Published online Aug 6, 2024. doi: 10.12998/wjcc.v12.i22.5208
Revised: May 29, 2024
Accepted: June 17, 2024
Published online: August 6, 2024
Processing time: 82 Days and 23.3 Hours
Infectious diseases are still one of the greatest threats to human health, and the etiology of 20% of cases of clinical fever is unknown; therefore, rapid identification of pathogens is highly important. Traditional culture methods are only able to detect a limited number of pathogens and are time-consuming; serologic detection has window periods, false-positive and false-negative problems; and nucleic acid molecular detection methods can detect several known pathogens only once. Three-generation nanopore sequencing technology provides new options for identifying pathogens.
Case 1: The patient was admitted to the hospital with abdominal pain for three days and cessation of defecation for five days, accompanied by cough and sputum. Nanopore sequencing of the drainage fluid revealed the presence of oral-like bacteria, leading to a clinical diagnosis of bronchopleural fistula. Cefope
Three-generation nanopore sequencing technology allows for rapid and accurate detection of pathogens in human infectious diseases.
Core Tip: Routine culture methods have traditionally been the primary clinical approach for detecting pathogenic microorganisms. However, this article reports on three cases of refractory infectious diseases caused by pathogens that are not easily detected through routine culture methods or are susceptible to inhibition by the growth of other normal flora during the culture process. Nanopore sequencing technology offers a novel approach to identifying the source of infection, achieving precise and prompt pathogen detection, and guiding the use of clinical antimicrobial drugs.
- Citation: Deng QM, Zhang J, Zhang YY, Jia M, Ding DS, Fang YQ, Wang HZ, Gu HC. Diagnosis and treatment of refractory infectious diseases using nanopore sequencing technology: Three case reports. World J Clin Cases 2024; 12(22): 5208-5216
- URL: https://www.wjgnet.com/2307-8960/full/v12/i22/5208.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i22.5208
Infectious diseases are a major burden on public health and the economic stability of societies around the world[1,2]. According to World Health Organization surveillance statistics, there will be approximately 62.3 million deaths caused by infectious diseases worldwide in 2022. The main cause of infection is the invasion of some foreign pathogenic bacteria, fungi, and mycoplasma when the patient’s immunity is weakened for various reasons. The main gold standard for the diagnosis of infectious patients is pathogen identification, i.e., collection of the patient's body fluids, such as sputum, thoracic and abdominal fluid, cerebrospinal fluid, secretions, and pathological tissues, for routine bacterial culture and identification. However, microbial detection using traditional culture methods is time-consuming and limited to culturable pathogens, which cannot quickly and accurately identify rare pathogens and easily leads to missed diagnoses of rare pathogens[3-6]. It is difficult to meet the needs of clinical diagnosis and treatment. Metagenomic second-generation sequencing [metagenomic next-generation sequencing (mNGS)] is a microbial detection method that does not require the cultivation of the pathogen and has rapid identification, broad coverage, the advantages of high sensitivity, and no specific amplification[7-9]. However, mNGS cannot analyze detection results based on short sequence combinations in real time, and bioinformatics analysis can only be performed after sequencing, which is slow and costly. Because the short reading length (50-300 bp) increases the difficulty of subsequent data analysis and genome splicing, it is difficult to parse the complex genome structure of microorganisms. In recent years, third-generation sequencing technologies, represented by Oxford Nanopore Technologies (ONT), have been able to generate longer reads in real time, with ultralong reads capable of fully assembling microbial genomes[10-14]. Compared with mNGS, nanopores have a shorter turnaround time, a wider detection range, and more accurate results[15-17]. The continuous updating of nanopore sequencing technology has resulted in an improvement in accuracy to 99% in detecting pathogen sequences, with a shorter turnaround time of less than six hours[18,19]. This is of critical importance for the rapid and accurate diagnosis of the source of infection. Our previous study evaluated the ability of nanopore sequencing technology to detect pathogens in immunocompromised cancer patients, and the sensitivity of pathogen detection increased from 44.6% to 83.9%. Nanopore sequencing technology was able to detect more samples with fastidious pathogen infections and coinfections than conventional culture methods and reduced the pathogen detection time to an average of 17.5 hours[20]. This technology has been employed for the rapid identification of pathogens in a variety of sample types, including blood, cerebrospinal fluid, alveolar lavage fluid, feces, and urine[3,21,22]. Its applications include the detection of infectious diseases such as central nervous system infections, bacterial lower respiratory tract infections, infective endocarditis, and artificial joint infections[14,23-26]. Antimicrobial resistance (AMR) is a prerequisite for the treatment of disease. Nanopore sequencing technology is capable of identifying known drug resistance gene targets and predicting the antimicrobial drug susceptibility of pathogens within 1-2 hours[27]. At present, third-generation sequencing technology based on nanopore technology is widely used in the study of animal, plant and microbial samples[28-31]. Its clinical applications mainly include rapid clinical diagnosis of the etiology of infectious diseases, detection of AMR, description of disease-related microbial communities, epidemic monitoring, diagnosis and treatment of tumors and genetic diseases[21,25,32-34].
