Published online Aug 16, 2024. doi: 10.12998/wjcc.v12.i23.5338
Revised: June 4, 2024
Accepted: June 20, 2024
Published online: August 16, 2024
Processing time: 82 Days and 22.4 Hours
Influenza A and B virus detection is pivotal in epidemiological surveillance and disease management. Rapid and accurate diagnostic techniques are crucial for timely clinical intervention and outbreak prevention. Quantum dot-encoded microspheres have been widely used in immunodetection. The integration of quantum dot-encoded microspheres with flow cytometry is a well-established technique that enables rapid analysis. Thus, establishing a multiplex detection method for influenza A and B virus antigens based on flow cytometry quantum dot microspheres will help in disease diagnosis.
To establish a codetection method of influenza A and B virus antigens based on flow cytometry quantum dot-encoded microsphere technology, which forms the foundation for the assays of multiple respiratory virus biomarkers.
Different quantum dot-encoded microspheres were used to couple the monoclonal antibodies against influenza A and B. The known influenza A and B antigens were detected both separately and simultaneously on a flow cytometer, and the detection conditions were optimized to establish the influenza A and B antigen codetection method, which was utilized for their detection in clinical samples. The results were compared with the fluorescence quantitative poly
The limits of detection of this method were 26.1 and 10.7 pg/mL for influenza A and B antigens, respectively, which both ranged from 15.6 to 250000 pg/mL. In the clinical sample evaluation, the proposed method well correlated with the fluorescent quantitative PCR method, with positive, negative, and overall compliance rates of 57.4%, 100%, and 71.6%, respectively.
A multiplex assay for quantitative detection of influenza A and B virus antigens has been established, which is characterized by high sensitivity, good specificity, and a wide detection range and is promising for clinical applications.
Core Tip: Respiratory viruses primarily target and affect the respiratory system, such as the influenza A and B viruses, highly contagious and can spread through various means. The detection of influenza A and B virus antigens is significant for the diagnosis, treatment, and prevention of influenza. In this study, a multiplex detective method for influenza A and B virus antigens was developed using flow cytometry quantum dot microspheres. The multiplex assay is characterized by high sensitivity, good specificity, and a broad detection range, making it a promising tool for clinical applications.
- Citation: Xia CJ, Li BH, Guo YN, Zhou XH, Zhang RL, Niu YN. Establishment and performance analysis of a new multiplex detection method for influenza an and B virus antigen. World J Clin Cases 2024; 12(23): 5338-5345
- URL: https://www.wjgnet.com/2307-8960/full/v12/i23/5338.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i23.5338
Infections caused by respiratory viruses and their prevention and control are common public health challenges worldwide. The coronavirus disease 2019 pandemic that occurred in recent years has increased the attention of the public, public health agencies, and policy makers worldwide to infections caused by respiratory viruses[1,2]. Because different respiratory viruses and different subtypes of the same virus may have different treatment modalities and preventive and control measures, rapid and accurate identification of these viruses and their subtypes provide not only a sufficient basis for disease diagnosis and treatment but also a solid foundation for epidemiological investigations and prevention and control of outbreaks[3,4]. Therefore, developing a rapid, sensitive, and multiplexed method for the detection of respiratory viruses is necessary. The bead array technology has a unique advantage in multiplexed detection because it can detect multiple viral proteins (antigens or antibodies) in a rapid, high-throughput, parallelized manner[5-7]. The core principle of bead array technology is to use specific molecules linked to differently labeled microspheres to recognize the corresponding detectors; thus, how to prepare encoded microspheres is the key to achieving high-throughput, high sensitivity, and multiplexed detection. Currently, the bead array technology in the market is mainly dominated by Luminex, which is expensive and requires expensive multichannel detection instruments. Compared with traditional dyes, quantum dots (QDs), which have the advantages of broad absorption spectrum, narrow excitation fluorescence spectrum, high luminescence efficiency, adjustable luminescence spectrum with particle size, and good photostability, have gradually become the focus of fluorescence coding particles in bead array[8-12].
In this study, we proposed to develop a multiplex flow cytometry (FCM) detection method for influenza A and Influenza B virus antigens, using QD-encoded microspheres as reaction carriers to establish a multiplex detection of influenza A and Influenza B virus antigens based on the FCM QD-encoded microsphere technology and lay the foundation for the multiplex detection of common respiratory viruses.
