Published online May 16, 2018. doi: 10.12998/wjcc.v6.i5.64
Peer-review started: January 15, 2018
First decision: January 22, 2018
Revised: February 9, 2018
Accepted: March 7, 2018
Article in press: March 7, 2018
Published online: May 16, 2018
Processing time: 121 Days and 12.2 Hours
Helicobacter pylori (H. pylori) is a model organism for understanding host-pathogen interactions and infection-mediated carcinogenesis. Gastric cancer and H. pylori colonization indicates the strong correlation. The progression and exacerbation of H. pylori infection are influenced by some factors of pathogen and host. Several virulence factors involved in the proper adherence and attenuation of immune defense to contribute the risk of emerging gastric cancer, therefore analysis of them is very important. H. pylori also modulates inflammatory and autophagy process to intensify its pathogenicity. From the host regard, different genetic factors particularly affect the development of gastric cancer. Indeed, epigenetic modifications, MicroRNA and long non-coding RNA received more attention. Generally, various factors related to pathogen and host that modulate gastric cancer development in response to H. pylori need more attention due to develop an efficacious therapeutic intervention. Therefore, this paper will present a brief overview of host-pathogen interaction especially emphases on bacterial virulence factors, interruption of host cellular signaling, the role of epigenetic modifications and non-coding RNAs.
Core tip: As Helicobacter pylori (H. pylori) is a model organism for understanding host-pathogen interactions and infection-mediated carcinogenesis, ongoing studies in this area should have broad relevance to these conditions. In this context, we tried to review salient host and pathogen factors that influence on gastric cancer in H. pylori infection with emphasis on bacterial virulence factors, interruption of host cellular signaling, the role of epigenetic modifications and non-coding RNAs.
- Citation: Vaziri F, Tarashi S, Fateh A, Siadat SD. New insights of Helicobacter pylori host-pathogen interactions: The triangle of virulence factors, epigenetic modifications and non-coding RNAs. World J Clin Cases 2018; 6(5): 64-73
- URL: https://www.wjgnet.com/2307-8960/full/v6/i5/64.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v6.i5.64
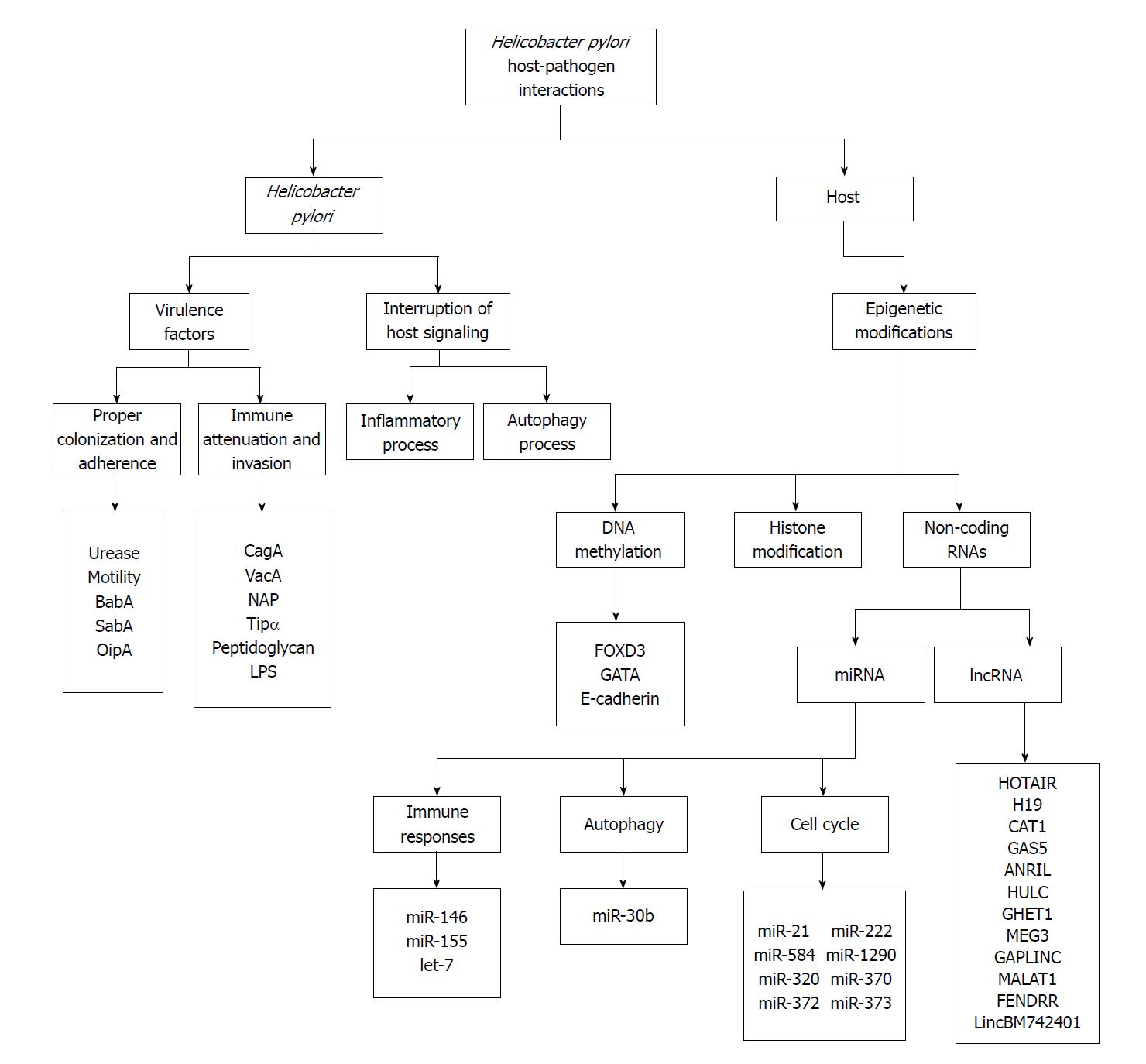
Helicobacter pylori (H. pylori) introduced as a pathogen colonized the gastric mucosa for at least 58000 years. The longtime of co-evolution between H. pylori and host can indicate that its virulence decreased over time[1]. But, this bacterium involved in the development of chronic gastritis, peptic ulcer, gastric carcinoma, colorectal cancers, and MALT lymphoma that can cause the high burden of morbidity and mortality[2,3]. Epidemiological studies have indicated that the half of the world’s population is infected with this organism and more than 10%-15% of infected individuals develop severe gastric diseases[4,5]. Numerous studies indicated H. pylori as a strong cause of gastric cancer[6]. This rod-shaped Gram-negative microorganism has classified as a carcinogen group I by the International Agency for Research on Cancer (IARC)[7]. Gastric cancer, the third reason for mortality because of cancer after lung and liver cancer, is diagnosed in over 950000 patients and caused more than 720000 deaths every year[8]. The risk of H. pylori on gastric cancer is estimated almost 74%[9]. However, carcinogenesis develop in only a very small proportion of H. pylori colonized individuals and just 1%-3% develops gastric cancer[10]. The reason for various virulence level between different H. pylori strains are unknown, but it can partly be explained by several factors such as H. pylori virulence or the bacterial genotypes, geographical regions, host genetic traits, and environmental influences[11]. The investigations indicated more virulent strains have more associated with the development of gastric cancer[12]. Overall, the virulence factors, signaling pathways, and some host genetic traits received more attention. Clearly, the progression and exacerbation of H. pylori infection are influenced by some factors of pathogen and host. However, the precise mechanisms related to pathogen and host that modulate gastric cancer development in response to H. pylori need more attention. Therefore, this paper will present a brief overview of host-pathogen interaction especially associated with gastric cancer development in four parts: H. pylori virulence factors, interruption of host signaling, the role of epigenetic modifications and non-coding RNAs. The diagram of mentioned parts in association with gastric cancer presented in the Figure 1.
Several virulence factor genes contribute the high heterogeneity of H. pylori[13]. Virulence factors increase the risk of emerging gastric carcinoma, therefore analysis of them in each region is very important[14]. H. pylori colonization in a niche near the surface of epithelial cells are known as the strongest but not sufficient risk factor, in this multi-factorial disease. The specific niche facilitates recognition of organisms by the immune system and H. pylori can modulate the inflammatory responses for its benefits[5]. Urease producing, motility and chemotaxis count as important properties of H. pylori to survive in the acidic environment of the stomach[12]. H. pylori able to produce a high level of urease into the stomach, which provides protection from gastric acidity. Urease produces NH3 and CO2 from urea and disrupts the epithelium by the production of ammonia. Ammonia interacts with neutrophil metabolites, induces the formation of carcinogenic agents and therefore increase the risk of gastric cancer. Besides, urease induces production of inflammatory cytokines[15]. Urease interacts with HLA class II molecules and CD74 on gastric cells. HLA class II molecules involved in regulation of immune responses and CD74 coordinate in antigen processing. Three genes code HLA class II molecules, HLA-DP, HLA-DQ, and HLA-DR, and more than 100 variant alleles have been detected. Some of these specific alleles are associated with gastric cancer[16]. Moreover, four to six polar flagella are essential for H. pylori motility[15].
In addition, H. pylori produces several effectors to intensify its pathogenicity. The CagA and specific alleles of VacA known as two best-studied virulence factors associated with the development of gastric cancer[1]. The CagA protein, encoded by the cytotoxin-associated genes (cag) pathogenicity island (PAI), carried by the most virulent H. pylori strains and the existence of cagA gene has a crucial role in the association of gastric cancer rates[17,18]. CagA introduced as the first bacterial oncoprotein. The type IV secretion system (T4SS) produced by the cagPAI and conveyed the CagA and some effector molecules like peptidoglycan into the cytoplasm of host’s gastric epithelial cells[19]. The imported CagA phosphorylated (and also remains un-phosphorylated) and initiates a signaling cascade that can perturb many host cell-signaling pathways and trigger pro-inflammatory responses, alteration of the cell polarity, disruption of the epithelial barrier and cytoskeletal rearrangements and cell elongation, induction of hummingbird phenotype, and promote the transformation of gastric epithelial cells[20]. Besides, CagA polymorphism is notable that may influence on severity of disease. The C-terminal of CagA contains specific repeated amino acids, including Glu-Pro-Ile-Tyr-Ala, that known as EPIYA motif[21]. The EPIYA motifs identified in 1993[22], including four different groups (named A, B, C, and D) based on the flanking amino acid arrangements and their position in CagA. The number and types of repeats in this region also influence on risk of carcinogenesis[23]. Overall, this virulence factor is well studied and much of the related process well described. Western CagA and East Asian CagA strains known as two main genotypes of CagA that distinguished based on the structure of its EPIYA motif profiles. Western CagA strains usually have the EPIYA-A, EPIYA-B and EPIYA-C sequences in the EPIYA repeat region and East Asian CagA strains have just EPIYA-A and EPIYA-B without the EPIYA-C segment, but they have a specific EPIYA-D segment[24]. Almost 70% and 90% of strains in some western and Asian countries presented the CagA virulence factor, respectively[25,26]. Infections with positive CagA isolates have a high risk of severe disorders in comparison to infections with negative ones[27]. Therefore, it can partly explain that why the highest incidence of gastric cancer reported from East Asia countries[28]. All H. pylori strains virtually produce a pore-forming protein, vacuolating cytotoxin (VacA) protein. The VacA uptakes by the host cells and forms vacuoles in the cytoplasm. The receptor protein tyrosine phosphatase (RPTP), RPTP-α and RPTP-β as the cell surface receptors, play an important role in recognition and secretion of VacA[29]. The function of VacA effectively depends on induction of membrane channel forming that influence on ion transport in the host cells, alteration in the cell membrane permeability, apoptosis, secretion of pro-inflammatory cytokines, modulation of immune cell function and induction of immunosuppressing[30]. The various regions of VacA include the s-type (signal region), m-type (middle region), i-type (intermediate region), and d-type (deletion region)[31]. The region of s, m, and i determine the formation of the channel, tropism to the host cells, and carcinogenic and vacuolating activity of VacA toxin, respectively. Deletion of 81 bp between the region of i and m known as d-type[32]. Each of these regions subdivided. Allelic diversity influences the level of pathogenicity by the mosaic recombination between two major alleles of them (s1, s2, i1, i2, m1, m2). For instance, vacuolating strains have mostly vacA s1/m1 and vacA s1/m2 with i1 alleles. While non-vacuolating strains have vacA s2/m2 and vacA s1/m2 with i2 alleles[15]. The vacA s1/m1 is strongly correlated with the development of gastric cancer. Actually, presence of strains with s1, m1, and i1 alleles are highly associated with the production of active toxins and gastric cancer[28,33]. In addition, all s1 strains almost belong to positive CagA isolates and all s2/m2 strains belong to negative CagA isolates[34].