In this study, we retrospectively analyzed the clinical data and nanopore sequencing results of 3 patients with infectious diseases who were hospitalized at Hefei Cancer Hospital, Chinese Academy of Sciences, from May 2022 to August 2023, and, combined with a review of previous literature, we discussed the application value of nanopore sequencing technology in the diagnosis and treatment of refractory infectious diseases.
Case 1: A 75-year-old male patient presented to our outpatient clinic with “abdominal pain for 3 days and cessation of bowel movement for 5 days”.
Case 2: A 37-year-old female patient presented to our outpatient clinic with nasal congestion, a runny nose for 2 days, and fever with headache for more than 4 hours.
Case 3: A 70-year-old female patient who presented to our outpatient clinic with “intermittent fever with an enlarged neck mass for more than half a month”.
Case 1: The patient was admitted to the hospital 5 days prior for “recurrent cough and sputum for more than 20 years, aggravated for 5 days”, and in the previous 3 days, he had persistent abdominal distension and pain and stopped defecation in the anus for 5 days. Intestinal obstruction was considered at the outside hospital.
Case 2: The patient presented with nasal congestion, coughing up phlegm, nausea, and vomiting after being exposed cold for two days prior to admission to the hospital. He then suddenly developed a severe explosive headache for the first four hours. Her body temperature was self-tested at 39 °C. After taking ibuprofen, the headache persisted, accompanied by emesis of the watery gastric contents. Consequently, the patient was transferred to the emergency department of our institution.
Case 3: Six months ago, the patient presented with fever and an enlarged neck mass without any obvious cause, and her symptoms were relieved after anti-infection treatment. During this period, the aforementioned symptoms were intermittent. In the past month, the symptoms worsened, and the patient was treated with levofloxacin at an outside hospital, which proved ineffective in treating the infection. He then presented to our hospital.
Case 1: The patient had been diagnosed with chronic obstructive pulmonary disease (COPD) for more than 20 years and denied a history of hypertension, diabetes mellitus, or cerebral infarction.
Case 2: The patient had meningitis three years prior, but no sequelae were observed. The patient had a history of ectopic pregnancy and otitis media surgery more than 10 years prior.
Case 3: The patient was in good health.
Case 1: The patient had been diagnosed with COPD for more than 20 years and denied a history of hypertension, diabetes mellitus, or cerebral infarction.
Case 2: The patient had a personal history of meningitis and denied a family history of the disease.
Case 3: The patient denied a family history of the disease.
Case 1: Body temperature, 36.9 °C; heart rate, 97 beats/min; respiratory rate, 20 breaths/min; blood pressure, 136/90 mmHg. Both lungs of the patient exhibited audible dry and wet rales, and the abdomen exhibited tenderness without rebound tenderness. The patient's abdominal bowel sounds exhibited slight hyperactivity, with a frequency of 6 beats per minute, mobile turbid sounds, and sounds of air passing through water.
Case 2: Body temperature, 38.5 °C; heart rate, 120 beats/min; respiratory rate, 19 breaths/min; blood pressure, 122/77 mmHg. The patient exhibited Kerning's sign (+), Brudzinski's sign (+), and no abnormalities on cardiopulmonary auscultation.
Case 3: Body temperature, 36.4 °C; heart rate, 95 beats/min; respiratory rate, 20 breaths/min; blood pressure, 134/94 mmHg. The patient had a subcutaneous mass palpated on the lower border of the sternocleidomastoid muscle on the left side of the neck.
Case 1: A routine blood examination revealed a white blood cell count of 16.12 × 109/L (normal value: 4-10 × 109/L), a neutrophil ratio of 95.82% (normal value: 50%-75%), a lymphocyte ratio of 1.9% (normal value: 20%-40%), a platelet counts of 336 × 109/L (normal value: 100-300 × 109/L), and a C-reactive protein (CRP) concentration > 300 mg/L (normal value: 0-10 mg/L). The plain culture of the pleural drainage fluid was negative.
Case 2: A routine blood examination revealed a white blood cell count of 17.17 × 109/L (normal value: 4-10 × 109/L), a neutrophil ratio of 86.95% (normal value: 50%-75%), a lymphocyte ratio of 4.88% (normal value: 20%-40%), and a CRP level of 176.90 mg/L (normal value: 0-10 mg/L). Cerebrospinal fluid (CSF) examination revealed the following results: transparency, cloudy; Pan's test, positive; CSF biochemistry, total protein, 220.00 mg/dL (normal value: 0-45 mg/dL); glucose, 1.81 mmol/L (normal value: 2.2-3.9 mmol/L); and chlorine, 124 mmol/L (normal value: 110-130 mmol/L). The culture results were negative.
Case 3: A routine blood examination revealed a white blood cell count of 3.64 × 109/L (normal value: 4-10 × 109/L), a neutrophil ratio of 80.97% (normal value: 50%-75%), a lymphocyte ratio of 10.77% (normal value: 20%-40%), a hemoglobin level of 108 g/L (normal value: 120-160 g/L), a red blood cell count of 3.75 × 1012/L (normal value: 4-5.5 × 1012/L), and a CPR level of 89.23 mg/L (normal value: 0-10 mg/L). The blood culture was negative.
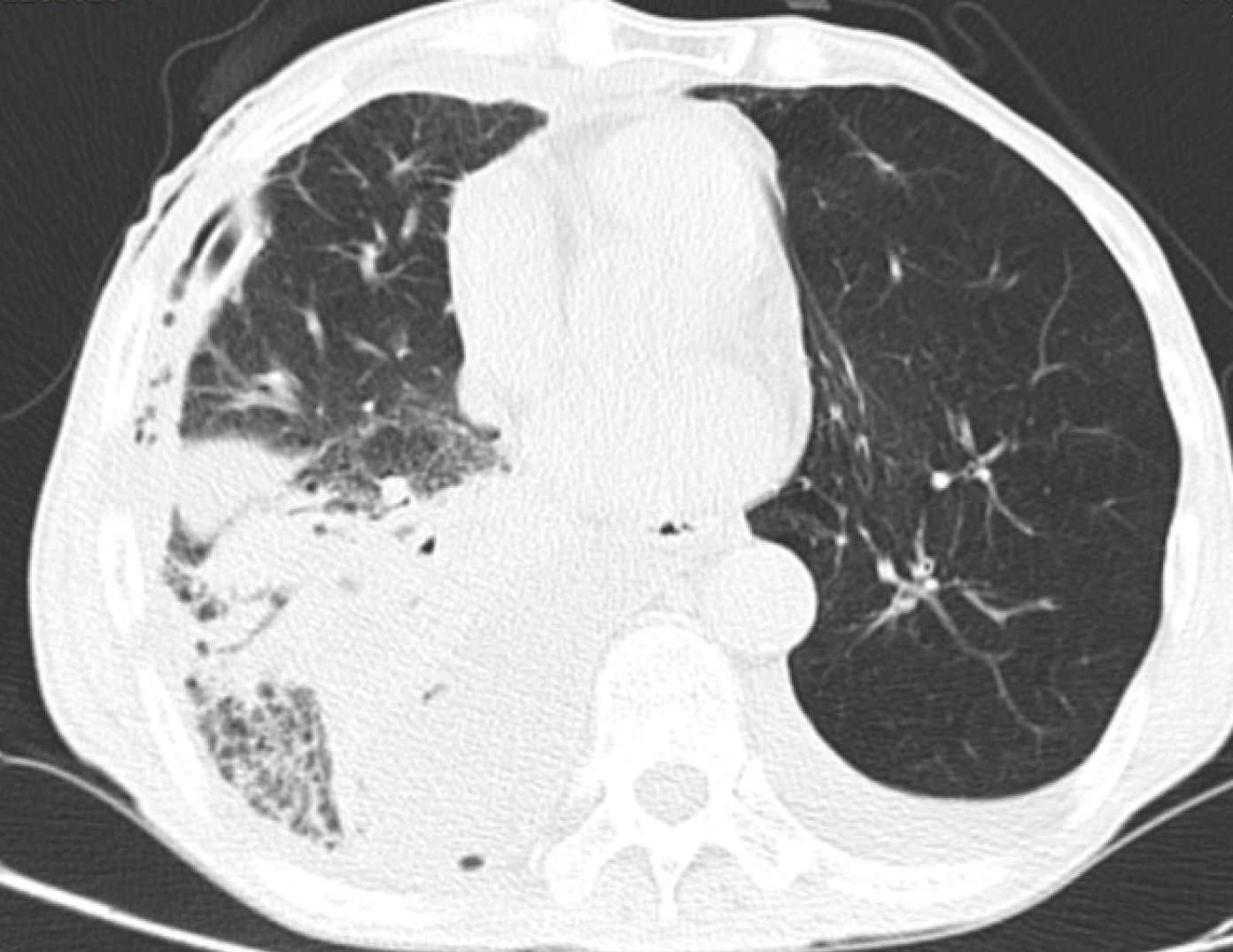
Case 1: Computed tomography (CT) scans of the chest, upper abdomen, lower abdomen, and pelvic cavity showed multiple high-density ground-glass opacities in the right lung, double lung fiber focus, two areas of emphysema, pulmonary bullosa, right pleural effusion and gas, and left pleural effusion (Figure 1).
Case 2: CT scans of the head and chest: CT scan of the brain parenchyma showed no obvious abnormalities. Bone density decreased in the parietal bone, with multiple low-density foci, multiple areas of inflammation in both lungs, and ethmoid sinusitis. Imaging suggested infectious fever and bacterial pneumonia.
Case 3: CT scans of the neck, chest, upper abdomen, lower abdomen, and pelvic cavity show multiple enlarged lymph nodes in the left neck, left clavicle region, bilateral armpits, mediastinum, right lung hilus, and abdominal cavity, and multiple small lymph nodes in the right cervical sheath area and submaxillary area.
Case 1: Sequencing of closed chest drainage fluid revealed Clostridium nucleatus (23, 355), Prevotella Barone (12924), Bacteroides heparinolyticus (4843), Porphyromonas endodontalis (3501), Listeria pneumoniae invasives (3276), Peptostreptococcus stomatitis (3123), and Mycoplasma pharyngopharynx (4671). Gene sequencing suggested more oral-like bacteria in the pleural drainage fluid, and bronchopleural fistula formation was considered.
Case 2: Nanopore sequencing of cerebrospinal fluid revealed Streptococcus pneumoniae (10608).
Case 3: Cervical lymph node cases with biopsies suggestive of hyperplasia are considered inflammatory lesions, although the possibility of T-cell lymphoma cannot be excluded. Sequencing of blood samples revealed the presence of Staphylococcus epidermidis (2) and Aspergillus scopularis (17).
Case 1: A bronchoscopy was performed, during which narrowing of the bronchial lumen in the basal segment of the lower lobe of the right lung was observed. This was confirmed by the use of an "indigo carmine mucous membrane stain" for lavage, which indicated the formation of a bronchopleural fistula in the right lower lung.
Case 2: Septic meningitis, bacterial pneumonia.
Case 3: Bacteremia, mixed-cell Hodgkin's lymphoma.
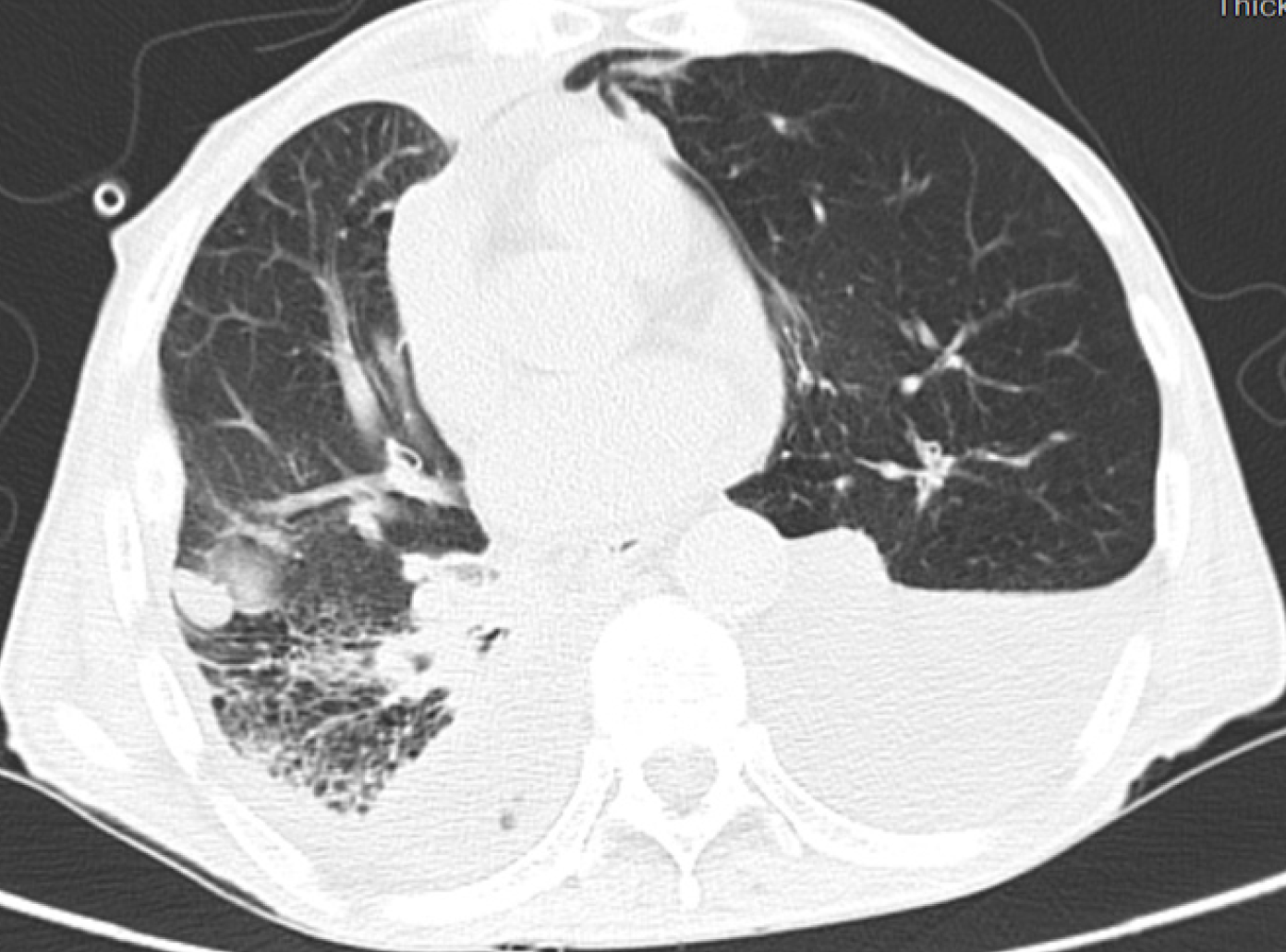
Case 1: Following 14 days of intravenous treatment with cefoperazone sodium sulbactam, the patient was discharged with progressive improvement in chest tightness (Figure 2).
Case 2: Following 14 days of ceftriaxone- and vancomycin-based treatment, the patient was discharged with notable improvement in symptoms and no fever or headache.
Case 3: After the patient was admitted to the hospital and empirically treated with azithromycin for 7 days, her fever improved. Therefore, azithromycin was discontinued, and the patient was discharged. Ten days after azithromycin was discontinued, the patient was admitted to the hospital with a recurrence of fever. Blood culture sequencing revealed Aspergillus scopularis. Voriconazole was administered intravenously for 7 days, the patient's infection symptoms were controlled, and she was discharged to continue oral voriconazole therapy.
Case 1: At the one-month follow-up, the patient's symptoms of chest tightness improved, and he was subsequently admitted to the hospital for further treatment of a bronchopleural fistula.
Case 2: At the eight-month follow-up, the patient exhibited no further fever or headache symptoms.
Case 3: At the 4-month follow-up, the patient was admitted to our oncology department for continued treatment without further intermittent fever due to bacteremia.
A search of the PubMed database using the keywords “Oxford Nanopore Technologies” and “infectious diseases” yielded six case reports on the topic of “case reports of nanopore sequencing technology for the diagnosis of infectious diseases”. After the literature review, four studies reported the use of different second and third-generation sequencing platforms (including ONT, Pacific Biosciences, Ion Torrent, and Illumina)[35-38]. Whole-genome sequencing and bioinformatic analysis of pathogens cultured from patients or special hospital wards (e.g., intensive care units and burn wards) were employed to explore resistance genes, virulence genes, and genetic properties of the infectious strains at the DNA level. Only two papers were “Case reports on nanopore sequencing-based diagnosis of refractory infectious diseases”. One such case was reported by Huang et al[39], a 67-year-old male patient was included. An abscess infection was triggered by a crab bite to the hand. Routine cultures of the wound pus were conducted over a 42-day period, during which no Mycobacterium bovis growth was observed. Targeted nanopore sequencing was then employed to rapidly and accurately identify Mycobacterium maritimus, which were observed to have grown within 16-17 h. The patient was treated with a combination of ethambutol, rifampicin, and isoniazid, and no recurrence was observed during the six-month follow-up period. Another case was reported by Bialasiewicz et al[40], a 62-year-old female patient was included. On the fourth day of blood culture, small and slow-growing colonies were observed. On the sixth day, Capnocytophaga canimorsus was detected by MALDI-TOF. Nanopore macrogenomic sequencing technology was employed to detect the presence of Capnocytophaga canimorsus within 19 h. After 14 days of symptomatic treatment with meropenem, the patient's infection symptoms were successfully controlled. The analogous characteristics of the medical records reported in this paper and the above two articles are as follows: (1) Both employed nanopore sequencing to identify caustic bacteria that cannot be cultured or grow slowly in infectious diseases; and (2) the infectious diseases are clinically progressive and aggressive.
Infection is a common disease with a high mortality rate. The immunity and age of the body are the main factors affecting infection, and from the perspective of pathogens, the number of infected pathogens and the variability of pathogens will affect the state of the body after infection. The most common pathogenic bacteria of various infections in clinical practice are mostly from external invasions, such as Staphylococcus aureus, and some infections are caused by colonizing bacteria of the body itself migrating to sterile tissues or organs, such as Escherichia coli[41,42].
Fernando et al[43] retrospectively studied 68 cases of infection and reported that the percentage of positive microbial cultures was low, at 27.9% (19/68). However, nanopore sequencing can detect bacteria and fungi in specific body fluids that are difficult to detect by conventional culture, which can compensate for the low positive rate of conventional culture. In this group, the oral bacteria that were difficult to identify by conventional culture were sequenced from the thoracic drainage fluid, which was of great help in investigating the etiology of the patient and formulating a follow-up treatment plan. In patient 2, the results of the routine CSF culture were negative, but Streptococcus pneumoniae was detected by CSF nanopore sequencing. Because cerebrospinal fluid is a unique body fluid, conventional bacterial culture results take a long time, and the positivity rate is low, resulting in delayed or even missed diagnoses of patients with cerebrospinal fluid bacterial infection. However, nanopore sequencing can quickly and accurately identify pathogens, which is highly helpful for clinical management. In case 3, nanopore sequencing of whole blood detected a fungus (Aspergillus scopularis) that often grows on indoor air, dust and low-water active substrates. Combined with the patient's life history, the infection was effectively controlled after treatment with voriconazole. Blood culture is still the "gold standard" for the etiological diagnosis of bloodstream infections, but blood culture results are negative in approximately 50% of cases. An increasing number of scholars have begun to use nanopore metagenomic sequencing for the etiological diagnosis of patients with bloodstream infections or as a supplement to routine detection methods such as blood culture[44,45]. The above three examples demonstrate the successful application of nanopore sequencing technology in the treatment of infectious diseases in our hospital, which has overcome the shortcomings of conventional bacterial culture. In particular, in patients with negative cultures, nanopore sequencing can identify difficult-to-diagnose pathogens in a relatively short time, providing a basis for the clinical diagnosis and treatment of patients and providing an effective supplement to traditional detection methods. Nanopore sequencing technology can obtain a rich set of sequence information by using a cost-effective real-time long-read sequencing strategy. Nanopore sequencing has been used to successfully identify more than 10 pathogens involved in the diagnosis of central infection pathogens. These include Streptococcus pneumoniae, Streptococcus suis, Neisseria meningitidis, Staphylococcus aureus, Klebsiella pneumoniae, Pseudomonas aeruginosa, Streptococcus Mitter, Cryptococcus neoformans, Aeromonas, and other rare pathogens. Based on the current limited research, the emergence of nanopore macrogene gene sequencing may provide us with a powerful tool for the diagnosis of infectious disease pathogens, which can achieve rapid and more accurate pathogen detection in human infectious diseases.
As an unbiased etiological diagnostic method for infectious diseases, nanopore sequencing has broad prospects for development. With the continuous development and improvement of nanopore sequencing technology, nanopore sequencing technology will be more widely used in clinical etiological diagnosis. Because of the unique technical advantages of nanopore sequencing, the detection of highly sensitive and rare pathogens can provide a new means for diagnosing clinically refractory infectious diseases to guide clinical antimicrobial therapy.
We are very grateful to the Medical Health Science and Technology for their support of our research. Thank you to all the members of our team for their hard work. We thank all the physicians, nurses and medical staff who cared for these patients.
| 1. | Lehrnbecher T, Averbuch D, Castagnola E, Cesaro S, Ammann RA, Garcia-Vidal C, Kanerva J, Lanternier F, Mesini A, Mikulska M, Pana D, Ritz N, Slavin M, Styczynski J, Warris A, Groll AH; 8th European Conference on Infections in Leukaemia. 8th European Conference on Infections in Leukaemia: 2020 guidelines for the use of antibiotics in paediatric patients with cancer or post-haematopoietic cell transplantation. Lancet Oncol. 2021;22:e270-e280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 81] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 2. | Zaorsky NG, Churilla TM, Egleston BL, Fisher SG, Ridge JA, Horwitz EM, Meyer JE. Causes of death among cancer patients. Ann Oncol. 2017;28:400-407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 446] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 3. | Lamy B, Sundqvist M, Idelevich EA; ESCMID Study Group for Bloodstream Infections, Endocarditis and Sepsis (ESGBIES). Bloodstream infections - Standard and progress in pathogen diagnostics. Clin Microbiol Infect. 2020;26:142-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 128] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 4. | Peri AM, Harris PNA, Paterson DL. Culture-independent detection systems for bloodstream infection. Clin Microbiol Infect. 2022;28:195-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 81] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 5. | Dippenaar A, Goossens SN, Grobbelaar M, Oostvogels S, Cuypers B, Laukens K, Meehan CJ, Warren RM, van Rie A. Nanopore Sequencing for Mycobacterium tuberculosis: a Critical Review of the Literature, New Developments, and Future Opportunities. J Clin Microbiol. 2022;60:e0064621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 41] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 6. | Noone JC, Helmersen K, Leegaard TM, Skråmm I, Aamot HV. Rapid Diagnostics of Orthopaedic-Implant-Associated Infections Using Nanopore Shotgun Metagenomic Sequencing on Tissue Biopsies. Microorganisms. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 7. | Hilt EE, Ferrieri P. Next Generation and Other Sequencing Technologies in Diagnostic Microbiology and Infectious Diseases. Genes (Basel). 2022;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 68] [Reference Citation Analysis (0)] |
| 8. | Gu W, Miller S, Chiu CY. Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection. Annu Rev Pathol. 2019;14:319-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 389] [Cited by in RCA: 839] [Article Influence: 119.9] [Reference Citation Analysis (0)] |
| 9. | Han D, Li Z, Li R, Tan P, Zhang R, Li J. mNGS in clinical microbiology laboratories: on the road to maturity. Crit Rev Microbiol. 2019;45:668-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 235] [Article Influence: 39.2] [Reference Citation Analysis (0)] |
| 10. | Ferreira FA, Helmersen K, Visnovska T, Jørgensen SB, Aamot HV. Rapid nanopore-based DNA sequencing protocol of antibiotic-resistant bacteria for use in surveillance and outbreak investigation. Microb Genom. 2021;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 11. | Hong M, Peng D, Fu A, Wang X, Zheng Y, Xia L, Shi W, Qian C, Li Z, Liu F, Wu Q. The application of nanopore targeted sequencing in the diagnosis and antimicrobial treatment guidance of bloodstream infection of febrile neutropenia patients with hematologic disease. J Cell Mol Med. 2023;27:506-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 12. | Zhu X, Yan S, Yuan F, Wan S. The Applications of Nanopore Sequencing Technology in Pathogenic Microorganism Detection. Can J Infect Dis Med Microbiol. 2020;2020:6675206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 13. | Martí-Carreras J, Carrasco M, Gómez-Ponce M, Noguera-Julián M, Fisa R, Riera C, Alcover MM, Roura X, Ferrer L, Francino O. Identification of Leishmania infantum Epidemiology, Drug Resistance and Pathogenicity Biomarkers with Nanopore Sequencing. Microorganisms. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 14. | Luo W, He Y, Xu J, Zhang S, Li C, Lv J, Shen Y, Ou Z, Dong H. Comparison of Third-Generation Sequencing Technology and Traditional Microbiological Detection in Pathogen Diagnosis of Lower Respiratory Tract Infection. Discov Med. 2023;35:332-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 15. | Petersen LM, Martin IW, Moschetti WE, Kershaw CM, Tsongalis GJ. Third-Generation Sequencing in the Clinical Laboratory: Exploring the Advantages and Challenges of Nanopore Sequencing. J Clin Microbiol. 2019;58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 151] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 16. | Zhao N, Cao J, Xu J, Liu B, Liu B, Chen D, Xia B, Chen L, Zhang W, Zhang Y, Zhang X, Duan Z, Wang K, Xie F, Xiao K, Yan W, Xie L, Zhou H, Wang J. Targeting RNA with Next- and Third-Generation Sequencing Improves Pathogen Identification in Clinical Samples. Adv Sci (Weinh). 2021;8:e2102593. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 39] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 17. | Jain M, Koren S, Miga KH, Quick J, Rand AC, Sasani TA, Tyson JR, Beggs AD, Dilthey AT, Fiddes IT, Malla S, Marriott H, Nieto T, O'Grady J, Olsen HE, Pedersen BS, Rhie A, Richardson H, Quinlan AR, Snutch TP, Tee L, Paten B, Phillippy AM, Simpson JT, Loman NJ, Loose M. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol. 2018;36:338-345. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1615] [Cited by in RCA: 1162] [Article Influence: 166.0] [Reference Citation Analysis (0)] |
| 18. | Sereika M, Kirkegaard RH, Karst SM, Michaelsen TY, Sørensen EA, Wollenberg RD, Albertsen M. Oxford Nanopore R10.4 long-read sequencing enables the generation of near-finished bacterial genomes from pure cultures and metagenomes without short-read or reference polishing. Nat Methods. 2022;19:823-826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 235] [Article Influence: 78.3] [Reference Citation Analysis (0)] |
| 19. | Gu W, Deng X, Lee M, Sucu YD, Arevalo S, Stryke D, Federman S, Gopez A, Reyes K, Zorn K, Sample H, Yu G, Ishpuniani G, Briggs B, Chow ED, Berger A, Wilson MR, Wang C, Hsu E, Miller S, DeRisi JL, Chiu CY. Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat Med. 2021;27:115-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 413] [Article Influence: 82.6] [Reference Citation Analysis (0)] |
| 20. | Deng Q, Cao Y, Wan X, Wang B, Sun A, Wang H, Wang Y, Wang H, Gu H. Nanopore-based metagenomic sequencing for the rapid and precise detection of pathogens among immunocompromised cancer patients with suspected infections. Front Cell Infect Microbiol. 2022;12:943859. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 21. | Sun X, Song J, Leng X, Li F, Wang H, He J, Zhai W, Wang Z, Wu Q, Li Z, Ruan X. A preliminary evaluation of targeted nanopore sequencing technology for the detection of Mycobacterium tuberculosis in bronchoalveolar lavage fluid specimens. Front Cell Infect Microbiol. 2023;13:1107990. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 22. | Afflerbach AK, Rohrandt C, Brändl B, Sönksen M, Hench J, Frank S, Börnigen D, Alawi M, Mynarek M, Winkler B, Ricklefs F, Synowitz M, Dührsen L, Rutkowski S, Wefers AK, Müller FJ, Schoof M, Schüller U. Classification of Brain Tumors by Nanopore Sequencing of Cell-Free DNA from Cerebrospinal Fluid. Clin Chem. 2024;70:250-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 35] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 23. | Noone JC, Ferreira FA, Aamot HV. Culture-Independent Genotyping, Virulence and Antimicrobial Resistance Gene Identification of Staphylococcus aureus from Orthopaedic Implant-Associated Infections. Microorganisms. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 24. | Cheng J, Hu H, Kang Y, Chen W, Fang W, Wang K, Zhang Q, Fu A, Zhou S, Cheng C, Cao Q, Wang F, Lee S, Zhou Z. Identification of pathogens in culture-negative infective endocarditis cases by metagenomic analysis. Ann Clin Microbiol Antimicrob. 2018;17:43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 25. | Charalampous T, Kay GL, Richardson H, Aydin A, Baldan R, Jeanes C, Rae D, Grundy S, Turner DJ, Wain J, Leggett RM, Livermore DM, O'Grady J. Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat Biotechnol. 2019;37:783-792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 437] [Cited by in RCA: 389] [Article Influence: 64.8] [Reference Citation Analysis (0)] |
| 26. | Zhang H, Wang M, Han X, Wang T, Lei Y, Rao Y, Xu P, Wang Y, Gu H. The application of targeted nanopore sequencing for the identification of pathogens and resistance genes in lower respiratory tract infections. Front Microbiol. 2022;13:1065159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 27. | Zhang LL, Zhang C, Peng JP. Application of Nanopore Sequencing Technology in the Clinical Diagnosis of Infectious Diseases. Biomed Environ Sci. 2022;35:381-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 28. | Ciuffreda L, Rodríguez-Pérez H, Flores C. Nanopore sequencing and its application to the study of microbial communities. Comput Struct Biotechnol J. 2021;19:1497-1511. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 112] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 29. | Zhu X, Zhao L, Huang L, Yang W, Wang L, Yu R. cgMSI: pathogen detection within species from nanopore metagenomic sequencing data. BMC Bioinformatics. 2023;24:387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 30. | Sheka D, Alabi N, Gordon PMK. Oxford nanopore sequencing in clinical microbiology and infection diagnostics. Brief Bioinform. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 31. | Simner PJ, Miller S, Carroll KC. Understanding the Promises and Hurdles of Metagenomic Next-Generation Sequencing as a Diagnostic Tool for Infectious Diseases. Clin Infect Dis. 2018;66:778-788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 242] [Cited by in RCA: 506] [Article Influence: 84.3] [Reference Citation Analysis (0)] |
| 32. | Lin Y, Dai Y, Zhang S, Guo H, Yang L, Li J, Wang K, Ni M, Hu Z, Jia L, Liu H, Li P, Song H. Application of nanopore adaptive sequencing in pathogen detection of a patient with Chlamydia psittaci infection. Front Cell Infect Microbiol. 2023;13:1064317. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 33. | Hall MB, Rabodoarivelo MS, Koch A, Dippenaar A, George S, Grobbelaar M, Warren R, Walker TM, Cox H, Gagneux S, Crook D, Peto T, Rakotosamimanana N, Grandjean Lapierre S, Iqbal Z. Evaluation of Nanopore sequencing for Mycobacterium tuberculosis drug susceptibility testing and outbreak investigation: a genomic analysis. Lancet Microbe. 2023;4:e84-e92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 41] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 34. | Zhang Y, Lu X, Tang LV, Xia L, Hu Y. Nanopore-Targeted Sequencing Improves the Diagnosis and Treatment of Patients with Serious Infections. mBio. 2023;14:e0305522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 35. | Roberts LW, Harris PNA, Forde BM, Ben Zakour NL, Catchpoole E, Stanton-Cook M, Phan MD, Sidjabat HE, Bergh H, Heney C, Gawthorne JA, Lipman J, Allworth A, Chan KG, Chong TM, Yin WF, Schembri MA, Paterson DL, Beatson SA. Integrating multiple genomic technologies to investigate an outbreak of carbapenemase-producing Enterobacter hormaechei. Nat Commun. 2020;11:466. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 36. | Muthuirulandi Sethuvel DP, Veeraraghavan B, Vasudevan K, Devanga Ragupathi NK, Murugan D, Walia K, Anandan S. Complete genome analysis of clinical Shigella strains reveals plasmid pSS1653 with resistance determinants: a triumph of hybrid approach. Gut Pathog. 2019;11:55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 37. | Mu X, Wang Y, Sun L, Zhao S, Jin X, Zhang J, Yu Y, Wu X. Invasive Infection With emm3/ST15 Streptococcus pyogenes: The First Case Report From China and Complete Genome Analysis. Front Med (Lausanne). 2022;9:861087. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 38. | Jost G, Schwendener S, Liassine N, Perreten V. Methicillin-resistant Macrococcus canis in a human wound. Infect Genet Evol. 2021;96:105125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 39. | Huang YY, Li QS, Li ZD, Sun AH, Hu SP. Rapid diagnosis of Mycobacterium marinum infection using targeted nanopore sequencing: a case report. Front Cell Infect Microbiol. 2023;13:1238872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 40. | Bialasiewicz S, Duarte TPS, Nguyen SH, Sukumaran V, Stewart A, Appleton S, Pitt ME, Bainomugisa A, Jennison AV, Graham R, Coin LJM, Hajkowicz K. Rapid diagnosis of Capnocytophaga canimorsus septic shock in an immunocompetent individual using real-time Nanopore sequencing: a case report. BMC Infect Dis. 2019;19:660. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 41. | Moller AG, Petit RA 3rd, Davis MH, Read TD. Development of an Amplicon Nanopore Sequencing Strategy for Detection of Mutations Conferring Intermediate Resistance to Vancomycin in Staphylococcus aureus Strains. Microbiol Spectr. 2023;11:e0272822. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 42. | Bird MT, Greig DR, Nair S, Jenkins C, Godbole G, Gharbia SE. Use of Nanopore Sequencing to Characterise the Genomic Architecture of Mobile Genetic Elements Encoding bla (CTX-M-15) in Escherichia coli Causing Travellers' Diarrhoea. Front Microbiol. 2022;13:862234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 43. | Fernando DT, Bhatt R, Saiganesh A, Schultz A, Gera P. Lung abscess: 14 years of experience in a tertiary paediatric hospital. ANZ J Surg. 2022;92:1850-1855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 44. | Taxt AM, Avershina E, Frye SA, Naseer U, Ahmad R. Rapid identification of pathogens, antibiotic resistance genes and plasmids in blood cultures by nanopore sequencing. Sci Rep. 2020;10:7622. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 45. | Harris PNA, Bauer MJ, Lüftinger L, Beisken S, Forde BM, Balch R, Cotta M, Schlapbach L, Raman S, Shekar K, Kruger P, Lipman J, Bialasiewicz S, Coin L, Roberts JA, Paterson DL, Irwin AD. Rapid nanopore sequencing and predictive susceptibility testing of positive blood cultures from intensive care patients with sepsis. Microbiol Spectr. 2024;12:e0306523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 11.0] [Reference Citation Analysis (0)] |