In this retrospective study, throat swab samples were collected from 81 patients with suspected respiratory viral infections attending the outpatient clinic of the University of Chinese Academy of Sciences, Shenzhen Hospital (Guangming) West Campus between May and August 2022. Of these patients, 43 were male and 38 were female, aged 1–63 years. Patient samples were clinically diagnosed with influenza A and B virus infections using the colloidal gold antigen detection kit (Guangzhou Wondfo), and the remaining pharyngeal swab samples were frozen at −80°C. Patients with clinical symptomatic manifestations of respiratory viral infections were included, whereas those who had taken anti-influenza medications within 1 month before sampling were excluded.
DxFlex flow cytometer (Beckman Coulter, CA, United States); ABI 7,500 real-time fluorescence quantitative polymerase chain reaction (PCR) instrument (Life Technologies, CA, United States); influenza A and B virus NP proteins, influenza A virus NP protein monoclonal antibodies (clone nos. FluA6 and FluA8), influenza B virus NP protein monoclonal antibodies (clone nos. FluB15, FluB66, and FluB79) were obtained from Nanjing Novozymes Medical Technology Co, Ltd. (China). Influenza A virus NP protein monoclonal antibodies (clone nos. FluB15, FluB66, and FluB79) are from Nanjing Novozymes Medical Technology Co., Ltd. Microspheres and QD reagents included fluorescence-encoded microspheres (Bangs Laboratories, Inc., IN, United States), polystyrene microspheres (Suzhou Institute of Nano-Tech and Nano-Bionics, China), CdSe/ZnS QD nanocrystals (Wuhan Jiayuan, China). N-hydroxysuccinimide N-hydroxysuccinimide (sulfo-NHS), 1-ethyl-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC), and 2-morpholine ethanesulfonic acid (MES) (Sigma Aldrich, MA, United States); biotin labeling kit (Thermo Fisher Scientific, MA, United States); flow cytometer quality control microspheres and flow cytometer sheath solution (Beckman Coulter); and fluorescence quantitative PCR reagents and consumables (Daan Gene Co., Ltd., China).
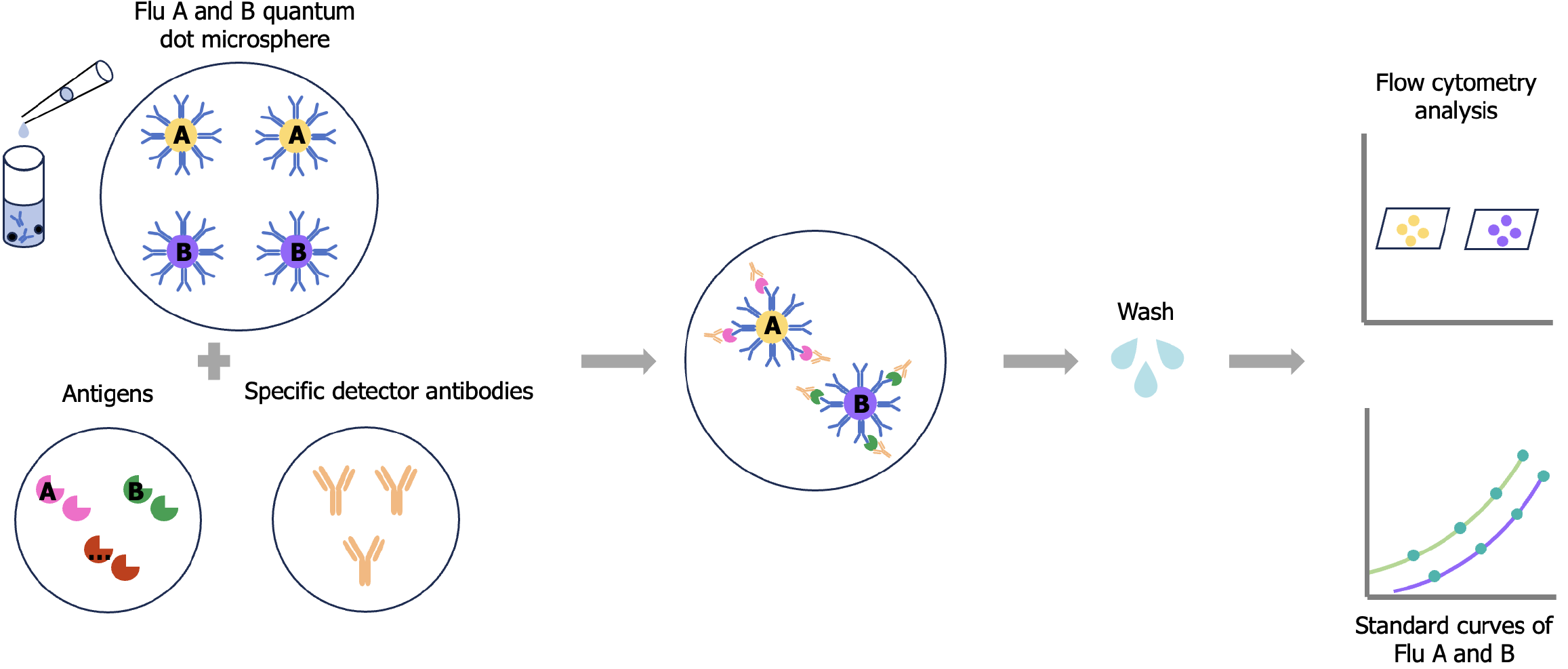
QD-encoded microsphere detection system construction: (1) For QD-encoded microsphere preparation, polystyrene microspheres were firstly dissolved, and then fluorescent staining was performed by adding different concentrations of CdSe/ZnS QDs in the dissolution system. The microspheres were transferred from the organic-phase dissolution system to the aqueous-phase preservation system after staining. By adjusting the concentration ratio of QDs and polystyrene microspheres, multi-peak QD-encoded microspheres with different brightness could be obtained. Because polystyrene microspheres have a carboxyl group modification on the surface, they can be used for subsequent antibody coupling; (2) For antibody coupling to microspheres, the microsphere concentration was first determined using a blood counting plate, then approximately one million microspheres were removed for activation, and the microspheres were washed to MES buffer as the activation system. After rapid addition of EDC (0.2 mg) and sulfo-NHS (0.2 mg) to the microspheres, they were vortexed rapidly for 10 s and placed on a turnover mixer to react for 20 min. The activated microspheres were washed twice using MES buffer, and the antibody was added, rapidly vortexed for 10 s, and placed on a turnover mixer for 2 hours. The coupled microspheres were washed, the concentration was determined, the yield was calculated, and coupling was confirmed. To compare the performance of different antibodies, different mouse anti-influenza A/B virus NP protein monoclonal antibodies (clone no. Flu A6, Flu A8, Flu B15, Flu B66, and Flu B79) were coupled to select the optimal combination of antibody pairs in subsequent tests; and (3) In the preparation of biotinylated antibodies, the ultrafiltration column was initially used to replace the influenza A and B antibodies into the phosphate-buffered saline solution, and then the concentrations of influenza A and B antibodies were measured. The biotinylated reagent was added to the solution-exchanged antibody, mixed upside down for 10 s, and then turned over and mixed for 1 h at room temperature to complete the biotin labeling of the antibody. After the reaction, the reaction product was purified using an ultrafiltration column to remove the unreacted biotinylated reagent, and the concentration of the biotinylated antibody was then determined using an enzyme marker. The concentration of biotin (B) bound to IgG was determined by the HABA reaction, and the molar ratio (B/P) of biotin to IgG antibody (P) was then calculated.
Performance testing of the single test for influenza A and B virus antigens: The development of the encoded microsphere-based multiple detection method for influenza A and B virus antigens requires the establishment and validation of a single test for influenza A and B antigens, and the single test must be subjected to cotesting of this single test method.
The influenza A antigen single test mainly consists of the following steps: (1) Preparation of standards: Dilute the influenza A antigen masterbatch to a series of concentrations such as 1 × 106, 2 × 105, 40000, 8000, 1600, 320, 64, 12.8, and 2.56 pg/mL by serial dilution; (2) Immunoassay: Add the diluent, standards/samples, and biotinylated antibody for detection of influenza A antigen, and capture the microspheres coupled to the antibody for the influenza A antigen to a 96-well filtration plate. After incubation for 2.5 hours away from light, add streptavidin-phycoerythrin and incubate for 0.5 hours away from light, then use a vacuum filtration device for filtration, and use a cleaning solution two times, and resuspend the microspheres into the cleaning solution for testing; (3) Testing and data processing: The flow cytometer was used to collect data from each microsphere, and the obtained median fluorescence intensity (MFI) data were imported into GraphPad Prism software for analysis; and (4) A calibration curve was drawn between a series of MFI values of the standards and the concentration of the standards, and the concentration of samples was deduced using the calibration curve. The steps for the single test for influenza B virus antigens were the same as those for influenza A virus, with different antibodies only. The initial performance evaluation of the single test method mainly included the measurement range and cross-reactivity. By observing the distribution of different concentrations of standards on the calibration curve, the measurement range of the method for influenza A and B antigens concentration was determined.
Performance test of influenza A and B virus antigens codetection: The codetection test of influenza A and B antigens is based on a single detection test in which two types of capture microspheres of influenza A and B viruses and two types of biotinylated detection antibodies of influenza A and B viruses are directly added to the same reaction system to establish the formation of the detection sandwich of influenza A and B virus antigens, respectively, at the same time, and ultimately output the fluorescence signal of the antigens on the flow cytometer using the encoded microspheres as the medium. The fluorescence signals of the influenza A and B detection microspheres were then used to obtain the results of the calibration curves and sample concentrations (Figure 1). Referring to the description of the relevant literature[12,13], a performance evaluation was performed on QD microsphere-based codetection methods, including detection limit, measurement range, and cross-reactivity.
Validation and method comparison of clinical samples on an FCM QD microsphere technology-based influenza A and B codetection platform: The clinical pharyngeal swab samples stored at −80 °C were thawed at room temperature for 30 minutes, then added with 500 μL of extraction solution, and rotated along the wall of the tube for approximately 10 times to dissolve the specimens into the extraction solution as much as possible. The extracted sample solution was divided into two: one was used for this method and the other for nucleic acid detection by real-time fluorescence quantitative PCR (the specific procedure was performed in accordance with the instructions for the use of the reagents). The detection limit of the influenza A and B virus antigen detection was used as the critical value for negative and positive judgments, and the negative and positive results were compared with those of PCR to analyze the clinical sensitivity and specificity.
The results were statistically analyzed using Microsoft Excel. Measurement information was expressed as mean ± SD and CV = 100% × s/x. The standard curves were plotted using GraphPad Prism 9 software.
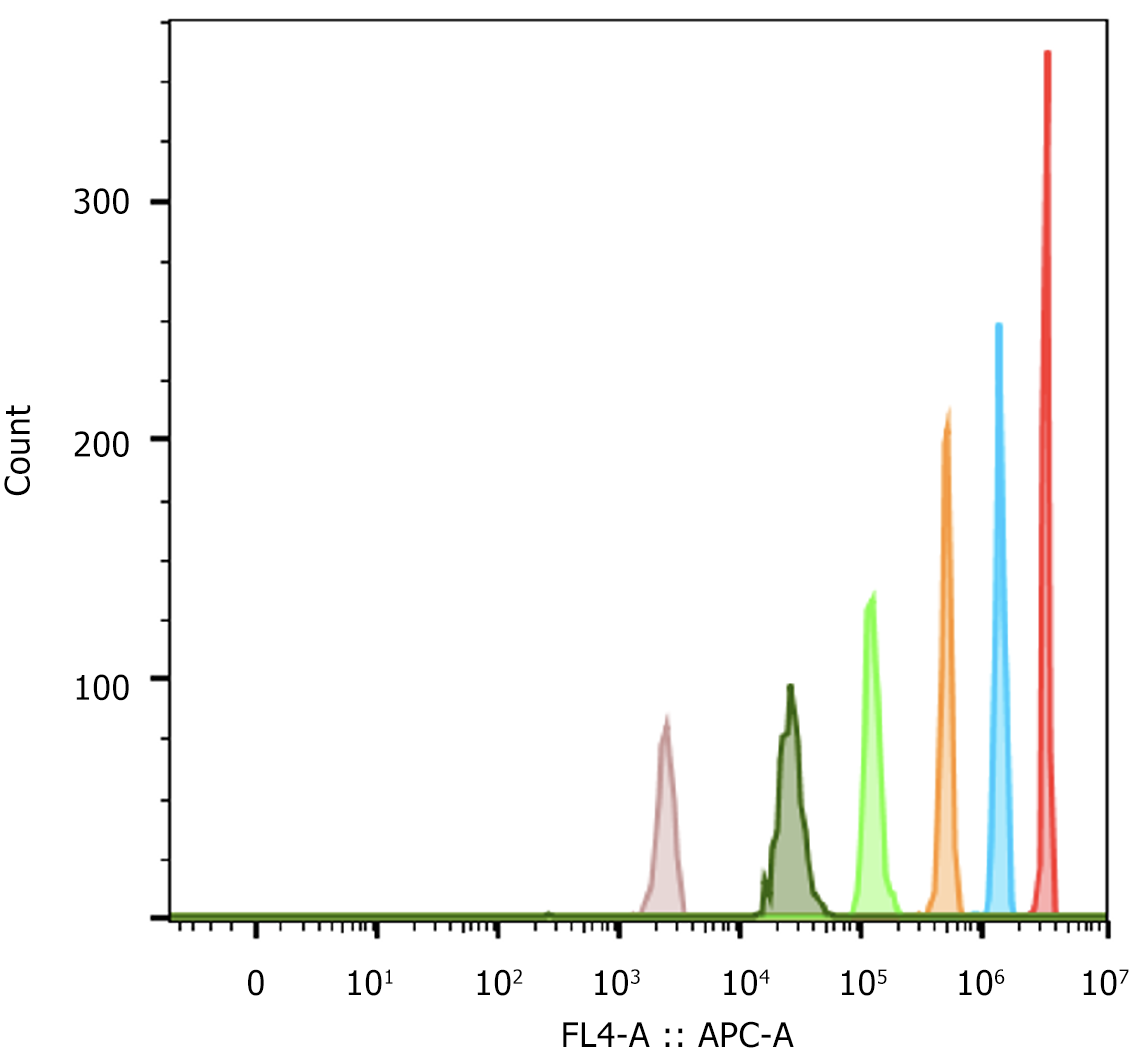
By the solvent-embedding method, a set of fluorescent microspheres based on QD encoding was obtained. FCM analyses showed that the fluorescent microspheres had good clustering performance; theoretically, the simultaneous detection of six analytes could be achieved (Figure 2). To demonstrate the feasibility of antibody coupling, conventional fluorescent dye-based encoded microspheres, BAL1 and BAL2, were first used to couple with influenza A and B antibodies, whereas a species-specific fluorescent-labeled antibody (FITC-Donkey-anti-mouse IgG at a concentration of 10 μg/mL) was used for labeling the coupled microspheres, and the relevant characterization parameters were determined (Table 1), which were consistent with previously reported coupling results[14,15]. On the basis of the results, QD-encoded microspheres were used for the labeling of antibodies against influenza A and B, and the relevant characterization parameters were also determined. As a result, the microsphere recovery and antibody coverage and coupling uniformity (CV%) after antibody coupling were good and could be used for subsequent detection. The overall recoveries of biotinylated antibodies before and after labeling were 55%–70%. The B/P of the biotinylated antibodies to influenza A and B viruses was between 4 and 6, which are consistent with the ideal labeling results reported previously[16], proving that these reagents can meet the needs of subsequent tests.
| Microsphere recovery, % | MEFL value | CV, % | |
| BAL1, BAL2 | > 80 | 2 × 105-6 × 105 | < 25 |
| Quantum dot microspheres | > 80 | 1 × 106-1.5 × 106 | < 25 |
In the early stage of the study, to explore the feasibility of the single detection technique for influenza A and B virus antigens, BAL1- and BAL2-encoded microspheres were initially used for the preliminary establishment of the performance, and parameters such as the antibody concentration, 1-Stearoyl-2-arachidonoyl-sn-glycero-3-phosphorylethanolamine dye quantity, incubation time, and incubation steps, were optimized, and analytical performances, such as the measurement range, and cross-reactivity, were also initially investigated. These research results laid the technical foundation of the microsphere multiplex immunoassay. In the preliminary study, the measurement range of influenza A and B combined assay was 7.81–1 × 106 pg/mL, and the signal generated by influenza A antigens on influenza B capture microspheres at a concentration of 2 × 105 pg/mL was 0.21%. Influenza B antigen at a concentration of 2 × 105 pg/mL produced a signal of 0.01% on influenza A capture microspheres (Table 2). These results demonstrate that the method can be used for the simultaneous detection of influenza A and B virus antigens. Thereafter, BAL1- and BAL2-encoded microspheres were replaced with QD-encoded microspheres, and a preliminary sensitivity assessment was performed for the single detection of influenza A and B virus antigens. The blank limits of the single test system using QD-encoded microspheres were 10.5 and 2.9 pg/mL for the detection of influenza A and B virus antigens, respectively.
| Microspheres | Proteins concentration (pg/mL) | Cross-reaction rate, % | |
| Influenza B NP proteins | Influenza A NP proteins | ||
| Influenza A | 2 × 105 | 0.01 | |
| 1 × 106 | 0.01 | ||
| Influenza B | 2 × 105 | 0.21 | |
| 1 × 106 | 0.16 | ||
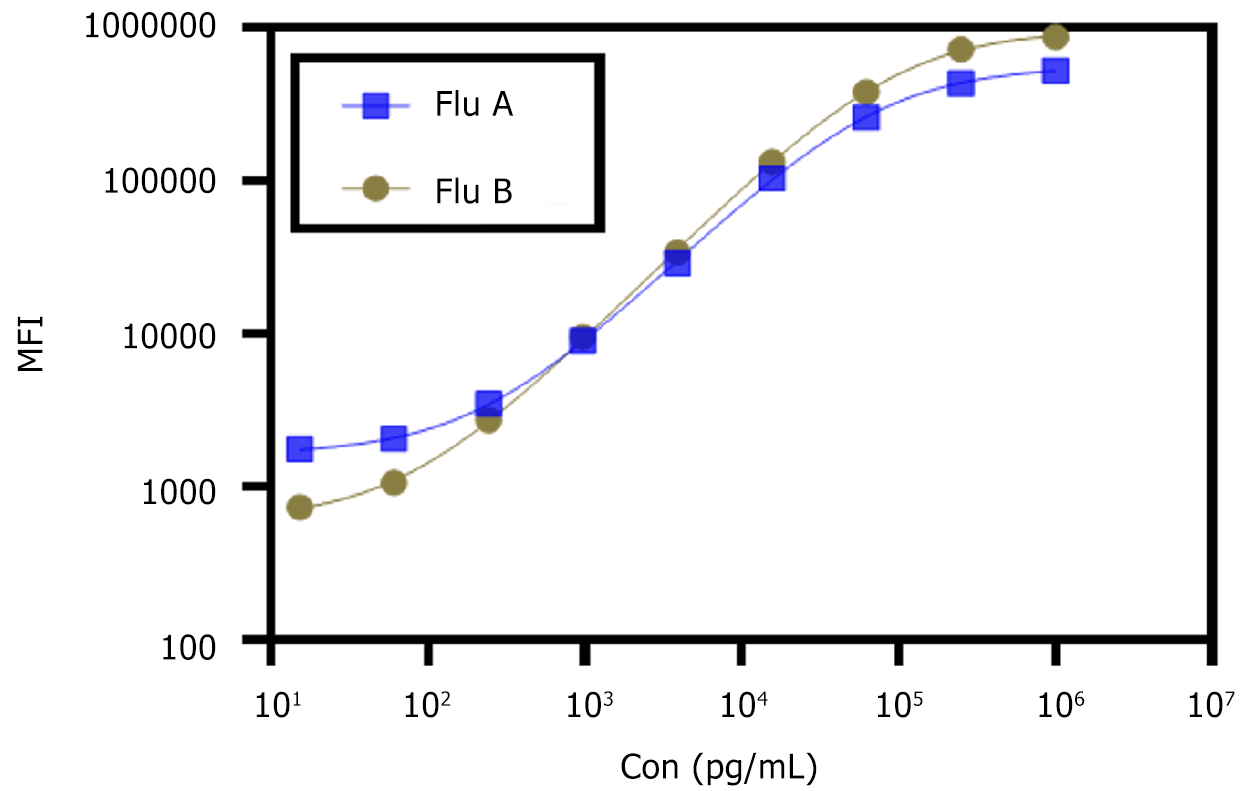
On the basis of the above preliminary study, a QD-encoded microsphere-based codetection method for influenza A and B virus antigens was established, and a systematic study of sensitivity and measurement range was then performed. The calibration curve of influenza A and B antigen codetection is shown in Figure 3. The detection limit of the multiplexed detection system using QD-encoded microspheres was 26.1 (range, 15.6–2.5 × 105 pg/mL) for the detection of influenza A antigens. The detection limit of the detection of influenza B antigens was 10.7 (range, 15.6–2.5 × 105) pg/mL.
Based on the analytical performance study, the results of the multiple detection of influenza A and B virus antigens were compared with those of fluorescent quantitative PCR influenza A and B nucleic acid detection. Among the 81 samples, 54 were positive in the influenza A PCR (Table 3), with CT values ranging from 25.59 to 39.12, and 27 samples were negative in the influenza A PCR, with CT values > 40. The results of the QD-encoded microsphere-based detection of influenza A virus antigens showed that 31 samples were positive for influenza A antigens, with concentrations ranging from 26.1 to 83,019.1 pg/mL, and 50 samples were negative for influenza A antigens, with detected concentrations < 26.1 pg/mL. Accordingly, a positive compliance rate (sensitivity) of 57.4%, negative compliance rate (specificity) of 100%, and overall compliance rate of 71.6% were calculated, which is superior to the currently commonly used clinical reagents for colloidal gold antigen detection, such as Wondfo’s positive compliance rate of 56.49% and a negative compliance rate of 99.75% (taken from the instructions for the reagent). Among the 81 samples, one was positive in the PCR for influenza B, with a CT value of 36.5, which was also positive in the multiplex method, with a concentration of 261.2 pg/mL of influenza B antigens, and the remaining 80 samples negative in the PCR for influenza B were also fully compatible with this multiplex method, with a sensitivity and specificity of 100%.
| Antigen multiplexing detection | q-PCR assay | Sensitivity, % | Idiosyncrasyy, % | |
| Positive | Negative | |||
| Influenza A virus | ||||
| Positive | 31 | 0 | 57.4 | 100 |
| Negative | 23 | 27 | ||
| Influenza B virus | ||||
| Positive | 1 | 0 | 100 | 100 |
| Negative | 0 | 80 | ||
In this study, a QD-encoded microsphere FCM immunoassay method for influenza A and B virus antigens was developed. Multiple QD-encoded microspheres, antibody coupled encoded microspheres, and biotinylated antibodies were successfully prepared, and their analytical performances, such as sensitivity, measurement range, and cross-reactivity, for the influenza A and B antigen detection were investigated. The results confirmed that the codetection method can be used for the subsequent testing of clinical samples. In response to the problems of QD leakage and fluorescence intensity fluctuations encountered during development and testing, QD-encoded microspheres with long-term stability were obtained by optimizing the solvent system, preservation conditions, and surface modification.
In a test of 81 clinical samples, 31 positive samples and 50 negative samples for influenza were measured using this method. When compared with the results of the fluorescent quantitative PCR, the negative and positive compliance rates were 100% and 57.4%, respectively. This indicates that this method is effective in avoiding false-positive results; however, for false-negative results, it suggests an inherent difference in the detection sensitivity between immunoassays and nucleic acid detection methods[3,17,18], which is due to the ability of PCR amplification to detect very low amounts of nucleic acid sequences and maybe because this method requires further optimization to improve the sensitivity. Subsequent optimization of the conditions can be referred to the protocol of multiple microspheres[19], where the sensitivity can be improved by decreasing the initial incubation volume of microspheres and samples, increasing the initial incubation time, decreasing the amount of capture antibody coupled to microspheres, and screening for antibodies with a higher affinity to be used as capture or detection antibodies to achieve increased sensitivity. On the contrary, PCR is used to determine viral nucleic acid sequences, which may also produce positive results for inactive viruses, with a certain false-positive rate. Quantitative immunological assays are more effective in assessing changes in viral activity status and viral load and have fewer false-positive results, making them valuable not only for diagnosis but also for determining disease severity and prognosis. In addition, the clinical samples used were secondary eluates performed on pharyngeal swab samples that had already undergone primary extract elution; thus, their results may have a greater likelihood of false-negative results relative to PCR, which also exceeds the sensitivity and specificity of the colloidal gold immunoassay when compared with detection by colloidal gold immunoassay after the first elution. This demonstrates the superiority of the method. Follow-up research work is needed to evaluate the antigen concentrations in real pharyngeal swab samples to obtain more complete data for performance studies. Meanwhile, very few influenza B virus-positive samples resulted in the evaluation of the sensitivity of this method for influenza B virus to be verified by more positive specimens; however, the specificity also reached 100% in comparison with PCR, which was consistent with the results for influenza A, proving the feasibility of this method in multiplex detection. As its main advantage, this method can be used for multiplex detection of respiratory viruses using the FCM detection platform, which is also the direction of our next research.
As study limitations, FCM QD microspheres are not sensitive enough compared with PCR; thus, more clinical samples are needed to validate the method.
This study demonstrated the feasibility of using QD-encoded microsphere for codetection of influenza A and B viruses. Therefore, we will continue to work on the development of immunoassays for various respiratory viruses and develop them into a rapid diagnostic technology platform that can be used for the immediate detection of multiple viruses by taking advantage of the unique advantages of QD-encoded microspheres to make rapid respiratory viruses testing more affordable, easy to use, and universally accessible.
| 1. | Qiu J. Covert coronavirus infections could be seeding new outbreaks. Nature. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 145] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 2. | Xu XL, Ge SW, Chen CN, Wei M, Fu JY. Analysis of etiology and epidemic characteristics of children respiratory tract virus infection in Shijiazhuang area. Xiandai Jianyan Yixue Zazhi. 2021;36:140-143. [DOI] [Full Text] |
| 3. | Zhang N, Wang L, Deng X, Liang R, Su M, He C, Hu L, Su Y, Ren J, Yu F, Du L, Jiang S. Recent advances in the detection of respiratory virus infection in humans. J Med Virol. 2020;92:408-417. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 243] [Cited by in RCA: 318] [Article Influence: 63.6] [Reference Citation Analysis (0)] |
| 4. | Lu X, Wang Q, Zhang Y. Detection of respiratory viruses by multiplex RT-PCR with a GeXP analyzer. Int J Infect Dis. 2014;21:444. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 5. | Graham H, Chandler DJ, Dunbar SA. The genesis and evolution of bead-based multiplexing. Methods. 2019;158:2-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 67] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 6. | Dincer C, Bruch R, Kling A, Dittrich PS, Urban GA. Multiplexed Point-of-Care Testing - xPOCT. Trends Biotechnol. 2017;35:728-742. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 288] [Cited by in RCA: 329] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 7. | Rho J, Jang W, Hwang I, Lee D, Lee CH, Chung TD. Multiplex immunoassays using virus-tethered gold microspheres by DC impedance-based flow cytometry. Biosens Bioelectron. 2018;102:121-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 8. | Bian F, Sun L, Cai L, Wang Y, Zhao Y. Quantum dots from microfluidics for nanomedical application. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2019;11:e1567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 9. | Cheng Y, Ling SD, Geng Y, Wang Y, Xu J. Microfluidic synthesis of quantum dots and their applications in bio-sensing and bio-imaging. Nanoscale Adv. 2021;3:2180-2195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 10. | Şahin S, Ünlü C, Trabzon L. Affinity biosensors developed with quantum dots in microfluidic systems. Emergent Mater. 2021;4:187-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 11. | Singh S, Dhawan A, Karhana S, Bhat M, Dinda AK. Quantum Dots: An Emerging Tool for Point-of-Care Testing. Micromachines (Basel). 2020;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 12. | Li J, Liu H, Han JX, Hou WJ, Sun ZZ, Fu Y, Yang BC, Shi ZH. Preparation and performance evaluation on novel corona virus (SARS-CoV-2) S1 antigen quantum dots fluorescence immunoassay reagent. Xiandai Jianyan Yixue Zazhi. 2022;37:148-152. [DOI] [Full Text] |
| 13. | Tan YH, Cao CL, Zhang RF, Pan XF, Li GC, She HJ, Liang TC, Feng JM. Establishment and performance evaluation of a time-resolved fluorescence immunoassay for the detection of placental growth Factor. Xiandai Jianyan Yixue Zazhi. 2023;38:112-116+146. [DOI] [Full Text] |
| 14. | Clotilde LM, Bernard C 4th, Salvador A, Lin A, Lauzon CR, Muldoon M, Xu Y, Lindpaintner K, Carter JM. A 7-plex microbead-based immunoassay for serotyping Shiga toxin-producing Escherichia coli. J Microbiol Methods. 2013;92:226-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 15. | de Jager W, te Velthuis H, Prakken BJ, Kuis W, Rijkers GT. Simultaneous detection of 15 human cytokines in a single sample of stimulated peripheral blood mononuclear cells. Clin Diagn Lab Immunol. 2003;10:133-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 307] [Cited by in RCA: 412] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 17. | Vemula SV, Zhao J, Liu J, Wang X, Biswas S, Hewlett I. Current Approaches for Diagnosis of Influenza Virus Infections in Humans. Viruses. 2016;8:96. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 206] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 18. | Zhang Y, Zhou ZQ, Wang ZX, Li Y, Xiao M, Xu YC, Wang H. Discussion on the clinical value of combined detection of SARS-CoV-2 nucleic acid, antigen and antibody. Xiandai Jianyan Yixue Zazhi. 2020;35:99-102, 109. [DOI] [Full Text] |
| 19. | Das S, Dunbar S. Multiplex Immunoassay Approaches Using Luminex® xMAP® Technology for the Study of COVID-19 Disease. Adv Exp Med Biol. 2023;1412:479-489. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |