbiota Profiles in Juvenile Idiopathic Arthritis. Associations with HLA-B27 Allele and Disease Status. Front Microbiol. 2016;7:1703. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 56] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 28. | Arvonen M, Berntson L, Pokka T, Karttunen TJ, Vähäsalo P, Stoll ML. Gut microbiota-host interactions and juvenile idiopathic arthritis. Pediatr Rheumatol Online J. 2016;14:44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 29. | Cao Y, Shen J, Ran ZH. Association between Faecalibacterium prausnitzii Reduction and Inflammatory Bowel Disease: A Meta-Analysis and Systematic Review of the Literature. Gastroenterol Res Pract. 2014;2014:872725. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 196] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 30. | Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottière HM, Doré J, Marteau P, Seksik P, Langella P. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731-16736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2747] [Cited by in RCA: 3181] [Article Influence: 187.1] [Reference Citation Analysis (0)] |
| 31. | van Dijkhuizen EHP, Del Chierico F, Malattia C, Russo A, Pires Marafon D, Ter Haar NM, Magni-Manzoni S, Vastert SJ, Dallapiccola B, Prakken B, Martini A, De Benedetti F, Putignani L; Model Driven Paediatric European Digital Repository Consortium. Microbiome Analytics of the Gut Microbiota in Patients With Juvenile Idiopathic Arthritis: A Longitudinal Observational Cohort Study. Arthritis Rheumatol. 2019;71:1000-1010. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 32. | Arvonen M, Virta LJ, Pokka T, Kröger L, Vähäsalo P. Repeated exposure to antibiotics in infancy: a predisposing factor for juvenile idiopathic arthritis or a sign of this group's greater susceptibility to infections? J Rheumatol. 2015;42:521-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 70] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 33. | Horton DB, Scott FI, Haynes K, Putt ME, Rose CD, Lewis JD, Strom BL. Antibiotic Exposure and Juvenile Idiopathic Arthritis: A Case-Control Study. Pediatrics. 2015;136:e333-e343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 106] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 34. | Davis-Richardson AG, Ardissone AN, Dias R, Simell V, Leonard MT, Kemppainen KM, Drew JC, Schatz D, Atkinson MA, Kolaczkowski B, Ilonen J, Knip M, Toppari J, Nurminen N, Hyöty H, Veijola R, Simell T, Mykkänen J, Simell O, Triplett EW. Bacteroides dorei dominates gut microbiome prior to autoimmunity in Finnish children at high risk for type 1 diabetes. Front Microbiol. 2014;5:678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 169] [Cited by in RCA: 209] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 35. | Li J, Zhao F, Wang Y, Chen J, Tao J, Tian G, Wu S, Liu W, Cui Q, Geng B, Zhang W, Weldon R, Auguste K, Yang L, Liu X, Chen L, Yang X, Zhu B, Cai J. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome. 2017;5:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 960] [Cited by in RCA: 1129] [Article Influence: 141.1] [Reference Citation Analysis (0)] |
| 36. | Martínez I, Lattimer JM, Hubach KL, Case JA, Yang J, Weber CG, Louk JA, Rose DJ, Kyureghian G, Peterson DA, Haub MD, Walter J. Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J. 2013;7:269-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 373] [Cited by in RCA: 408] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 37. | Badsha H. Role of Diet in Influencing Rheumatoid Arthritis Disease Activity. Open Rheumatol J. 2018;12:19-28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |