Published online Feb 16, 2024. doi: 10.12998/wjcc.v12.i5.951
Peer-review started: November 3, 2023
First decision: December 6, 2023
Revised: December 14, 2023
Accepted: January 22, 2024
Article in press: January 22, 2024
Published online: February 16, 2024
Processing time: 88 Days and 15.4 Hours
Helicobacter pylori (H. pylori) infection is a major risk factor for chronic gastritis, affecting approximately half of the global population. H. pylori eradication is a popular treatment method for H. pylori-positive chronic gastritis, but its mecha
To elucidate the urinary metabolic profiles during H. pylori eradication in patients with chronic gastritis.
We applied LC–MS-based metabolomics and network pharmacology to in
Our study revealed the different urinary metabolic profiles of H. pylori-positive chronic gastritis before and after H. pylori eradication. The metabolites regulated by H. pylori eradication therapy include cis-aconitic acid, isocitric acid, citric acid, L-tyrosine, L-phenylalanine, L-tryptophan, and hippuric acid, which were involved in four metabolic pathways: (1) Phenylalanine metabolism; (2) phenylalanine, tyrosine, and tryptophan biosynthesis; (3) citrate cycle; and (4) glyoxylate and dicarboxylate metabolism. Integrated metabolomics and network pharmacology revealed that MPO, COMT, TPO, TH, EPX, CMA1, DDC, TPH1, and LPO were the key proteins involved in the biological progress of H. pylori eradication in chronic gastritis.
Our research provides a new perspective for exploring the significance of urinary metabolites in evaluating the treatment and prognosis of H. pylori-positive chronic gastritis patients.
Core Tip: Urinary metabolomics has been used to elucidate the mechanisms of gastric disease treatment, whereas no clinical study is conducted on metabolomics of chronic gastritis. In this manuscript, we carried out LC-MS-based metabolomics to investigate urinary metabolites changes in Helicobacter pylori (H. pylori)-positive chronic gastritis treatment. Our study revealed the urinary metabolic profiles of H. pylori-positive chronic gastritis after H. pylori eradication. Integrated metabolomics and network pharmacology revealed the key proteins involved in H. pylori eradication of chronic gastritis. Our research provides a new perspective for exploring the significance of urinary metabolites in evaluating the treatment and prognosis of H. pylori-positive chronic gastritis.
- Citation: An WT, Hao YX, Li HX, Wu XK. Urinary metabolic profiles during Helicobacter pylori eradication in chronic gastritis. World J Clin Cases 2024; 12(5): 951-965
- URL: https://www.wjgnet.com/2307-8960/full/v12/i5/951.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i5.951
Chronic gastritis is a common digestive system disease affecting approximately half of the global population[1]. Chronic gastritis is also the most important risk factor for gastric cancer, the fifth most commonly diagnosed cancer and the fourth leading cause of cancer-related mortality[1,2]. Chronic gastritis can be classified into two major stages, non-atrophic and atrophic, according to the phenotypes of the gastric mucosa[3]. Chronic non-atrophic gastritis will develop into chronic atrophic gastritis if left untreated. A 16-year follow-up study revealed that up to 2% of patients with chronic atrophic gastritis develop gastric cancer annually[4]. Additionally, 24% of gastric cancer patients are first diagnosed with chronic atrophic gastritis[5]. Thus, managing chronic gastritis is an important approach for preventing gastric cancer develop
Helicobacter pylori (H. pylori) infection, a major risk factor for chronic gastritis, infects approximately 50% of the global population[6,7]. A portion of infected people will develop various degrees of gastrointestinal disease, such as dyspepsia (5%–10%), chronic gastritis (90%), peptic ulcers (15%–20%), and gastric malignancies (1%)[8]. H. pylori has been described as a first-class carcinogen for gastric cancer by the World Health Organization since 1994 and accounts for 16.1% of gastric cancer cases[9,10]. A 26.5-year follow-up report indicated that H. pylori eradication might confer long-term protection against gastric cancer in high-risk populations[11]. Therefore, eradication of H. pylori is recommended to reduce the occurrence of gastric diseases[8]. Numerous double, triple, and quadruple therapies have been proposed as first-line empiric treatments for H. pylori infection[12]. However, the molecular mechanisms underlying these treatment regimens are complicated and remain unclear[13].
Urinary metabolomics has been gradually applied to mine metabolic profiles for diagnosis, prognostic evaluation, and research of treatment mechanisms in gastric diseases. NMR-based urinary metabolomics revealed that urine metabolite levels were changed during oncogenesis in gastric cancer, and 4-hydroxyphenylacetate, alanine, phenylacetylglycine, mannitol, glycolate, and arginine are potential metabolic biomarkers for effectively diagnosing gastric cancer[14,15]. NMR- and UPLC-Q/TOF MS-mediated urinary metabolomics revealed that a traditional Chinese medicine, Huangqi Jianzhong Tang, treated chronic atrophic gastritis by balancing energy consumption, inhibiting inflammation, improving the immune system, and reducing oxidative stress in rats[16]. UPLC-Q-TOF/MS-based urinary metabolomics has also been applied to investigate the therapeutic effect and potential mechanism of berberine on chronic atrophic gastritis[17] and the therapeutic mechanism of palmatine in chronic atrophic gastritis induced by H. pylori[18]. However, no clinical study has been conducted on urinary metabolomics in chronic gastritis.
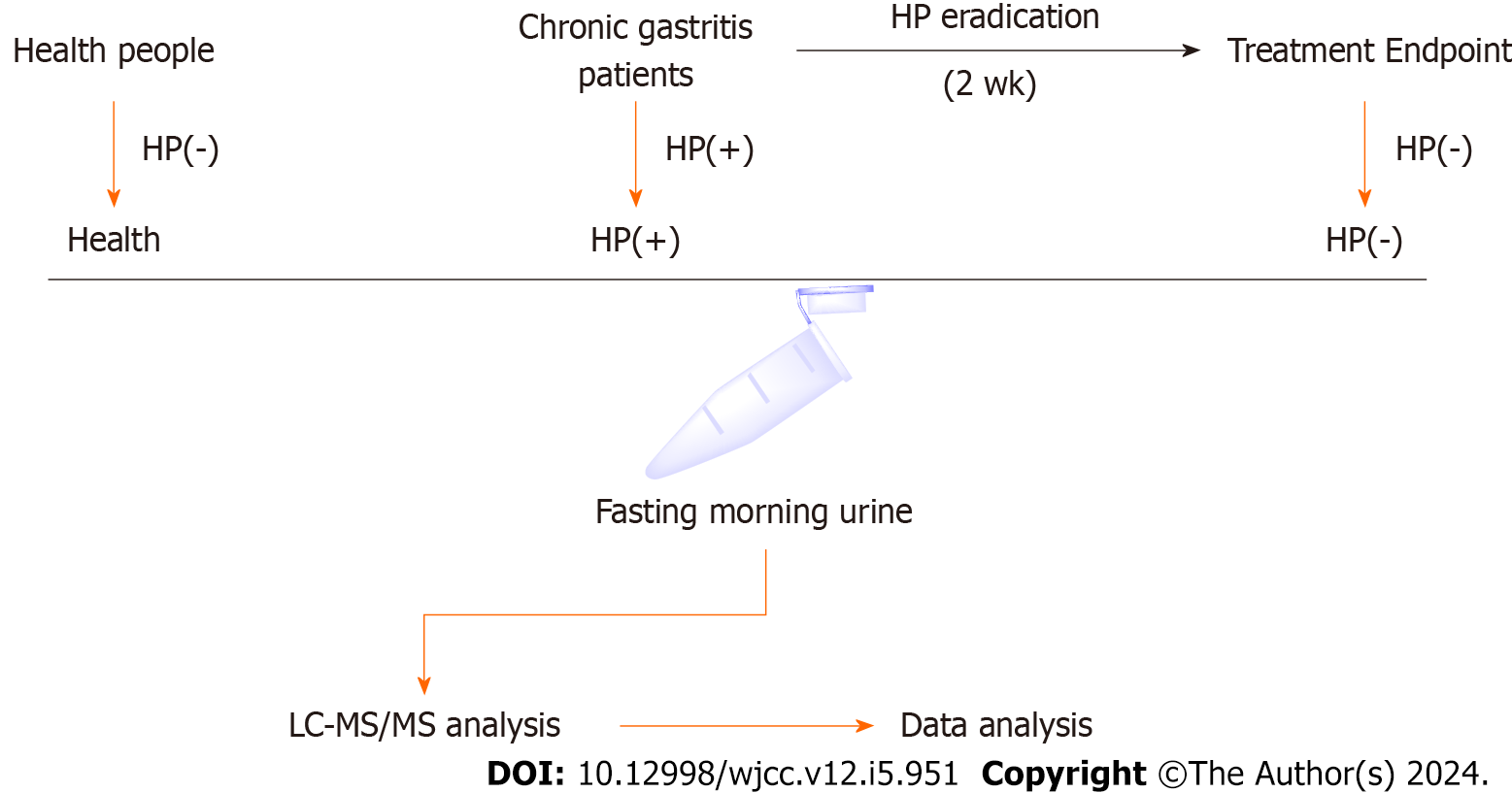
This study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of Shanxi Provincial People’s Hospital (Grant No. 2022-167). Patients with H. pylori-positive chronic gastritis who were hospitalized or outpatients in the Gastroenterology Department of Shanxi Provincial People’s Hospital were selected as the research participants. Patients with H. pylori-positive chronic gastritis were enrolled. These patients were diagnosed with chronic gastritis by endoscopy and pathological examination. For the diagnostic criteria of H. pylori infection, refer to the "Fifth National Consensus Report on the Treatment of H. pylori Infection." Those with a positive 14C or 13C urea breath test (UBT) were diagnosed with H. pylori infection. Fasting morning urine was collected from patients diagnosed with H. pylori-positive chronic gastritis and marked as “HP(+)”. Subsequently, the patients were treated with therapeutic strategies of H. pylori eradication for 2 wk (Figure 1). H. pylori eradication was conducted using conventional quadruple therapy,
| Parameters | HP(+) and HP(−) | Health |
| Case | 17 | 20 |
| Sex | ||
| Male | 11 | 12 |
| Female | 7 | 8 |
| Age (yr) | ||
| 31–40 | 2 | 2 |
| 41–50 | 8 | 9 |
| 51–60 | 6 | 7 |
| > 60 | 1 | 2 |
| Average age (yr) | 49.35 | 48.65 |
| Treatment | ||
| Omeprazole/amoxicillin/furazolidone/bismuth pectin | 3 | - |
| Ilaprazole/amoxicillin/furazolidone/bismuth pectin | 6 | - |
| Pantoprazole/amoxicillin/furazolidone/bismuth pectin | 8 | - |
In this study, approximately 180 patients were diagnosed with H. pylori-positive chronic gastritis, and their urine samples were collected at the first diagnosis. However, only 17 patients met the clinical assessment inclusion criteria and were willing to be reexamined for H. pylori infection after treatment (Table 1). These patients were treated with quadruple therapy strategies for H. pylori eradication. After 2 wk of treatment, 17 patients in our study were H. pylori-negative and discontinued treatment.
The first morning urine was collected from fasting patients and healthy individuals and stored at −80℃. The frozen urine samples were thawed in an ice bath and centrifuged at a low temperature for 10 min (10000 ×g, 4°C). The supernatant was transferred to a new 1.5 mL EP tube. The proteins were precipitated by adding methanol-acetonitrile (2:1). Then, the mixtures were subjected to ultrasonic extraction in an ice water bath for 10 min and centrifuged for 20 min (10000 ×g, 4°C). The supernatant was filtered through a 0.22 μm organic phase pinhole filter and transferred to an LC sample vial. Samples were stored at 4℃ until the LC–MS analysis. In addition, 10 μL of urine from each group was taken, mixed, and prepared as QC samples according to the sample preparation method.
The mobile phases were A (0.1% formic acid water) and B (0.1% formic acid acetonitrile). Elution was conducted according to the following gradient: 0–2 min, 2% B; 2–3 min, 2%–35% B; 3–17 min, 35%–70% B; 17–18 min, 70% B; 18–29 min, 70%–98% B; 29–31 min, 98% B; 31–33 min, 98%–2% B; and 33–35 min, 2% B. A Waters ACQUITYUPLC HSS T3 (2.1 × 100 mm, 1.7 μm) chromatographic column with a 5 μL injection volume, 0.2 mL/min flow rate, and 40°C temperature was then used for the liquid chromatographic analysis.
The mass spectrometry profiles of the urine metabolome were obtained on a UPLC (ExionLC AD) coupled with a Triple TOF 5600+ mass spectrometer (AB Sciex). The mass spectrometry conditions were set as follows: Electronspray ionization (ESI); mode: positive and negative ion scanning; mass scanning range: 50–1500 Da; atomizing gas pressure (GS1) and auxiliary gas pressure (GS2): 0.55 kPa; atomizing gas temperature: 550℃; spray voltage: +5500 V in positive ion mode and −4500 V in negative ion mode; curtain pressure: 0.3 kPa; and cluster fragmentation voltage: 100 V. Data were collected in information association mode, collision energy was ± 35 eV, and the collision energy rolling interval was (35 ± 15) eV.
The raw data from UHPLC-Q-TOF/MS were imported into One-MAP software, and all metabolite names, peak areas, retention times, and other information were calculated. The results were exported as Excel files, and the total peak area data of each group of metabolites were normalized to obtain the peak-normalized data per metabolite.
The above peak-normalized data were imported into Simca-P 14.1. A principal component analysis (PCA) was used for the exploratory analysis to determine possible clusters and outliers, and partial least square discriminant analysis (PLS–DA) and orthogonal partial least square discriminant analysis (OPLS–DA) were performed to explore different metabolites with metabolic profile changes, combining VIP > 1 and a t test (P < 0.05) in the S-plot to screen different metabolites.
The identification of metabolites was performed by importing the m/z values of metabolites into the One-map database
The Pathway Analysis module in MetaboAnalyst 5.0 (https://www.metaboanalyst.ca/) was used to perform metabolic pathway enrichment analyses on differential metabolites, and the pathway with an impact value (impact) greater than or equal to 0.1 was considered to be the main metabolism path.
The area under the receiver operating characteristic (ROC) curve was used to evaluate the quality and the predictive ability of the classification models. Univariate ROC analysis was conducted, and the area under curve (AUC) and P values of each ROC curve were used to evaluate the predictability. Then, to improve the discriminatory accuracy, multivariate ROC curves were plotted with false-positive and true-positive rates using a combination of significant metabolites with AUC > 0.5 (P value < 0.05).
Integrated metabolomics and network pharmacology were applied to reveal the regulatory network of the identified differential metabolites. First, a metabolite-related network construction was performed by importing the identified differential metabolites into Cytoscape 3.7.2 (Cytoscape Consortium, San Diego, CA, USA) equipped with MetScape. This network was constructed to visualize the interactions among the metabolites, pathways, enzymes, and genes. The key metabolites and proteins were recognized by combining the metabolite-reaction-enzyme-gene network with hub genes and metabolic pathways. Then, the candidate targets of H. pylori-positive chronic gastritis were acquired by taking the intersection of targets between chronic gastritis and H. pylori infection. The targets of chronic gastritis and H. pylori infection were obtained by searching the keywords “chronic gastritis” and “Helicobacter pylori” in the Genecards database (https://www.genecards.org/), respectively. Finally, the key proteins involved in regulating the identified differential metabolites were obtained by matching the H. pylori-positive chronic gastritis-related targets with the differential metabolite-related targets.
GraphPad Prism 8 software was used for generating figures and statistical analyses. Data are presented as the mean ± SD. The normality of data distribution was analyzed with SPSS software. Comparisons between two groups were performed using independent-sample t tests; comparisons between multiple groups were performed by one-way ANOVA. P < 0.05 was considered to represent significance.
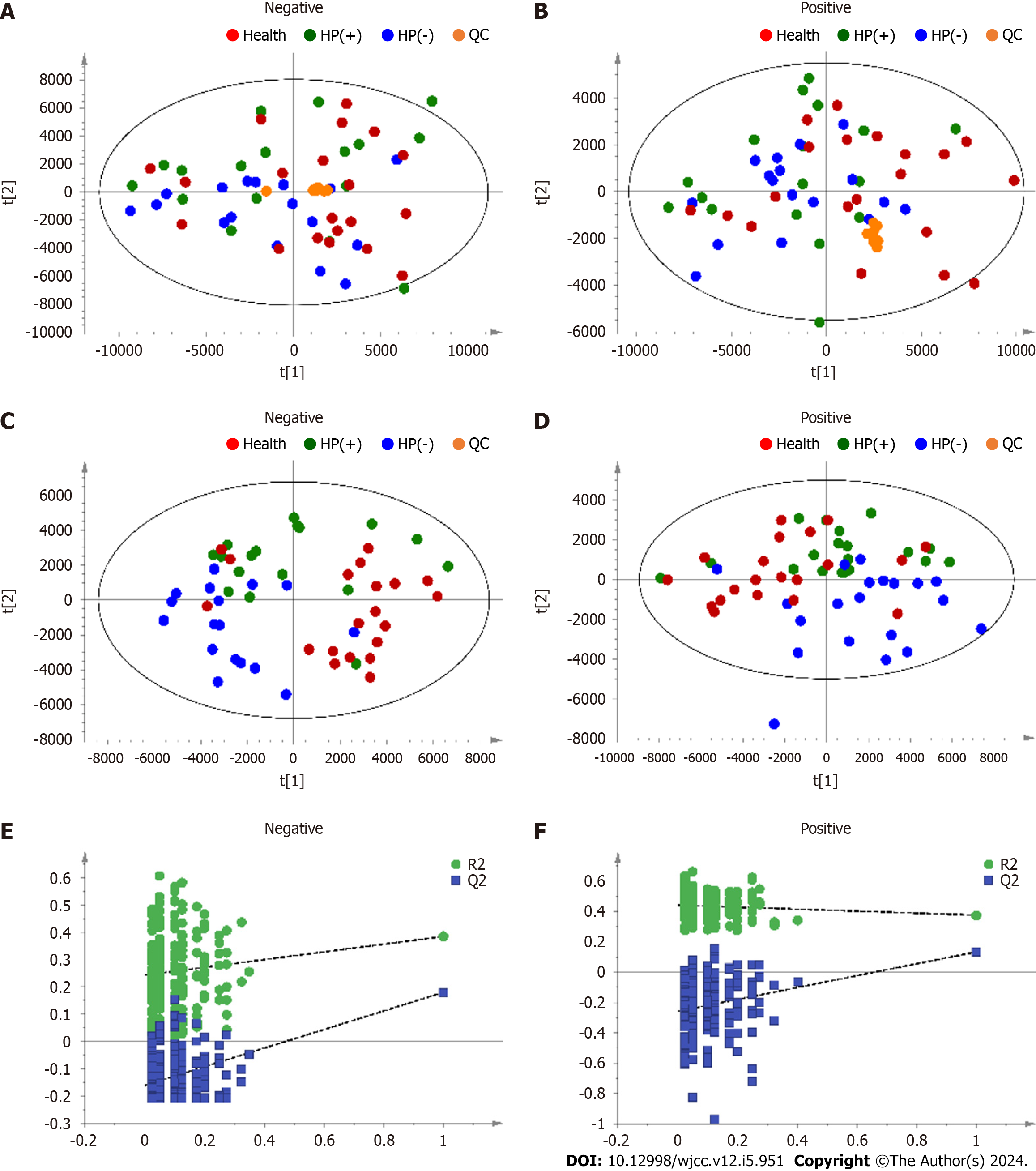
Metabolic profiling of urine samples was performed in positive and negative ion modes in an unsupervised model without grouping conditions using PCA. The QC samples were clustered, indicating good system stability. The results show that the samples Health, HP(+), and HP(−) cannot be effectively separated (Figure 2A and B). Therefore, PLS–DA was performed to reduce the dimensionality of the complex data obtained from the Health, HP(+), and HP(−) urine samples to distinguish the differences between groups. The results are shown in Figure 2C–F. The Health, HP(+), and HP(−) samples were significantly separated in ESI- and ESI+ modes, indicating that 2 wk of drug treatment alters urinary metabolic disorders in patients with chronic gastritis. In summary, the untargeted metabolomics analysis indicated that the urinary metabolic profiles changed during H. pylori eradication.
The OPLS–DA model was used to further evaluate the changes in metabolic profile in the sixth week after drug treatment. The CV-ANOVA diagnostics, in which the
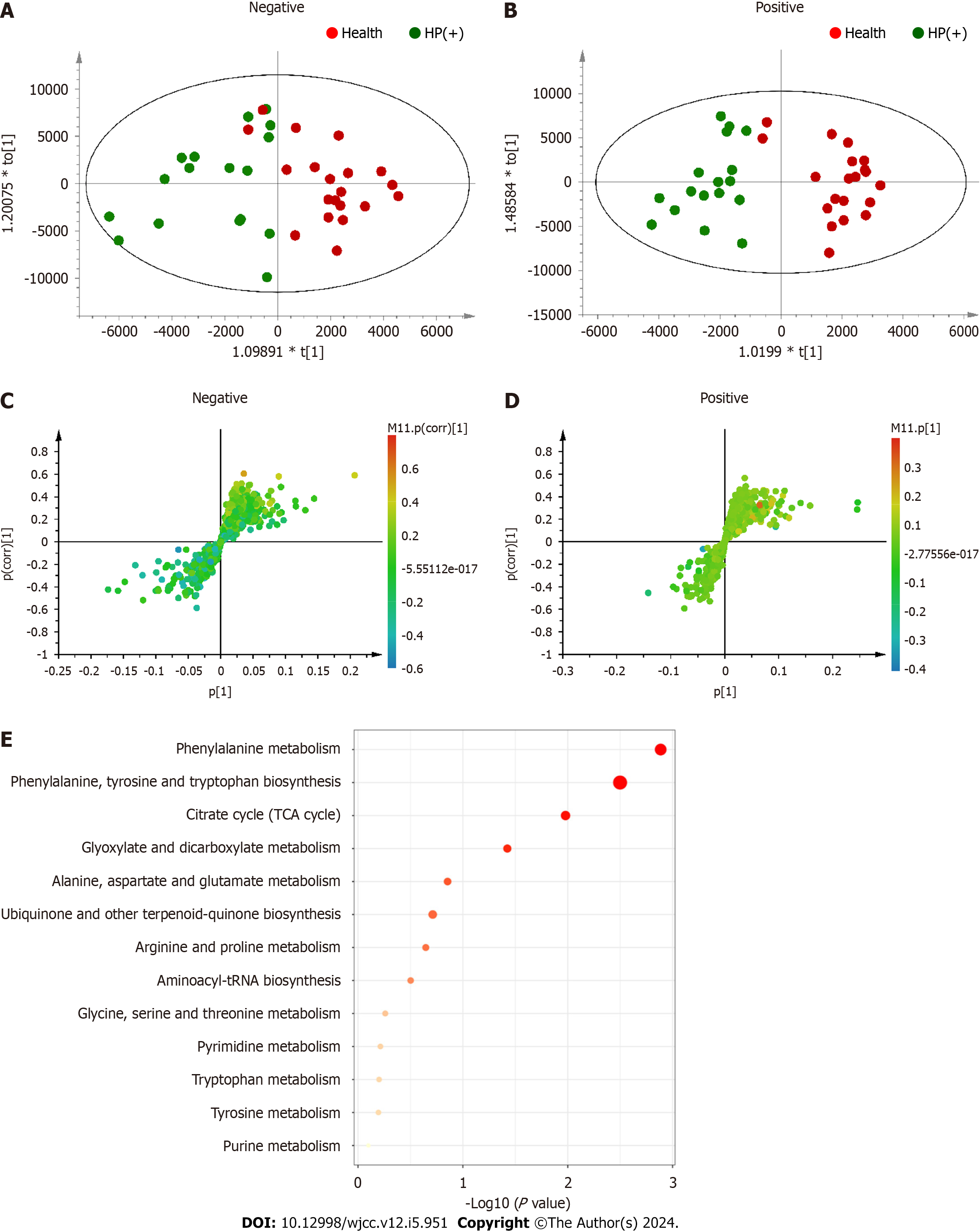
The differential metabolites were imported into the Pathway Analysis module in MetaboAnalyst5.0, and a metabolic pathway analysis was conducted to find the 10 metabolic pathways most related to H. pylori eradication: (1) Phenylalanine metabolism; (2) phenylalanine, tyrosine, and tryptophan biosynthesis; (3) citrate cycle; (4) glyoxylate and dicarboxylate metabolism; (5) alanine, aspartate, and glutamate metabolism; (6) ubiquinone and other terpenoid-quinone biosynthesis; (7) arginine and proline; (8) aminoacyl-tRNA biosynthesis; (9) glycine, serine, and threonine metabolism; and (10) pyrimidine metabolism (Figure 3E).
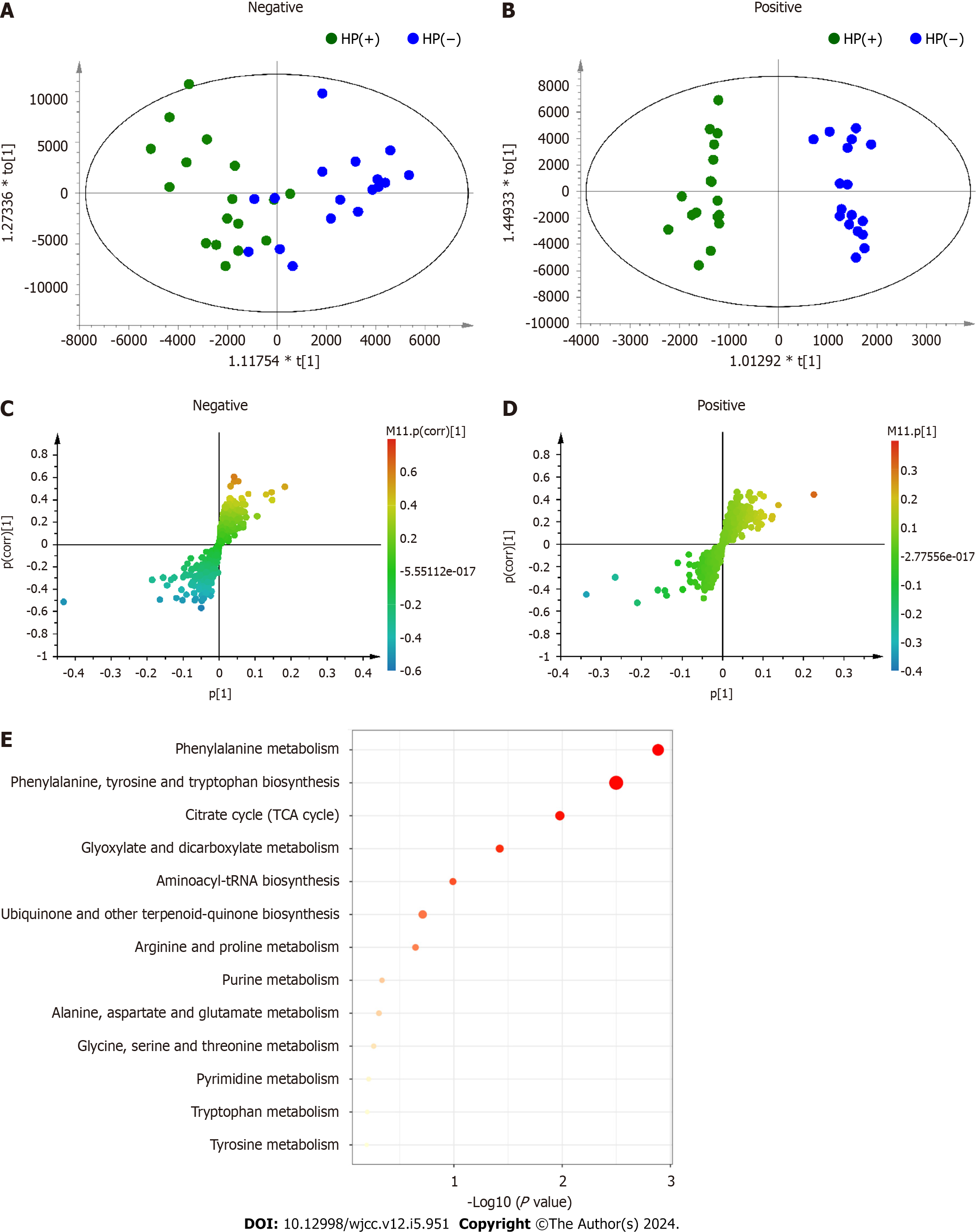
In both positive and negative ion modes, the OPLS–DA model was used to evaluate the effects of H. pylori eradication on metabolic profiles after 2 wk of treatment. The CV-ANOVA diagnostics, in which the
The differential metabolites were imported into MetaboAnalyst 5.0 software for pathway enrichment analysis to find the 10 most relevant metabolic pathways (pathway impact > 0.1) related to H. pylori eradication: (1) Phenylalanine metabolism; (2) phenylalanine, tyrosine, and tryptophan biosynthesis ; (3) citrate cycle; (4) glyoxylate and dicarboxylate metabolism; (5) aminoacyl-tRNA biosynthesis; (6) ubiquinone and other terpenoid-quinone biosynthesis; (7) arginine and proline metabolism; (8) purine metabolism; (9) alanine, aspartate, and glutamate metabolism; and (10) glycine, serine, and threonine metabolism (Figure 4E).
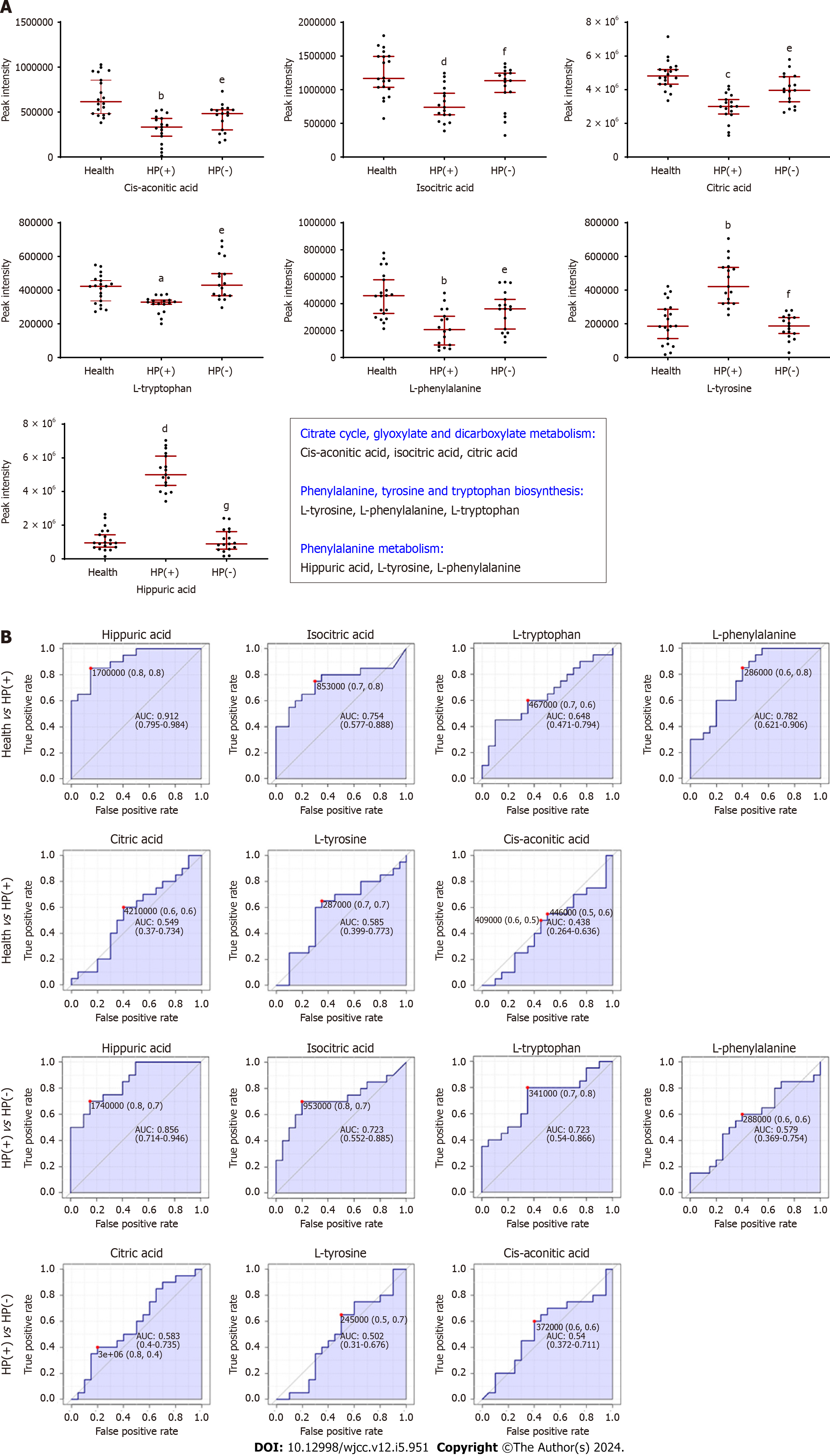
A joint pathway analysis was performed on the differential metabolites between the HP(+) and HP(−) groups and the Health and HP(−) groups using the Joint-pathway Analysis module in MetaboAnalyst to screen the key metabolic pathways. The top four metabolic pathways co-regulated in Health vs HP(+) and HP(+) vs HP(−) are as follows: (1) Phenylalanine metabolism; (2) phenylalanine, tyrosine, and tryptophan biosynthesis; (3) citrate cycle; and (4) glyoxylate and dicarboxylate. Seven differential metabolites related to these four metabolic pathways were found, and the results are shown in Figure 5A. Cis-aconitic acid, isocitric acid, and citric acid were involved in the citrate cycle, glyoxylate, and dicarboxylate metabolism; L-tyrosine, L-phenylalanine, and L-tryptophan were involved in phenylalanine, L-tyrosine, and L-tryptophan biosynthesis; and hippuric acid, L-tyrosine, and L-phenylalanine were involved in phenylalanine metabolism.
Next, the levels of these differential metabolites were investigated by assessing the peak intensity of ions (Figure 5A). Compared with the Health group, in the HP(+) group, the levels of cis-aconitic acid, isocitric acid, citric acid, L-tyrosine, and L-phenylalanine were decreased, while L-tyrosine and hippuric acid levels were increased. The levels of these seven metabolites returned to normal after H. pylori eradication.
We performed a univariate ROC curve analysis using the prominent seven metabolites to confirm the discriminative accuracy of individual metabolites between groups. The X-axis is the false-positive rate; the closer the X-axis value is to zero, the higher the accuracy is. The Y-axis is the true-positive rate; the larger the Y-axis value is, the higher the accuracy is. The result shows that the false-positive and true-positive rates in the Health vs HP(+) groups were higher than those in the HP(+) vs HP(−) groups, indicating that these urinary metabolites could reveal the treatment and prognosis progression of chronic gastritis with H. pylori infection (Figure 5B).
The criteria for assessing the accuracy of the signature based on AUC were summarized into a single metric, the ROC curve. According to the Swets criterion, AUC < 0.5 indicates that the test has no diagnostic value; AUC 0.5–0.7 indicates that the diagnostic test has low accuracy; AUC 0.7–0.9 indicates that the diagnostic test has good accuracy; and AUC > 0.9 indicates that the diagnostic test has high accuracy. In HP(+) vs HP(−) groups, the AUC values of hippuric acid, isocitric acid, L-tryptophan, L-phenylalanine, citric acid, L-tyrosine, and cis-aconitic acid were 0.856, 0.723, 0.723, 0.579, 0.583, 0.502, and 0.54, respectively (Figure 5B). In the Health vs HP(+) groups, the AUC values of these metabolites were 0.912, 0.754, 0.648, 0.782, 0.549, 0.585, and 0.438 (Figure 5B). Thus, hippuric acid, isocitric acid, L-tryptophan, and L-phenylalanine were the most related to the treatment effect and prognosis of chronic gastritis patients with H. pylori infection.
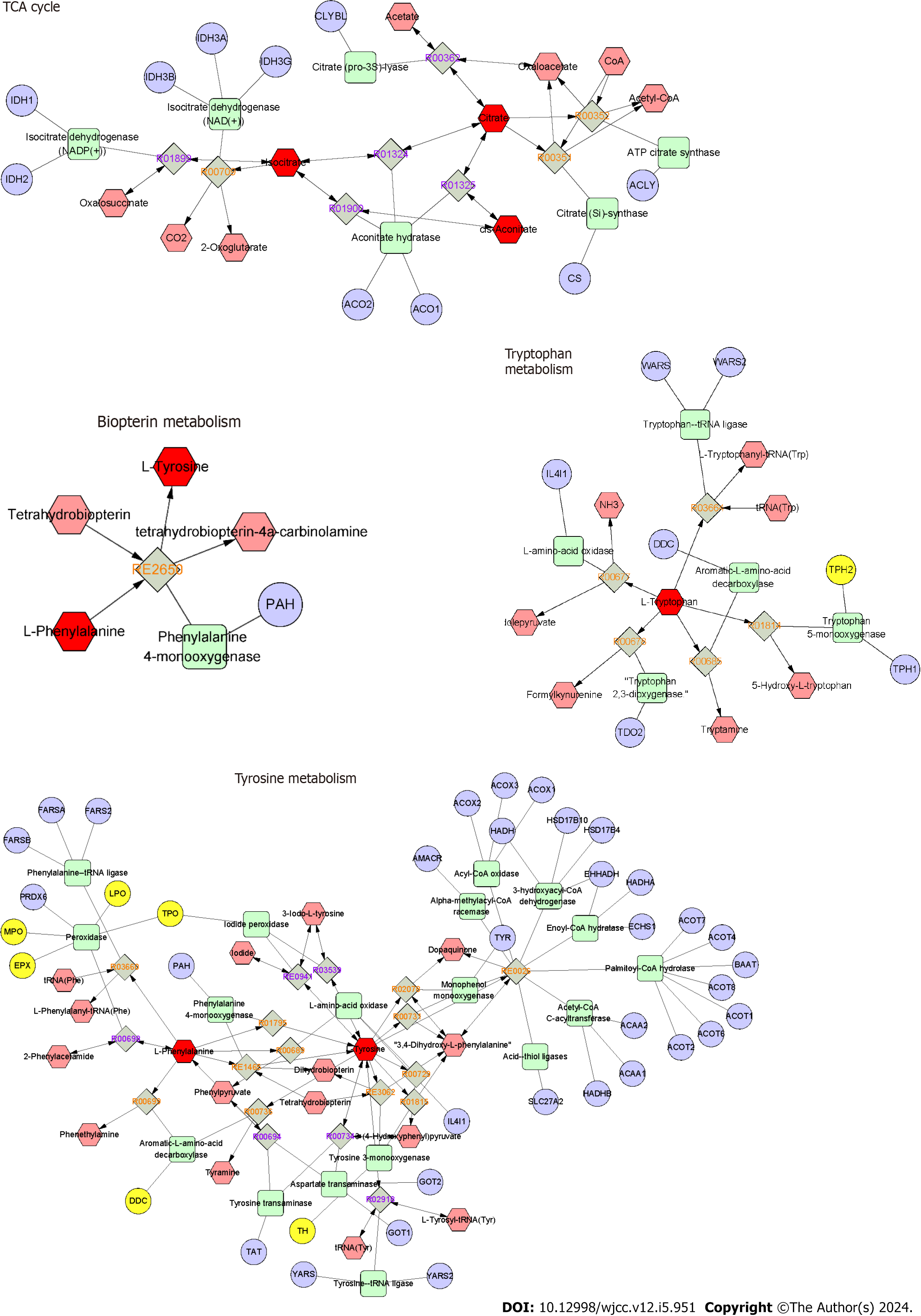
We constructed an interaction network based on metabolomics and network pharmacology to further explore the relationships between metabolite changes and H. pylori eradication in chronic gastritis patients. First, differential metabolites were imported into the MetScape plugin in Cytoscape to collect the metabolite-reaction-enzyme-gene networks. By analyzing the identified metabolites in MetScape analysis, we gathered 60 targets of the significant differential metabolites and found four key metabolism pathways, namely the TCA cycle, tryptophan metabolism, biopterin metabolism, and tyrosine metabolism (Figure 6). Then, we performed network pharmacology to explore the key proteins involved in the regulatory mechanism of the identified differential metabolites in H. pylori-positive chronic gastritis. We collected 1313 targets in H. pylori-positive chronic gastritis from the Genecards database. After matching the H. pylori-positive chronic gastritis-related targets with the significant metabolite-related targets, nine targets were identified as potential key proteins involved in the biological progress of H. pylori eradication in chronic gastritis. These nine targets were MPO, COMT, TPO, TH, EPX, CMA1, DDC, TPH1, and LPO, which may play essential roles in the therapeutic effect in H. pylori-chronic gastritis.
Here, to the best of our knowledge, urinary metabolomics of patients with H. pylori-positive chronic gastritis was investigated for the first time. A therapeutic follow-up was conducted to collect urine from patients during H. pylori eradication and evaluate the changes in urinary metabolic profiles between H. pylori-positive and -negative patients. Urinary metabolic profiles were altered during H. pylori eradication. The metabolic pathways involved in H. pylori eradication in H. pylori-positive chronic gastritis patients included: (1) Phenylalanine metabolism; (2) phenylalanine, tyrosine, and tryptophan biosynthesis; (3) citrate cycle; and (4) glyoxylate and dicarboxylate. The decrease in hippuric acid and the increase in isocitric acid, L-tryptophan, and L-phenylalanine were mostly related to the treatment and prognosis of H. pylori-positive chronic gastritis patients. Our results provide a new perspective for evaluating the prognosis of H. pylori-positive chronic gastritis patients: the analysis of urinary metabolites.
The citrate cycle was found to be a vital urinary metabolic pathway related to the prognosis of H. pylori-positive chronic gastritis patients. After H. pylori eradication, the levels of three citrate cycle intermediates, namely cis-aconitic acid, isocitric acid, and citric acid, were elevated in the urine of cured patients with H. pylori-positive chronic gastritis. These results were partly consistent with those from H. pylori-infected experimental animals reported in the literature. UPLC-Q-TOF/MS-based urinary metabolomics revealed that the citrate cycle was involved in the pathogenesis, development, and prognosis of H. pylori-positive chronic gastritis in a rat model[18]. H. pylori infection reduced the levels of oxalosuccinate in rat urine, and the cure of chronic gastritis elevated the levels of oxalosuccinate[18]. 1H NMR-based urinary metabolomics showed that H. pylori infection disturbs the citrate cycle by elevating the levels of cis-aconitate in H. pylori-infected gerbil chronic gastritis models[19]. Indeed, many studies have shown that H. pylori infection disrupts the citrate cycle in the stomach. GC/MS-based metabolomics revealed that H. pylori infection disturbs the citrate cycle of gastric epithelial cells by elevating the levels of citric acid and isocitric acid[20]. GC-TOF-MS-based metabolomics revealed that H. pylori infection disturbs the citrate cycle of the gastric mucosa by elevating the levels of citric, malic, and fumaric acid[21]. Overall, the urinary metabolomics results obtained from patients and animals with H. pylori-positive chronic gastritis indicate that urinary metabolites in the citrate cycle are involved in the pathogenesis, development, and treatment of H. pylori-positive chronic gastritis. However, the mechanisms underlying the disruption of the citrate cycle by H. pylori in patients with chronic gastritis required further exploration.
In this study, hippuric acid was the most differentially expressed urinary metabolite related to the prognosis of H. pylori-positive chronic gastritis patients, with AUC values of 0.856 [HP(+) vs HP(−)] and 0.912 [(Health vs HP(+)]. H. pylori eradication decreased the levels of hippuric acid in the urine. These results are consistent with those observed in previously reported chronic gastritis model animals. 1H NMR- and UPLC-Q/TOF MS-based urinary metabolomics revealed that hippuric acid increased in the urine of sodium deoxycholate/ammonia-induced chronic atrophic gastritis rats and decreased in the urine of rats cured by a celebrated traditional Chinese medicine, Huangqi Jianzhong Tang[16,22]. In addition, in another analysis of 1H NMR-based metabolomics, compared with control rats and rats cured by electroacupuncture stimulation, hippuric acid concentrations were increased in the urine of chronic atrophic gastritis rats[23]. Overall, our results and the previous literature indicate that the levels of hippuric acid are increased in the urine of patients with chronic gastritis, providing a potential urinary biomarker for evaluating the pathogenesis, development, and prognosis of chronic gastritis. However, the relationships between hippuric acid and chronic gastritis need to be further investigated.
Currently, the standard diagnostic method for the detection of chronic gastritis is gastroscopy, which is relatively invasive and is associated with poor patient compliance[24-27]. In addition, the UBT has been used for almost 30 years to test for H. pylori infection in the diagnosis of chronic gastritis; however, this approach also has drawbacks[28,29]. 14C UBT is not suitable for children and pregnant women as it emits higher radiation levels than 13C UBT[7]. H2 receptor antagonists, antibiotics, and bleeding impair the sensitivity of UBT[7,25]. No single method can be considered the gold standard for diagnosing chronic gastritis. Thus, investigations into potential and novel biomarkers of H. pylori-positive chronic gastritis have clinical significance for the diagnosis of chronic gastritis. As urine is a completely non-invasive and inexpensive sample, urine biomarkers are promising for clinical application in gastric diseases. One case-control study revealed a novel urinary protein biomarker panel for the early diagnosis of gastric cancer[30]. A follow-up study of gastric cancer patients after curative surgery demonstrated that urinary metabolic profiles are an effective early screening tool for gastric cancer[14]. Urinary 5-hydroxyindoleacetic acid levels are significantly higher in gastric cancer patients than in chronic gastritis patients or normal individuals[31]. Indeed, rapid urine tests that apply antibodies to detect H. pylori-specific IgG are convenient for screening for H. pylori infection[32-34]. Nevertheless, no urinary biomarkers have been used for the clinical diagnosis of H. pylori-positive chronic gastritis. This is a groundbreaking original clinical of the urinary biomarkers of H. pylori-positive chronic gastritis. According to our findings and previous literature, the levels of hippuric acid and metabolites in the citrate cycle in the urine are promising biomarkers for the better diagnosis and management of H. pylori-positive chronic gastritis. However, some issues still require attention, such as the false-positive results of non-targeted metabolomics. Therefore, future experiments should aim to confirm the roles of hippuric acid and metabolites of the citrate cycle as pivotal urinary biomarkers of H. pylori-positive chronic gastritis.
Integrated metabolomics and network pharmacology revealed that MPO, COMT, TPO, TH, EPX, CMA1, DDC, TPH1, and LPO were the key proteins involved in the biological progress of H. pylori eradication in chronic gastritis. Many researchers have reported that MPO protein levels are reduced during H. pylori eradication. In H. pylori-infected gerbils, MPO activity of stomach tissues decreased approximately tenfold[35]. In C57BL/6 mouse, H. pylori infection induced substantially higher MPO activity in the submucosa and the lamina propria of the stomach[36]. In one clinical study, MPO serum levels were significantly higher in H. pylori-positive chronic gastritis patients than in H. pylori-negative controls[37]. However, little research has been conducted on the relationship between proteins other than MPO and H. pylori eradication or infection in chronic gastritis. To the best of our knowledge, we are the first to demonstrate that COMT, TPO, TH, EPX, CMA1, DDC, TPH1, and LPO may be related to the therapeutic effect of H. pylori eradication in chronic gastritis patients.
In summary, this is the first clinical research that dissected the relationships between urinary metabolites and the therapy of H. pylori-positive chronic gastritis. Although this is a groundbreaking original clinical study of H. pylori-positive chronic gastritis, it is limited in that the results still require confirmation in further studies, such as targeted metabolomics, larger patient sample size, and animal experimental studies. Through further study, we expect to develop hippuric acid and metabolites of the citrate cycle as faster urinary biomarkers for evaluating the pathogenesis, development, and prognosis of H. pylori-positive chronic gastritis.
LC–MS-based metabolomics revealed that the major metabolites regulated by H. pylori eradication therapy include cis-aconitic acid, isocitric acid, citric acid, L-tyrosine, L-phenylalanine, L-tryptophan, and hippuric acid, which were involved in four metabolic pathways: (1) Phenylalanine metabolism; (2) phenylalanine, tyrosine, and tryptophan biosynthesis; (3) citrate cycle; and (4) glyoxylate and dicarboxylate metabolism. MPO, COMT, TPO, TH, EPX, CMA1, DDC, TPH1, and LPO were the key proteins involved in the biological process of H. pylori eradication in chronic gastritis. Hence, our research provides a new perspective for exploring the clinical significance of urinary metabolites in chronic gastritis.
Helicobacter pylori (H. pylori) infection is a major risk factor of chronic gastritis, which perhaps influence approximately one-half of global population. H. pylori eradication is a popular treatment method for H. pylori-positive chronic gastritis, but its mechanism is far from clear.
Urinary metabolomics is gradually applied to mine the treatment mechanism of gastric diseases. However, there is no clinical study on urinary metabolomics of chronic gastritis.
This article aimed to investigate metabolic profiles of urine obtained during H. pylori eradication from patients with chronic gastritis.
In this article, we applied LC-MS-based metabolomics and network pharmacology to investigate the relationships between urinary metabolites and H. pylori-positive chronic gastritis via a clinical follow-up study.
Our study revealed the different urinary metabolic profiles of H. pylori-positive chronic gastritis before and after H. pylori eradication. The metabolites regulated by H. pylori eradication include: cis-aconitic acid, isocitric acid, citric acid, L-tyrosine, L-phenylalanine, L-tryptophan and hippuric acid, which were involved in four metabolic pathways: (1) Phenylalanine metabolism; (2) phenylalanine, tyrosine and tryptophan biosynthesis; (3) citrate cycle; and (4) glyoxylate and dicarboxylate metabolism. Integrated metabolomics and network pharmacology revealed that MPO, COMT, TPO, TH, EPX, CMA1, DDC, TPH1 and LPO were the key proteins involved in the involved the biological progress of H. pylori eradication in chronic gastritis.
Our research provides a new perspective for exploring the clinical significance of urinary metabolites in chronic gastritis.
Although this is a groundbreaking original clinical study of H. pylori-positive chronic gastritis, it is limited in that the results still require confirmation in further studies, such as targeted metabolomics, larger patient sample size, and animal experimental studies.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Sato T, Japan S-Editor: Wang JL L-Editor: A P-Editor: Zheng XM
| 1. | Sipponen P, Maaroos HI. Chronic gastritis. Scand J Gastroenterol. 2015;50:657-667. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 126] [Cited by in RCA: 187] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 2. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64684] [Article Influence: 16171.0] [Reference Citation Analysis (177)] |
| 3. | Rugge M, Genta RM. Staging and grading of chronic gastritis. Hum Pathol. 2005;36:228-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 189] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 4. | Lahner E, Esposito G, Pilozzi E, Purchiaroni F, Corleto VD, Di Giulio E, Annibale B. Occurrence of gastric cancer and carcinoids in atrophic gastritis during prospective long-term follow up. Scand J Gastroenterol. 2015;50:856-865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 73] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 5. | de Vries AC, van Grieken NC, Looman CW, Casparie MK, de Vries E, Meijer GA, Kuipers EJ. Gastric cancer risk in patients with premalignant gastric lesions: a nationwide cohort study in the Netherlands. Gastroenterology. 2008;134:945-952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 483] [Cited by in RCA: 586] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 6. | Maluf S, Salgado JV, Cysne DN, Camelo DMF, Nascimento JR, Maluf BVT, Silva LDM, Belfort MRC, Silva LA, Guerra RNM, Salgado Filho N, Nascimento FRF. Increased Glycated Hemoglobin Levels in Patients With Helicobacter pylori Infection Are Associated With the Grading of Chronic Gastritis. Front Immunol. 2020;11:2121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 7. | Yang H, Hu B. Diagnosis of Helicobacter pylori Infection and Recent Advances. Diagnostics (Basel). 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 8. | Ding SZ, Du YQ, Lu H, Wang WH, Cheng H, Chen SY, Chen MH, Chen WC, Chen Y, Fang JY, Gao HJ, Guo MZ, Han Y, Hou XH, Hu FL, Jiang B, Jiang HX, Lan CH, Li JN, Li Y, Li YQ, Liu J, Li YM, Lyu B, Lu YY, Miao YL, Nie YZ, Qian JM, Sheng JQ, Tang CW, Wang F, Wang HH, Wang JB, Wang JT, Wang JP, Wang XH, Wu KC, Xia XZ, Xie WF, Xie Y, Xu JM, Yang CQ, Yang GB, Yuan Y, Zeng ZR, Zhang BY, Zhang GY, Zhang GX, Zhang JZ, Zhang ZY, Zheng PY, Zhu Y, Zuo XL, Zhou LY, Lyu NH, Yang YS, Li ZS; National Clinical Research Center for Digestive Diseases (Shanghai), Gastrointestinal Early Cancer Prevention & Treatment Alliance of China (GECA), Helicobacter pylori Study Group of Chinese Society of Gastroenterology, and Chinese Alliance for Helicobacter pylori Study. Chinese Consensus Report on Family-Based Helicobacter pylori Infection Control and Management (2021 Edition). Gut. 2022;71:238-253. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 117] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 9. | Navashenaq JG, Shabgah AG, Banach M, Jamialahmadi T, Penson PE, Johnston TP, Sahebkar A. The interaction of Helicobacter pylori with cancer immunomodulatory stromal cells: New insight into gastric cancer pathogenesis. Semin Cancer Biol. 2022;86:951-959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 10. | Zhang W, Cui N, Ye J, Yang B, Sun Y, Kuang H. Curcumin's prevention of inflammation-driven early gastric cancer and its molecular mechanism. Chin Herb Med. 2022;14:244-253. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (1)] |
| 11. | Yan L, Chen Y, Chen F, Tao T, Hu Z, Wang J, You J, Wong BCY, Chen J, Ye W. Effect of Helicobacter pylori Eradication on Gastric Cancer Prevention: Updated Report From a Randomized Controlled Trial With 26.5 Years of Follow-up. Gastroenterology. 2022;163:154-162.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 121] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 12. | Rokkas T, Gisbert JP, Malfertheiner P, Niv Y, Gasbarrini A, Leja M, Megraud F, O'Morain C, Graham DY. Comparative Effectiveness of Multiple Different First-Line Treatment Regimens for Helicobacter pylori Infection: A Network Meta-analysis. Gastroenterology. 2021;161:495-507.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 120] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 13. | Li H, Wang R, Sun H. Systems Approaches for Unveiling the Mechanism of Action of Bismuth Drugs: New Medicinal Applications beyond Helicobacter Pylori Infection. Acc Chem Res. 2019;52:216-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 80] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 14. | Jung J, Jung Y, Bang EJ, Cho SI, Jang YJ, Kwak JM, Ryu DH, Park S, Hwang GS. Noninvasive diagnosis and evaluation of curative surgery for gastric cancer by using NMR-based metabolomic profiling. Ann Surg Oncol. 2014;21 Suppl 4:S736-S742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 84] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 15. | Kwon HN, Lee H, Park JW, Kim YH, Park S, Kim JJ. Screening for Early Gastric Cancer Using a Noninvasive Urine Metabolomics Approach. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 16. | Liu Y, Jin Z, Qin X, Zheng Q. Urinary metabolomics research for Huangqi Jianzhong Tang against chronic atrophic gastritis rats based on (1) H NMR and UPLC-Q/TOF MS. J Pharm Pharmacol. 2020;72:748-760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 17. | Tong Y, Zhao X, Wang R, Li R, Zou W, Zhao Y. Therapeutic effect of berberine on chronic atrophic gastritis based on plasma and urine metabolisms. Eur J Pharmacol. 2021;908:174335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 18. | Chen X, Zhang J, Wang R, Liu H, Bao C, Wu S, Wen J, Yang T, Wei Y, Ren S, Tong Y, Zhao Y. UPLC-Q-TOF/MS-Based Serum and Urine Metabonomics Study on the Ameliorative Effects of Palmatine on Helicobacter pylori-Induced Chronic Atrophic Gastritis. Front Pharmacol. 2020;11:586954. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 19. | Gao XX, Ge HM, Zheng WF, Tan RX. NMR-based metabonomics for detection of Helicobacter pylori infection in gerbils: which is more descriptive. Helicobacter. 2008;13:103-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 20. | Matsunaga S, Nishiumi S, Tagawa R, Yoshida M. Alterations in metabolic pathways in gastric epithelial cells infected with Helicobacter pylori. Microb Pathog. 2018;124:122-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 21. | Son SY, Lee CH, Lee SY. Different Metabolites of the Gastric Mucosa between Patients with Current Helicobacter pylori Infection, Past Infection, and No Infection History. Biomedicines. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 22. | Liu Y, Xu W, Wang G, Qin X. Material basis research for Huangqi Jianzhong Tang against chronic atrophic gastritis rats through integration of urinary metabonomics and SystemsDock. J Ethnopharmacol. 2018;223:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 23. | Xu J, Zheng X, Cheng KK, Chang X, Shen G, Liu M, Wang Y, Shen J, Zhang Y, He Q, Dong J, Yang Z. NMR-based metabolomics Reveals Alterations of Electro-acupuncture Stimulations on Chronic Atrophic Gastritis Rats. Sci Rep. 2017;7:45580. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 24. | Rahman I, Afzal NA, Patel P. The role of magnetic assisted capsule endoscopy (MACE) to aid visualisation in the upper GI tract. Comput Biol Med. 2015;65:359-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 25. | Wang YK, Kuo FC, Liu CJ, Wu MC, Shih HY, Wang SS, Wu JY, Kuo CH, Huang YK, Wu DC. Diagnosis of Helicobacter pylori infection: Current options and developments. World J Gastroenterol. 2015;21:11221-11235. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 218] [Cited by in RCA: 264] [Article Influence: 26.4] [Reference Citation Analysis (8)] |
| 26. | Domșa AT, Gheban D, Lazăr C, Pop B, Borzan CM. Particular Morphological Features in the Diagnosis of Pediatric Helicobacter pylori Gastritis: A Morphometry-Based Study. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 27. | Uematsu J, Sugimoto M, Hamada M, Iwata E, Niikura R, Nagata N, Fukuzawa M, Itoi T, Kawai T. Efficacy of a Third-Generation High-Vision Ultrathin Endoscope for Evaluating Gastric Atrophy and Intestinal Metaplasia in Helicobacter pylori-Eradicated Patients. J Clin Med. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 28. | Alzoubi H, Al-Mnayyis A, Al Rfoa I, Aqel A, Abu-Lubad M, Hamdan O, Jaber K. The Use of (13)C-Urea Breath Test for Non-Invasive Diagnosis of Helicobacter pylori Infection in Comparison to Endoscopy and Stool Antigen Test. Diagnostics (Basel). 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 29. | Cardos AI, Maghiar A, Zaha DC, Pop O, Fritea L, Miere Groza F, Cavalu S. Evolution of Diagnostic Methods for Helicobacter pylori Infections: From Traditional Tests to High Technology, Advanced Sensitivity and Discrimination Tools. Diagnostics (Basel). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 41] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 30. | Shimura T, Dayde D, Wang H, Okuda Y, Iwasaki H, Ebi M, Kitagawa M, Yamada T, Hanash SM, Taguchi A, Kataoka H. Novel urinary protein biomarker panel for early diagnosis of gastric cancer. Br J Cancer. 2020;123:1656-1664. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 31. | Mokhtari M, Rezaei A, Ghasemi A. Determination of urinary 5-hydroxyindoleacetic acid as a metabolomics in gastric cancer. J Gastrointest Cancer. 2015;46:138-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 32. | Mabe K, Kikuchi S, Okuda M, Takamasa M, Kato M, Asaka M. Diagnostic accuracy of urine Helicobacter pylori antibody test in junior and senior high school students in Japan. Helicobacter. 2017;22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 33. | Aumpan N, Vilaichone RK, Chotivitayatarakorn P, Pornthisarn B, Cholprasertsuk S, Bhanthumkomol P, Kanokwanvimol A, Siramolpiwat S, Mahachai V. High Efficacy of Rapid Urine Test for Diagnosis of Helicobacter pylori Infection in Thai People. Asian Pac J Cancer Prev. 2019;20:1525-1529. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 34. | Syam AF, Miftahussurur M, Uwan WB, Simanjuntak D, Uchida T, Yamaoka Y. Validation of Urine Test for Detection of Helicobacter pylori Infection in Indonesian Population. Biomed Res Int. 2015;2015:152823. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 35. | Chang CC, Chen SH, Lien GS, Lou HY, Hsieh CR, Fang CL, Pan S. Eradication of Helicobacter pylori significantly reduced gastric damage in nonsteroidal anti-inflammatory drug-treated Mongolian gerbils. World J Gastroenterol. 2005;11:104-108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 8] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 36. | Shiomi S, Toriie A, Imamura S, Konishi H, Mitsufuji S, Iwakura Y, Yamaoka Y, Ota H, Yamamoto T, Imanishi J, Kita M. IL-17 is involved in Helicobacter pylori-induced gastric inflammatory responses in a mouse model. Helicobacter. 2008;13:518-524. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 77] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 37. | Rautelin HI, Oksanen AM, Veijola LI, Sipponen PI, Tervahartiala TI, Sorsa TA, Lauhio A. Enhanced systemic matrix metalloproteinase response in Helicobacter pylori gastritis. Ann Med. 2009;41:208-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 65] [Article Influence: 4.1] [Reference Citation Analysis (0)] |