Other virulence factors produced by H. pylori, including BabA, SabA, OipA, NAP, Tipα, peptidoglycan, and LPS, are associated with the progression of infection. The blood-group antigen binding adhesion (BabA) and sialic acid binding protein A (SabA) count as two major outer membrane proteins (OMPs) that can serve as adhesion. BabA was found as a prominent adhesion of H. pylori. BabA binds to fucosylated glycoconjugates containing ABO/Lewis b blood group antigens on the gastric cells. SabA also is known as a key adhesion of H. pylori. SabA binds to specific components appeared in inflammation manner, a sialyl-Lewis x antigen on inflamed cells and MUC5B, which expressed only in a diseased manner. The attachment of this virulence factor increases survival chance of organism, helps to escape from sites with high bactericidal effects and increases accessibility to nutrients[35]. Overall, the efficient binding of H. pylori in different conditions provides a proper situation for efficient delivery of effector molecules and modulation of host cells[16]. Another major OMP in H. pylori are known as Outer inflammatory protein (OipA). According to Yamaoka’s research, OipA is associated with IL-8 levels[36]. OipA induces secretion of IL-8 from gastric cells and plays a proinflammatory role in H. pylori infection[15]. OipA also involves in bacterial colonization[37]. The neutrophil-activating protein (NAP) modulates the oxidative burst in neutrophils and induces inflammation during the H. pylori infection[15]. Tipα (TNF-α inducing protein) is a novel effector related to carcinogenesis. Tipα ligates to a cell surface receptor encoded by NCL gene, nucleolin, and transfer to the nucleus. Nucleolin performs several functions in the nucleous, including rRNA processing, chromatin remodeling, mRNA stabilization, DNA recombination, which may increase the risk of gastric cancer[38]. In the nucleus of host cells, Tipα also binds to a different form of DNA and induces the expression of TNF-α and chemokines[16]. In addition to proper adherence, attenuation of immune defense is a key step in the development of gastric cancer. The peptidoglycan and lipopolysaccharide (LPS), two the most important microbial associated molecular patterns (MAMPs) of H. pylori, ligate to pattern recognition receptors (PRRs). Thereby, H. pylori preserved from immune detection by various mechanisms such as phase variation, structural modification, molecular mimicry, and morphological transition that increased its persistence[39]. Indeed, H. pylori virulence factors play a central role in the intensification of infection. Already, the impact of these virulence factors has been reviewed thoroughly[28,40,41].
In addition to virulence factors, H. pylori induces some modifications in host signaling cascades to intensify pathogenicity. In this regard, modulation of inflammatory and autophagy process are so interesting. H. pylori produces several inflammatory mediators, such as peptidoglycan, NAP, and LPS that increase the risk of oncogenesis[12]. Environmental factors, including smoking, alcoholism, high intake of salt and low intake of fruits and vegetables can effect on activation of inflammatory signaling and secretion of cytokines and chemokines. Several PRRs recognize the MAMPs and damage associated molecular patterns (DAMP) that lead to the initiation of inflammatory responses. PRR generates signals to activate activator protein 1 (AP-1) and nuclear factor kappa-B (NF-κB) that finally produces related cytokines and chemokines. In addition, signaling of PRR leads to activation of some mechanisms for clearance of H. pylori. Furthermore, signaling of PRR plays both useful and harmful roles in hosts, clearance of pathogens and induction of carcinogenesis[12]. Followed by the production of pro-inflammatory cytokines and chemokines, macrophages and granulocytes attract to the infection site and produce reactive nitrogen species (RNS) and reactive oxygen species (ROS). Moreover, H. pylori can directly induces production of ROS in gastric cells[42]. These factors induce DNA damage and lead to oncogenic mutations[43]. The inflammation initially caused by hypergastrinemia and destruction of D-cells. If inflammation persists, tissue damage because of gastrin level and hypochlorhydria occurs[44] that can directly induce carcinogenic effects. Increasing gastrin level due to inflammation can directly heighten the risk of carcinogenesis. Gastrin binds to the cholecystokinin-2 receptor (CCK-2R), activates phosphoinositide3 kinase (PI3K)/Akt and JAK-STAT3 signaling pathways, and effect on adherence of host cells[45]. CCK-2R upregulates in different types of cancers[46]. This event cause loss of acid producing cells and deregulates expression of some growth factors that influence on cell differentiation[47]. This deregulation may influence on the development of carcinogenesis[12].
In addition, autophagy as a self-degrading mechanism counts significant in H. pylori pathogenicity. In autophagy process, cytoplasmic components are degraded in the lysosomes[48]. In the autophagy process, ULK1 kinase complex (containing ULK1-mAtg13, FIP200, and ATG101) formed by induction of IRGM to form autophagy. Then, autophagosome formed by ATG9 and the ATG2-WIPI1/2 complex function. The vesicles assembly by the class III phosphatidylinositol 3-kinase complex[12,49]. Autophagy count as an essential process in function of antigen-presenting cells and activation of inflammatory responses[50]. Although autophagy has useful effects for the host, H. pylori can use some mechanisms to modulate it. VacA induces autophagy and this event can lead to interference of the autophagy process and production of defective autophagosome particles[51]. On the other hand, autophagy degrades the CagA[52]. Therefore, VacA help to CagA for the promotion of carcinogenesis by disruption in the autophagy[53]. Generally, autophagy plays a complex role in the carcinogenesis. Prolonged or forced autophagy formation increase the risk of host cell death and carcinogenesis[51].
From the side of host, various significant changes create in H. pylori infection. Numerous studies indicated, different polymorphisms in the host genes have been associated with differing risk of gastric cancer[54,55]. This paper provides a brief of some host genetic factors that particularly affect the development of gastric cancer. Many of host factors related to the severity of H. pylori infection have been discussed elsewhere[41,56]. Indeed, epigenetic modifications, MicroRNA and long non-coding RNA, which has influence on the severity of H. pylori infection and gastric cancer development, will be discussed.
A growing area of interest is the investigation of epigenetic modifications in the pathogenicity of H. pylori. Epigenetic mechanisms regulate gene expression independent of direct modification of DNA sequence. Several types of epigenetic mechanisms are identified. First, methylation of DNA in cytosine or adenosine nucleotides by DNA methyltransferases. DNA methylation predominantly occurs on CpG islands and is associated with gene silencing. Second, histone modifications by phosphorylation, methylation, or acetylation regulate the accessibility of DNA to transcriptional factors and gene expression. Finally, chromatin remodeling and non-coding RNAs (ncRNA) which recently count as other major levels in epigenetic control; ncRNA is separately addressed in this review[57,58]. Modification of transcriptional profile of the host cells has been found as pathogen strategies to modulate host cells by various mechanisms to their benefits. Chronic exposure to H. pylori enhances DNA methylation and histone modifications particularly in the promoter region of tumor suppressor genes and oncogenes, that leads to silencing of them[57,59]. Induction of these epigenetic modifications by H. pylori is linked with oncogenesis and development of gastric cancer[60]. As mentioned, various types of cytokines, ROS, and RNS generate upon H. pylori infection that probably induces activation of DNA methyltransferases, may induce gene silencing[61]. Several studies have been published associating H. pylori infection with abnormally methylated genes in gastric cancer cases[62]. The more important of them include genes associated with cell growth, apc, p14(ARF), and p16(INK4a); the E-cadherin genes as cell adherence, cdh1, flnc, hand1, lox, hrasls, thbd, and p41ARC; genes associated DNA repair, brca1, mgmt, and hMLH1; and several other genes with unknown correlation with H. pylori infection[58,60,61,63]. For example, aberrant methylation in FOXD3, a fork head transcription factor with a tumor suppressor function, correlated with gastric cancer development. FOXD3 normally regulates transcription of proapoptotic factors. Therefore, hypermethylation of related promoter in gastric cancer inhibits activation FOXD3 and suppresses apoptosis[64]. Additional, GATA is known as a transcriptional family with six members that involved in host cells development. GATA2 is crucial for the development of hematopoietic cells whereas GATA6 is essential for the differential of gastrointestinal. Interestingly, hypermethylation of GATA2 repressed its expression while GATA6 overexpressed in gastric cancer cells[65]. Methylation-dependent silencing of the E-cadherin gene, a tumor suppressor gene, through IL-1β also known as main epigenetic changes in gastric cancer development. IL-1β stimulates NF-κB pathway that leads to activation of DNA methyltransferases and methylation of E-cadherin gene[66]. Besides, histone modifications and chromatin accessibility to transcription factors have been found significant in the progression of gastric cancer. Wide ranges of histone modifications that influence on tumor suppressor genes and oncogenes in response to H. pylori infection have been explored[58,67]. Modification in the phosphorylation status of H3 is associated with cell cycle arrest which induced by H. pylori. It can cause to inhibition of gastric cell renewal[68]. H. pylori also induces dephosphorylation of H3S10 and inhibits expression of NF-κB responsive genes[69]. Epigenetic change of the tumor suppressor protein p27 is linked with chronic H. pylori infection. Induction of histone acetylation of p27 gene by stimulation of a specific G-protein leads to the reduction of its expression in gastric cancer[67]. A study has shown that nucleosome repositioning in CpG island p16 occurs in response to gastric cancer which induced by H. pylori. Nevertheless, interpretation of relevance of this data is difficult due to lack of more genome-wide investigations[70].
As mentioned, one of the most interesting fields in host-pathogen interaction is gene expression regulation, especially by RNA. RNA counted as a key regulatory molecule and non-coding RNAs (ncRNA) recently added to the list of epigenetic regulators. Recognition by base-pairing allows one single ncRNA to bind multiple targets, and thereby to regulate several pathways simultaneously[71]. The ncRNA classified into two classes, microRNAs (miRNA) and long non-coding RNAs (lncRNA). The miRNA and lncRNA typically include 21-24 and more than 200 transcribed nucleotides by RNA polymerase II, respectively. To date, several distinct miRNA and lncRNA have been introduced that involved in gene expression regulation in different manners[72,73]. Some lncRNAs introduced which act in the response against bacterial infections. In bacterial infection, host cells employed miRNA and lncRNA to adjust gene expression program. Equally, pathogens employed various strategies, particularly by targeting ncRNAs, to overcome host defense mechanisms. Various pathogens activate expression of specific miRNAs in the host cells[74]. The bacteria manipulate the defense mechanisms of host cells by particularly modulating production of miRNA in order to increase pathogen survival. The main processes manipulated by down regulation or up regulation of various miRNAs including immune responses, autophagy, cell cycle and apoptosis[75]. The most important of these miRNAs include miR-146, miR-155, and let-7 family, which influence on immune responses in order to bacterial clearance[76]. The miR-146 and miR-155 are two main NF-κB-dependent miRNAs. The PRR activates NF-κB pathway by sensing of MAMPs and induced production of miR-155 and miR-146 that regulate distinct genes during infections[77,78]. The miR-146 counts as an anti-inflammatory regulator and targets IRAK1 (IL-1R-associated kinase 1) and TRAF6 (TNF Receptor-associated factor 6) in the NF-κB pathway that increases tolerance to a low dose of LPS[78]. The role of miR-146 is also crucial for intestinal microbiota due to the prevention of inappropriate inflammation[79]. The miR-155 induces expression of pro-inflammatory cytokines, including TNF-α, IL-6, IL-1β, IL-8, and IL-12, and acts as an essential factor against infections[39,78]. Besides, miR-155 involved in the development of T helper cells and autophagy by suppressing the mTOR pathway[80]. This miRNA represses some genes, NIK, IKKε, and TAB; encoding proteins involved in the inflammatory pathway and limits the inflammation. Interestingly, NOD2 receptor stimulated the expression of miR-155. Generally, miR-155 acts as a negative regulator to adjust inflammatory responses in H. pylori infection[75]. Another miRNA, which targets various genes in immunity, is let-7. Upon infections, let-7 repressed by activation of NF-κB pathway because of exposure to LPS[76]. The NF-κB pathway induces expression of Lin-28B, which inhibit maturation of let-7. In addition, some bacteria directly repress this miRNA[81,82].
H. pylori induces expression of miR-30b, which influences on the transcript of proteins that involved in the formation of autophagosomes and so its effects on autophagy process[83]. In the following, up regulation or down regulation of specific miRNAs involved in different steps of cell cycle described. Overexpression of miR-21 and miR-222 that target RECK, a tumor suppressor, induce proliferation of gastric cells[84,85]. CagA induces the expression of miR-584 and miR-1290 that influence on epithelial-mesenchymal transition by targeting of FOXA1, a related negative regulator[86]. In addition, down regulation of miR-320 and miR-370 in a CagA dependent manner also noticed in H. pylori infections. The miR-320 induces expression of MCL1, an anti-apoptotic gene. The miR-370 down regulates expression of FoxM1 and subsequently activates p27KIP1, a cell cycle inhibitor. Therefore, down regulation of miR-320 and miR-370 can lead to tumor suppression through decreasing apoptosis and increasing cell proliferation[87,88]. CagA of H. pylori activates NF-κB pathway and downregulates expression of miR-372 and miR-373, which inhibit renewal of gastric epithelium by blocking of cell cycle progression in the G1-S checkpoint[89]. H. pylori also induces expression of miR-1289 in a CagA dependent manner, which finally decrease gastric acidity and increase the possibility of H. pylori colonization[90]. These data suggested a link between H. pylori infection and gastric cancer development.
Most investigations focused on the role of lncRNAs in host-pathogen interaction on viral infections, but its role in inflammatory responses has recently elucidated. The regulatory lncRNAs participate in almost every part of gene expression and can interact with DNA, RNA, and protein. Therefore, disruption in the expression of lncRNAs and subsequently alteration of cellular pathways have been discovered in gastric cancer studies[91]. In the following, briefly described some of the most important oncogenic lncRNAs involved in cell proliferation, apoptosis, and metastasis processes in gastric cancer. For instance, the aberrant expression GAPLINC (gastric adenocarcinoma predictive long intergenic noncoding RNA) markedly correlated with alteration of CD44 and thereby increased proliferation and angiogenesis of cancer cells. GAPLINC is known as a decoy molecule to protect CD44 from degradation[92]. Upregulation of HOTAIR, ANRIL, and GHET1 has been found in gastric cancer. This upregulation directly related to cell proliferation, invasion, and progression of cancer[93-95]. The lncRNA H19 overexpressed in gastric cancer. The production of miR-675 related to H19 that directly silences certain tumor suppressor, RUNX1, and increases cell proliferation and inhibits apoptosis[96]. H19 directly inhibit the function of p53, a tumor suppressor molecule, and leading to cell proliferation[97]. CCAT1 overexpressed in gastric cancer and promoted cell proliferation[98]. Overexpression of MALAT1 induced localization of SF2/ASF proteins in the nucleus that involved in splicing. Therefore, MALAT1 may partly modulate cell proliferation by regulation of SF2/ASF expression[99]. Overexpression of HULC well described in hepatocellular carcinoma, but the high level of it also shown in gastric cancer and related to proliferation[100]. Downregulation of FENDRR (FOXF1 adjacent non-coding developmental regulatory RNA) is associated with cell invasion and migration in gastric cancer[101]. Downregulation of GAS5 (growth arrest-specific transcript 5) plays an important role in the proliferation of gastric cells. GAS5 interacts with YBX1, a transcriptional activator, and subsequently induces expression of p21. Therefore, downregulation of GAS5 eventually leads to abolishing of cell cycle arrest[91]. Downregulation of MEG3 (maternally expressed gene 3) is correlated with cell proliferation and inhibition of apoptosis in gastric cancer cells[102]. Downregulation of LincBM742401 has been closely associated with cell metastasis[103]. To achieve better understanding of the importance of lncRNAs in H. pylori infection more investigations are inevitable[104,105]. In fact, accumulating data indicate participation and collaboration of miRNAs and lncRNAs to modulate gene expression and gastric cancer development.
In summary, nowadays, the knowledge of factors involved in H. pylori disease pathogenesis continues to be elucidated and refined. We tried to review salient host and pathogen factors that influence on gastric cancer in H. pylori infection. Ultimately, the development of efficacious therapeutic interventions will likely need to switch host-pathogen interactions science to translational research for enhancing host immunity and circumvent bacterial evasion strategies.
We tender our apologies to those authors whose deserving research was not cited in this manuscript.
Manuscript source: Invited manuscript
Specialty type: Medicine, research and experimental
Country of origin: Iran
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): C, C, C, C
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: Bernal G, Bugaj AM, Buzas GM, Dinc T, Engin AB, Niu ZS, Paoluzi OA, Zhu YL S- Editor: Cui LJ L- Editor: A E- Editor: Wang CH
| 1. | Amieva M, Peek RM Jr. Pathobiology of Helicobacter pylori-Induced Gastric Cancer. Gastroenterology. 2016;150:64-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 633] [Cited by in RCA: 635] [Article Influence: 70.6] [Reference Citation Analysis (0)] |
| 2. | Miftahussurur M, Yamaoka Y, Graham DY. Helicobacter pylori as an oncogenic pathogen, revisited. Expert Rev Mol Med. 2017;19:e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 3. | Engin AB, Karahalil B, Karakaya AE, Engin A. Helicobacter pylori and serum kynurenine-tryptophan ratio in patients with colorectal cancer. World J Gastroenterol. 2015;21:3636-3643. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 36] [Cited by in RCA: 49] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 4. | Pachathundikandi SK, Lind J, Tegtmeyer N, El-Omar EM, Backert S. Interplay of the Gastric Pathogen Helicobacter pylori with Toll-Like Receptors. Biomed Res Int. 2015;2015:192420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 5. | Lind J, Backert S, Hoffmann R, Eichler J, Yamaoka Y, Perez-Perez GI, Torres J, Sticht H, Tegtmeyer N. Systematic analysis of phosphotyrosine antibodies recognizing single phosphorylated EPIYA-motifs in CagA of East Asian-type Helicobacter pylori strains. BMC Microbiol. 2016;16:201. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 6. | Cover TL. Helicobacter pylori Diversity and Gastric Cancer Risk. MBio. 2016;7:e01869-e01815. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 146] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 7. | Schistosomes, liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7-14 June 1994. IARC Monogr Eval Carcinog Risks Hum. 1994;61:1-241. [PubMed] |
| 8. | Stewart B, Wild CP. World cancer report 2014. Health. 2017;. |
| 9. | Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030-3044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1946] [Cited by in RCA: 1970] [Article Influence: 103.7] [Reference Citation Analysis (0)] |
| 10. | Giorgio A, Iacoviello G, D’Avino A. Biology of the “Salmonella wien” (author’s transl). Ann Sclavo. 1976;18:563-573. [PubMed] |
| 11. | Li Q, Liu J, Gong Y, Yuan Y. Association of CagA EPIYA-D or EPIYA-C phosphorylation sites with peptic ulcer and gastric cancer risks: A meta-analysis. Medicine (Baltimore). 2017;96:e6620. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 12. | Mommersteeg MC, Yu J, Peppelenbosch MP, Fuhler GM. Genetic host factors in Helicobacter pylori-induced carcinogenesis: Emerging new paradigms. Biochim Biophys Acta. 2018;1869:42-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 13. | Bagheri N, Azadegan-Dehkordi F, Rafieian-Kopaei M, Rahimian G, Asadi-Samani M, Shirzad H. Clinical relevance of Helicobacter pylori virulence factors in Iranian patients with gastrointestinal diseases. Microb Pathog. 2016;100:154-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 14. | Vaziri F, Najar Peerayeh S, Alebouyeh M, Mirzaei T, Yamaoka Y, Molaei M, Maghsoudi N, Zali MR. Diversity of Helicobacter pylori genotypes in Iranian patients with different gastroduodenal disorders. World J Gastroenterol. 2013;19:5685-5692. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 34] [Cited by in RCA: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 15. | Roesler BM, Rabelo-Gonçalves EM, Zeitune JM. Virulence Factors of Helicobacter pylori: A Review. Clin Med Insights Gastroenterol. 2014;7:9-17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 101] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 16. | He C, Chen M, Liu J, Yuan Y. Host genetic factors respond to pathogenic step-specific virulence factors of Helicobacter pylori in gastric carcinogenesis. Mutat Res Rev Mutat Res. 2014;759:14-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 17. | Yamaoka Y, Kodama T, Kashima K, Graham DY, Sepulveda AR. Variants of the 3’ region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases. J Clin Microbiol. 1998;36:2258-2263. [PubMed] |
| 18. | Vaziri F, Peerayeh SN, Alebouyeh M, Maghsoudi N, Azimzadeh P, Siadat SD, Zali MR. Novel effects of Helicobacter pylori CagA on key genes of gastric cancer signal transduction: a comparative transfection study. Pathog Dis. 2015;73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 19. | Hatakeyama M. Structure and function of Helicobacter pylori CagA, the first-identified bacterial protein involved in human cancer. Proc Jpn Acad Ser B Phys Biol Sci. 2017;93:196-219. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 156] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 20. | Backert S, Tegtmeyer N, Selbach M. The versatility of Helicobacter pylori CagA effector protein functions: The master key hypothesis. Helicobacter. 2010;15:163-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 180] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 21. | Backert S, Blaser MJ. The Role of CagA in the Gastric Biology of Helicobacter pylori. Cancer Res. 2016;76:4028-4031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 66] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 22. | Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993;90:5791-5795. [PubMed] |
| 23. | Vaziri F, Najar Peerayeh S, Alebouyeh M, Molaei M, Maghsoudi N, Zali MR. Determination of Helicobacter pylori CagA EPIYA types in Iranian isolates with different gastroduodenal disorders. Infect Genet Evol. 2013;17:101-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 24. | Vianna JS, Ramis IB, Halicki PC, Gastal OL, Silva RA, Junior JS, Dos Santos DM, Chaves AL, Juliano CR, Jannke HA. Detection of Helicobacter pylori CagA EPIYA in gastric biopsy specimens and its relation to gastric diseases. Diagn Microbiol Infect Dis. 2015;83:89-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 25. | Ferreira RM, Pinto-Ribeiro I, Wen X, Marcos-Pinto R, Dinis-Ribeiro M, Carneiro F, Figueiredo C. Helicobacter pylori cagA Promoter Region Sequences Influence CagA Expression and Interleukin 8 Secretion. J Infect Dis. 2016;213:669-673. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 26. | Sayehmiri F, Kiani F, Sayehmiri K, Soroush S, Asadollahi K, Alikhani MY, Delpisheh A, Emaneini M, Bogdanović L, Varzi AM. Prevalence of cagA and vacA among Helicobacter pylori-infected patients in Iran: a systematic review and meta-analysis. J Infect Dev Ctries. 2015;9:686-696. [PubMed] |
| 27. | Argent RH, Kidd M, Owen RJ, Thomas RJ, Limb MC, Atherton JC. Determinants and consequences of different levels of CagA phosphorylation for clinical isolates of Helicobacter pylori. Gastroenterology. 2004;127:514-523. [PubMed] |
| 28. | Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat Rev Gastroenterol Hepatol. 2010;7:629-641. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 500] [Cited by in RCA: 458] [Article Influence: 30.5] [Reference Citation Analysis (1)] |
| 29. | Sewald X, Fischer W, Haas R. Sticky socks: Helicobacter pylori VacA takes shape. Trends Microbiol. 2008;16:89-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 30. | Isomoto H, Moss J, Hirayama T. Pleiotropic actions of Helicobacter pylori vacuolating cytotoxin, VacA. Tohoku J Exp Med. 2010;220:3-14. [PubMed] |
| 31. | Cid TP, Fernández MC, Benito Martínez S, Jones NL. Pathogenesis of Helicobacter pylori infection. Helicobacter. 2013;18 Suppl 1:12-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 32. | Chmiela M, Karwowska Z, Gonciarz W, Allushi B, Stączek P. Host pathogen interactions in Helicobacter pylori related gastric cancer. World J Gastroenterol. 2017;23:1521-1540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 79] [Cited by in RCA: 91] [Article Influence: 11.4] [Reference Citation Analysis (1)] |
| 33. | Rhead JL, Letley DP, Mohammadi M, Hussein N, Mohagheghi MA, Eshagh Hosseini M, Atherton JC. A new Helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology. 2007;133:926-936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 283] [Cited by in RCA: 312] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 34. | Atherton JC, Cao P, Peek RM Jr, Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771-17777. [PubMed] |
| 35. | Talarico S, Whitefield SE, Fero J, Haas R, Salama NR. Regulation of Helicobacter pylori adherence by gene conversion. Mol Microbiol. 2012;84:1050-1061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 36. | Yamaoka Y, Kodama T, Graham DY, Kashima K. Search for putative virulence factors of Helicobacter pylori: the low-molecular-weight (33-35 K) antigen. Dig Dis Sci. 1998;43:1482-1487. [PubMed] |
| 37. | Dossumbekova A, Prinz C, Mages J, Lang R, Kusters JG, Van Vliet AH, Reindl W, Backert S, Saur D, Schmid RM. Helicobacter pylori HopH (OipA) and bacterial pathogenicity: genetic and functional genomic analysis of hopH gene polymorphisms. J Infect Dis. 2006;194:1346-1355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 116] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 38. | Tajrishi MM, Tuteja R, Tuteja N. Nucleolin: The most abundant multifunctional phosphoprotein of nucleolus. Commun Integr Biol. 2011;4:267-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 184] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 39. | Billeter AT, Hellmann J, Roberts H, Druen D, Gardner SA, Sarojini H, Galandiuk S, Chien S, Bhatnagar A, Spite M. MicroRNA-155 potentiates the inflammatory response in hypothermia by suppressing IL-10 production. FASEB J. 2014;28:5322-5336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 40. | Peek RM Jr, Fiske C, Wilson KT. Role of innate immunity in Helicobacter pylori-induced gastric malignancy. Physiol Rev. 2010;90:831-858. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 184] [Cited by in RCA: 175] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 41. | Sutton P, Mitchell H. Helicobacter pylori in the 21st Century. CABI. 2010;. |
| 42. | Kawahara T, Kohjima M, Kuwano Y, Mino H, Teshima-Kondo S, Takeya R, Tsunawaki S, Wada A, Sumimoto H, Rokutan K. Helicobacter pylori lipopolysaccharide activates Rac1 and transcription of NADPH oxidase Nox1 and its organizer NOXO1 in guinea pig gastric mucosal cells. Am J Physiol Cell Physiol. 2005;288:C450-C457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 120] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 43. | Kawanishi S, Ohnishi S, Ma N, Hiraku Y, Oikawa S, Murata M. Nitrative and oxidative DNA damage in infection-related carcinogenesis in relation to cancer stem cells. Genes Environ. 2017;38:26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 70] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 44. | Malfertheiner P. The intriguing relationship of Helicobacter pylori infection and acid secretion in peptic ulcer disease and gastric cancer. Dig Dis. 2011;29:459-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 63] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 45. | Smith JP, Nadella S, Osborne N. Gastrin and Gastric Cancer. Cell Mol Gastroenterol Hepatol. 2017;4:75-83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 46. | Roy J, Putt KS, Coppola D, Leon ME, Khalil FK, Centeno BA, Clark N, Stark VE, Morse DL, Low PS. Assessment of cholecystokinin 2 receptor (CCK2R) in neoplastic tissue. Oncotarget. 2016;7:14605-14615. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 47. | Koh TJ, Chen D. Gastrin as a growth factor in the gastrointestinal tract. Regul Pept. 2000;93:37-44. [PubMed] |
| 48. | Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861-2873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2692] [Cited by in RCA: 3065] [Article Influence: 170.3] [Reference Citation Analysis (0)] |
| 49. | Chauhan S, Mandell MA, Deretic V. IRGM governs the core autophagy machinery to conduct antimicrobial defense. Mol Cell. 2015;58:507-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 189] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 50. | Shibutani ST, Saitoh T, Nowag H, Münz C, Yoshimori T. Autophagy and autophagy-related proteins in the immune system. Nat Immunol. 2015;16:1014-1024. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 429] [Article Influence: 42.9] [Reference Citation Analysis (0)] |
| 51. | Terebiznik MR, Raju D, Vázquez CL, Torbricki K, Kulkarni R, Blanke SR, Yoshimori T, Colombo MI, Jones NL. Effect of Helicobacter pylori’s vacuolating cytotoxin on the autophagy pathway in gastric epithelial cells. Autophagy. 2009;5:370-379. [PubMed] |
| 52. | Jones KR, Whitmire JM, Merrell DS. A Tale of Two Toxins: Helicobacter Pylori CagA and VacA Modulate Host Pathways that Impact Disease. Front Microbiol. 2010;1:115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 126] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 53. | Júnior MF, Batista SA, Barbuto RC, Gomes AD, Queiroz DM, Araújo ID, Caliari MV. CagA-positive Helicobacter pylori strain containing three EPIYA C phosphorylation sites produces increase of G cell and decrease of D cell in experimentally infected gerbils (Meriones unguiculatus). Adv Med Sci. 2016;61:231-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 54. | Ma J, Wu D, Hu X, Li J, Cao M, Dong W. Associations between cytokine gene polymorphisms and susceptibility to Helicobacter pylori infection and Helicobacter pylori related gastric cancer, peptic ulcer disease: A meta-analysis. PLoS One. 2017;12:e0176463. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 55. | Melchiades JL, Zabaglia LM, Sallas ML, Orcini WA, Chen E, Smith MAC, Payão SLM, Rasmussen LT. Polymorphisms and haplotypes of the interleukin 2 gene are associated with an increased risk of gastric cancer. The possible involvement of Helicobacter pylori. Cytokine. 2017;96:203-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 56. | Fulurija A, Lundin S, Ee H, Kumarasinghe MP, Marshall BJ. Unravelling the Immune Response to Helicobacter Pylori: Results from a Human Challenge Study. Gastroenterology. 2017;152:667-668. [DOI] [Full Text] |
| 57. | Silmon de Monerri NC, Kim K. Pathogens hijack the epigenome: a new twist on host-pathogen interactions. Am J Pathol. 2014;184:897-911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 98] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 58. | Ding SZ, Goldberg JB, Hatakeyama M. Helicobacter pylori infection, oncogenic pathways and epigenetic mechanisms in gastric carcinogenesis. Future Oncol. 2010;6:851-862. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 128] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 59. | Gorrell R, Kwok T. The Helicobacter pylori Methylome: Roles in Gene Regulation and Virulence. Curr Top Microbiol Immunol. 2017;400:105-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 60. | Sitaraman R. Helicobacter pylori DNA methyltransferases and the epigenetic field effect in cancerization. Front Microbiol. 2014;5:115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 61. | Nardone G, Compare D, De Colibus P, de Nucci G, Rocco A. Helicobacter pylori and epigenetic mechanisms underlying gastric carcinogenesis. Dig Dis. 2007;25:225-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 35] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 62. | Santos JC, Ribeiro ML. Epigenetic regulation of DNA repair machinery in Helicobacter pylori-induced gastric carcinogenesis. World J Gastroenterol. 2015;21:9021-9037. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 13] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (1)] |
| 63. | Maekita T, Nakazawa K, Mihara M, Nakajima T, Yanaoka K, Iguchi M, Arii K, Kaneda A, Tsukamoto T, Tatematsu M. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006;12:989-995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 474] [Cited by in RCA: 478] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 64. | Cheng AS, Li MS, Kang W, Cheng VY, Chou JL, Lau SS, Go MY, Lee CC, Ling TK, Ng EK. Helicobacter pylori causes epigenetic dysregulation of FOXD3 to promote gastric carcinogenesis. Gastroenterology. 2013;144:122-133.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 112] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 65. | Song SH, Jeon MS, Nam JW, Kang JK, Lee YJ, Kang JY, Kim HP, Han SW, Kang GH, Kim TY. Aberrant GATA2 epigenetic dysregulation induces a GATA2/GATA6 switch in human gastric cancer. Oncogene. 2018;37:993-1004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 66. | Huang FY, Chan AO, Rashid A, Wong DK, Cho CH, Yuen MF. Helicobacter pylori induces promoter methylation of E-cadherin via interleukin-1β activation of nitric oxide production in gastric cancer cells. Cancer. 2012;118:4969-4980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 86] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 67. | Byun SW, Chang YJ, Chung IS, Moss SF, Kim SS. Helicobacter pylori decreases p27 expression through the delta opioid receptor-mediated inhibition of histone acetylation within the p27 promoter. Cancer Lett. 2012;326:96-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 68. | Fehri LF, Rechner C, Janssen S, Mak TN, Holland C, Bartfeld S, Brüggemann H, Meyer TF. Helicobacter pylori-induced modification of the histone H3 phosphorylation status in gastric epithelial cells reflects its impact on cell cycle regulation. Epigenetics. 2009;4:577-586. [PubMed] |
| 69. | Ding SZ, Fischer W, Kaparakis-Liaskos M, Liechti G, Merrell DS, Grant PA, Ferrero RL, Crowe SE, Haas R, Hatakeyama M. Helicobacter pylori-induced histone modification, associated gene expression in gastric epithelial cells, and its implication in pathogenesis. PLoS One. 2010;5:e9875. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 66] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 70. | Lu ZM, Zhou J, Wang X, Guan Z, Bai H, Liu ZJ, Su N, Pan K, Ji J, Deng D. Nucleosomes correlate with in vivo progression pattern of de novo methylation of p16 CpG islands in human gastric carcinogenesis. PLoS One. 2012;7:e35928. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 71. | Peterson SM, Thompson JA, Ufkin ML, Sathyanarayana P, Liaw L, Congdon CB. Common features of microRNA target prediction tools. Front Genet. 2014;5:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 269] [Cited by in RCA: 315] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 72. | Quan M, Chen J, Zhang D. Exploring the secrets of long noncoding RNAs. Int J Mol Sci. 2015;16:5467-5496. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 92] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 73. | Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5833] [Cited by in RCA: 6530] [Article Influence: 384.1] [Reference Citation Analysis (0)] |
| 74. | Jin W, Ibeagha-Awemu EM, Liang G, Beaudoin F, Zhao X, Guan le L. Transcriptome microRNA profiling of bovine mammary epithelial cells challenged with Escherichia coli or Staphylococcus aureus bacteria reveals pathogen directed microRNA expression profiles. BMC Genomics. 2014;15:181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 131] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 75. | Maudet C, Mano M, Eulalio A. MicroRNAs in the interaction between host and bacterial pathogens. FEBS Lett. 2014;588:4140-4147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 132] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 76. | Duval M, Cossart P, Lebreton A. Mammalian microRNAs and long noncoding RNAs in the host-bacterial pathogen crosstalk. Semin Cell Dev Biol. 2017;65:11-19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 59] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 77. | Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481-12486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3121] [Cited by in RCA: 3536] [Article Influence: 186.1] [Reference Citation Analysis (0)] |
| 78. | Schulte LN, Westermann AJ, Vogel J. Differential activation and functional specialization of miR-146 and miR-155 in innate immune sensing. Nucleic Acids Res. 2013;41:542-553. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 149] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 79. | Duerr CU, Hornef MW. The mammalian intestinal epithelium as integral player in the establishment and maintenance of host-microbial homeostasis. Semin Immunol. 2012;24:25-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 53] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 80. | Wang J, Yang K, Zhou L, Minhaowu , Wu Y, Zhu M, Lai X, Chen T, Feng L, Li M. MicroRNA-155 promotes autophagy to eliminate intracellular mycobacteria by targeting Rheb. PLoS Pathog. 2013;9:e1003697. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 187] [Cited by in RCA: 208] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 81. | Kumar M, Sahu SK, Kumar R, Subuddhi A, Maji RK, Jana K, Gupta P, Raffetseder J, Lerm M, Ghosh Z. MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-κB pathway. Cell Host Microbe. 2015;17:345-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 206] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 82. | Matsushima K, Isomoto H, Inoue N, Nakayama T, Hayashi T, Nakayama M, Nakao K, Hirayama T, Kohno S. MicroRNA signatures in Helicobacter pylori-infected gastric mucosa. Int J Cancer. 2011;128:361-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 160] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 83. | Tang B, Li N, Gu J, Zhuang Y, Li Q, Wang HG, Fang Y, Yu B, Zhang JY, Xie QH. Compromised autophagy by MIR30B benefits the intracellular survival of Helicobacter pylori. Autophagy. 2012;8:1045-1057. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 112] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 84. | Li N, Tang B, Zhu ED, Li BS, Zhuang Y, Yu S, Lu DS, Zou QM, Xiao B, Mao XH. Increased miR-222 in H. pylori-associated gastric cancer correlated with tumor progression by promoting cancer cell proliferation and targeting RECK. FEBS Lett. 2012;586:722-728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 76] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 85. | Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W, Xiao S, Lu H. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest. 2008;88:1358-1366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 339] [Cited by in RCA: 375] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 86. | Zhu Y, Jiang Q, Lou X, Ji X, Wen Z, Wu J, Tao H, Jiang T, He W, Wang C. MicroRNAs up-regulated by CagA of Helicobacter pylori induce intestinal metaplasia of gastric epithelial cells. PLoS One. 2012;7:e35147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 87. | Feng Y, Wang L, Zeng J, Shen L, Liang X, Yu H, Liu S, Liu Z, Sun Y, Li W. FoxM1 is overexpressed in Helicobacter pylori-induced gastric carcinogenesis and is negatively regulated by miR-370. Mol Cancer Res. 2013;11:834-844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 80] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 88. | Noto JM, Piazuelo MB, Chaturvedi R, Bartel CA, Thatcher EJ, Delgado A, Romero-Gallo J, Wilson KT, Correa P, Patton JG. Strain-specific suppression of microRNA-320 by carcinogenic Helicobacter pylori promotes expression of the antiapoptotic protein Mcl-1. Am J Physiol Gastrointest Liver Physiol. 2013;305:G786-G796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 89. | Belair C, Baud J, Chabas S, Sharma CM, Vogel J, Staedel C, Darfeuille F. Helicobacter pylori interferes with an embryonic stem cell micro RNA cluster to block cell cycle progression. Silence. 2011;2:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 53] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 90. | Zhang YM, Noto JM, Hammond CE, Barth JL, Argraves WS, Backert S, Peek RM Jr, Smolka AJ. Helicobacter pylori-induced posttranscriptional regulation of H-K-ATPase α-subunit gene expression by miRNA. Am J Physiol Gastrointest Liver Physiol. 2014;306:G606-G613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 91. | Zhao J, Liu Y, Huang G, Cui P, Zhang W, Zhang Y. Long non-coding RNAs in gastric cancer: versatile mechanisms and potential for clinical translation. Am J Cancer Res. 2015;5:907-927. [PubMed] |
| 92. | Hu Y, Wang J, Qian J, Kong X, Tang J, Wang Y, Chen H, Hong J, Zou W, Chen Y. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014;74:6890-6902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 223] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 93. | Lee NK, Lee JH, Park CH, Yu D, Lee YC, Cheong JH, Noh SH, Lee SK. Long non-coding RNA HOTAIR promotes carcinogenesis and invasion of gastric adenocarcinoma. Biochem Biophys Res Commun. 2014;451:171-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 61] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 94. | Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, Xu TP, Xia R, Yang JS, De W. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5:2276-2292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 259] [Cited by in RCA: 307] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 95. | Yang F, Xue X, Zheng L, Bi J, Zhou Y, Zhi K, Gu Y, Fang G. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS J. 2014;281:802-813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 190] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 96. | Zhuang M, Gao W, Xu J, Wang P, Shu Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem Biophys Res Commun. 2014;448:315-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 170] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 97. | Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159-3165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 376] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 98. | Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y, Fang G. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol. 2013;139:437-445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 214] [Cited by in RCA: 225] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 99. | Wang J, Su L, Chen X, Li P, Cai Q, Yu B, Liu B, Wu W, Zhu Z. MALAT1 promotes cell proliferation in gastric cancer by recruiting SF2/ASF. Biomed Pharmacother. 2014;68:557-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 139] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 100. | Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol Rep. 2014;31:358-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 221] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 101. | Xu TP, Huang MD, Xia R, Liu XX, Sun M, Yin L, Chen WM, Han L, Zhang EB, Kong R. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol. 2014;7:63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 181] [Cited by in RCA: 225] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 102. | Sun M, Xia R, Jin F, Xu T, Liu Z, De W, Liu X. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol. 2014;35:1065-1073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 236] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 103. | Park SM, Park SJ, Kim HJ, Kwon OH, Kang TW, Sohn HA, Kim SK, Moo Noh S, Song KS, Jang SJ. A known expressed sequence tag, BM742401, is a potent lincRNA inhibiting cancer metastasis. Exp Mol Med. 2013;45:e31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 104. | Zhou X, Chen H, Zhu L, Hao B, Zhang W, Hua J, Gu H, Jin W, Zhang G. Helicobacter pylori infection related long noncoding RNA (lncRNA) AF147447 inhibits gastric cancer proliferation and invasion by targeting MUC2 and up-regulating miR-34c. Oncotarget. 2016;7:82770-82782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 105. | Zhu H, Wang Q, Yao Y, Fang J, Sun F, Ni Y, Shen Y, Wang H, Shao S. Microarray analysis of Long non-coding RNA expression profiles in human gastric cells and tissues with Helicobacter pylori Infection. BMC Med Genomics. 2015;8:84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |