INTRODUCTION
Irritable bowel syndrome (IBS) is classified as a functional gastrointestinal (GI) disorder with a global prevalence that varies from 9% to 23%, influenced by the diagnostic criteria and research methodologies in use[1,2]. IBS is characterized by a diverse clinical spectrum and is associated with a significant disease burden. The etiology and progression of IBS involve complex, multifactorial processes that are not yet fully comprehended[3]. Emerging research has provided novel insights into the pathophysiology of IBS, with hypotheses spanning intestinal immunity, the brain-gut-microbiota axis[4], visceral hypersensitivity, intestinal dysbiosis[5], inflammation[6], post-infectious factors[7], food sensitivities[8], genetic factors[9], and psychosocial dysfunctions[10]. These insights have spurred the development of a variety of therapeutic strategies.
Particularly noteworthy is the brain-gut-microbiota axis[11], which has introduced a conceptual shift in neuroscience and has emerged as a novel target for IBS treatment. The immune system within the gastrointestinal tract maintains physiological equilibrium in response to environmental triggers such as allergens, dietary antigens, or pathogens[12]. The variability in luminal contents, anatomical structures, and physiological roles along the gut is mirrored by the diverse immune cell populations and immune responses found in each intestinal segment[13].
Immune cells and their cytokines play a crucial role in combating infections, regulating inflammation, and facilitating communication between the brain and the immune system[10]. In IBS, alterations in lymphocyte populations, including changes in B and T lymphocyte counts and activation levels, have been documented. These immune changes are linked to increased eosinophil counts in the duodenum of patients with functional dyspepsia (FD) and elevated colonic mast cell numbers in those with IBS[14]. Additionally, elevated levels of α4+β7+ gut-homing T cells in the bloodstream are implicated in the pathophysiology of FD and IBS. The homeostatic balance within the GI tract can be disrupted in gastrointestinal disorders, with enhanced mucosal infiltration of mast cells and T cells, their activation state, cytokine production, and genetic variations all potentially influencing bowel function and symptom manifestation in IBS. Meta-analyses have reported increased colonic mast cell[15] and T cell (CD3+, CD4+, or CD8+ T cells) infiltration in IBS patients. B cell-activating factor is thought to regulate immune responses involving B and T cells and is associated with inflammatory activities in autoimmune conditions and certain B cell cancers[16,17].
Recent research has increasingly supported the role of immune cells in IBS pathogenesis. For instance, alterations in intestinal mucosal immune cells may compromise intestinal barrier function, leading to infections and dysbiosis. Furthermore, immune cells can influence the intestinal nervous system by modulating gut microbiota metabolites, thereby contributing to IBS symptoms. Thus, immune cell research holds significant value for IBS, as it can uncover disease pathogenesis, identify new therapeutic targets, and assess disease severity and progression to guide clinical practice.
Mendelian randomization (MR) is an analytical method used in epidemiology to infer causal relationships in disease etiology, based on the principles of Mendelian genetics[18,19]. The MR approach, considered a form of "natural" randomized controlled trial, can reduce common biases in observational studies and provide robust causal evidence between exposures and outcomes through genetic variants[20,21]. Prior observational studies have identified numerous associations between immune cell characteristics and IBS, reinforcing the link between these elements[16]. In this study, we conduct a comprehensive analysis using a two-sample MR approach to investigate the potential causal effects of immune cells on IBS. Our goal is to determine if immune cells could serve as potent predictors of this disease, thereby contributing to the prediction and management of IBS.
MATERIALS AND METHODS
Study design
In our study, we utilized a two-sample MR analysis to assess the causal relationship between 731 immune cell markers, categorized into seven groups, and IBS. The MR technique employs genetic variants as instrumental variables (IVs) to infer causality. To be considered credible, these IVs must satisfy three critical conditions: (1) A direct association between the genetic variants and the exposure (immune cell markers); (2) no correlation with confounders that could distort the relationship between exposure and outcome; and (3) no impact on the outcome through pathways independent of the exposure. We ensured that all studies incorporated into our analysis were approved by the appropriate institutional review boards and that all participants provided informed consent, after being thoroughly informed about the study's objectives, procedures, potential risks, and benefits. This process upholds rigorous ethical standards and protects the rights and well-being of the participants.
Genome-Wide Association Study Data Sources for IBS
In our investigation of the genetic underpinnings of IBS, we leveraged genome-wide association study (GWAS) summary statistics from the United Kingdom Biobank[22,23]. This extensive analysis involved 53400 individuals diagnosed with IBS and 433201 controls, with subsequent validation performed using data from 23 and Me, which included 205252 cases and 1384055 controls[22,23]. The research team successfully identified and replicated associations at six genetic loci that confer an increased risk for IBS, pinpointing the genes NCAM1, CADM2, PHF2/FAM120A, DOCK9, CKAP2/TPTE2P3, and BAG6 as being of particular interest. Notably, the first four of these genes have established connections to mood and anxiety disorders, with expression profiles in the nervous system or, in some instances, dual roles within the nervous system and other tissues. In parallel, the study uncovered a strong, genome-wide association between IBS risk and measures of anxiety, neuroticism, and depression, with a correlation coefficient (rg) exceeding 0.5. Subsequent analysis suggests that this relationship is likely rooted in shared pathogenic mechanisms rather than a direct causality from anxiety to abdominal symptoms[24]. These findings warrant further exploration to better understand the disrupted brain-gut interactions that are a hallmark of IBS and to identify potential therapeutic targets for this complex disorder.
Immunity-wide GWAS data sources
The GWAS Catalog serves as a repository for GWAS summary statistics on a wide array of immune traits, with accession numbers spanning from GCST0001391 to GCST0002121[25]. This comprehensive dataset encompasses 731 immunological phenotypes, including absolute cell (AC) counts (n = 118), median fluorescence intensities (MFI) indicative of surface antigen levels (n = 389), morphological parameters (MP) (n = 32), and relative cell (RC) counts (n = 192). The MFI, AC, and RC traits incorporate a diverse range of immune cell types, such as B cells, conventional dendritic cells (CDCs), mature T cells, monocytes, myeloid cells, T cells, B cells, natural killer cells (TBNK), and regulatory T cells (Tregs). The MP trait is specifically focused on CDCs and TBNK panels.
The inaugural GWAS aimed at immune traits was conducted using data from 3,757 European individuals, with no overlap between the cohorts included in this analysis. This methodology facilitated a meticulous investigation into the genetic determinants of immune-related phenotypes. To enhance the precision of the genetic analysis, a high-density array was employed to genotype approximately 22 million single nucleotide polymorphisms (SNPs). The data obtained were analyzed using a reference panel based on the Sardinian sequence to estimate genetic variations.
After accounting for potential confounders such as gender, age, and two-year age groups, the study conducted an in-depth analysis of the associations between the genotyped SNPs and the immune traits under investigation. By considering the modifying effects of age and gender, the study provided a more nuanced characterization of the genetic factors influencing immune-related phenotypes. This adjustment for confounders was crucial in clarifying the genetic variants associated with immune traits, thereby bolstering the reliability and broader applicability of the findings.
Selection of IVs
In alignment with recent advancements[26,27] in genetic epidemiology, we have established a significance threshold of 1 × 10-5 for IVs correlated with each immune trait. This threshold standardizes the identification of genetic factors that may influence immune-related phenotypes. To enhance the precision of SNP selection and to filter out those in high linkage disequilibrium (LD) with proximate SNPs, thereby simplifying the genetic data and increasing the accuracy of subsequent analyses, we employed PLINK software (version 1.90). This software utilizes a clustering program with an LD r2 threshold of less than 0.1 within a 500 kb window to effectively reduce genetic complexity.
For the IBS trait, the significance level was more conservatively adjusted to 5 × 10-8. To evaluate the strength of the IVs and to counteract the potential effects of weak instrumental variables, we computed the proportion of phenotypic variation explained and the F statistic for each IV. These metrics are essential for ensuring the robustness of our genetic instruments and for minimizing the risk of biased causal estimates in our Mendelian randomization analysis.
Statistical analysis
All statistical analyses were performed using R 3.5.3 software, which is accessible through the R Project website (http://www.Rproject.org). To evaluate the causal relationship between the 731 immunological phenotypes and IBS, we utilized the inverse variance weighting (IVW) method[28], as well as weighted median[29] and mode-based approaches[30]. These methods were implemented using the 'Mendelian Randomization' package (version 0.4.3)[31].
The IVW method operates under the principle of adjusting the contribution of each data point according to its statistical significance, akin to the way ingredients are proportioned in a recipe based on their impact on the final dish. By doing so, it ensures that every data point is given appropriate weight, mitigating the influence of those with a more pronounced effect and maintaining a balanced analysis.
To assess the consistency of our selected IVs, we employed Cochran's Q statistic and its corresponding P value. These tools help to identify whether there are genuine, non-random differences among the IVs. If the null hypothesis of heterogeneity is rejected, we switch from fixed-effects to random-effects IVW to account for variability.
Considering the potential for horizontal pleiotropy, we applied the MR-egger method[32], which serves as a detector for data points that may unduly influence the analysis. Furthermore, we utilized the MR-PRESSO method[33] to identify and exclude outlier genetic variants that could significantly skew our results. These outliers, which can impact multiple outcomes, are crucial to remove to prevent distortion of our conclusions.
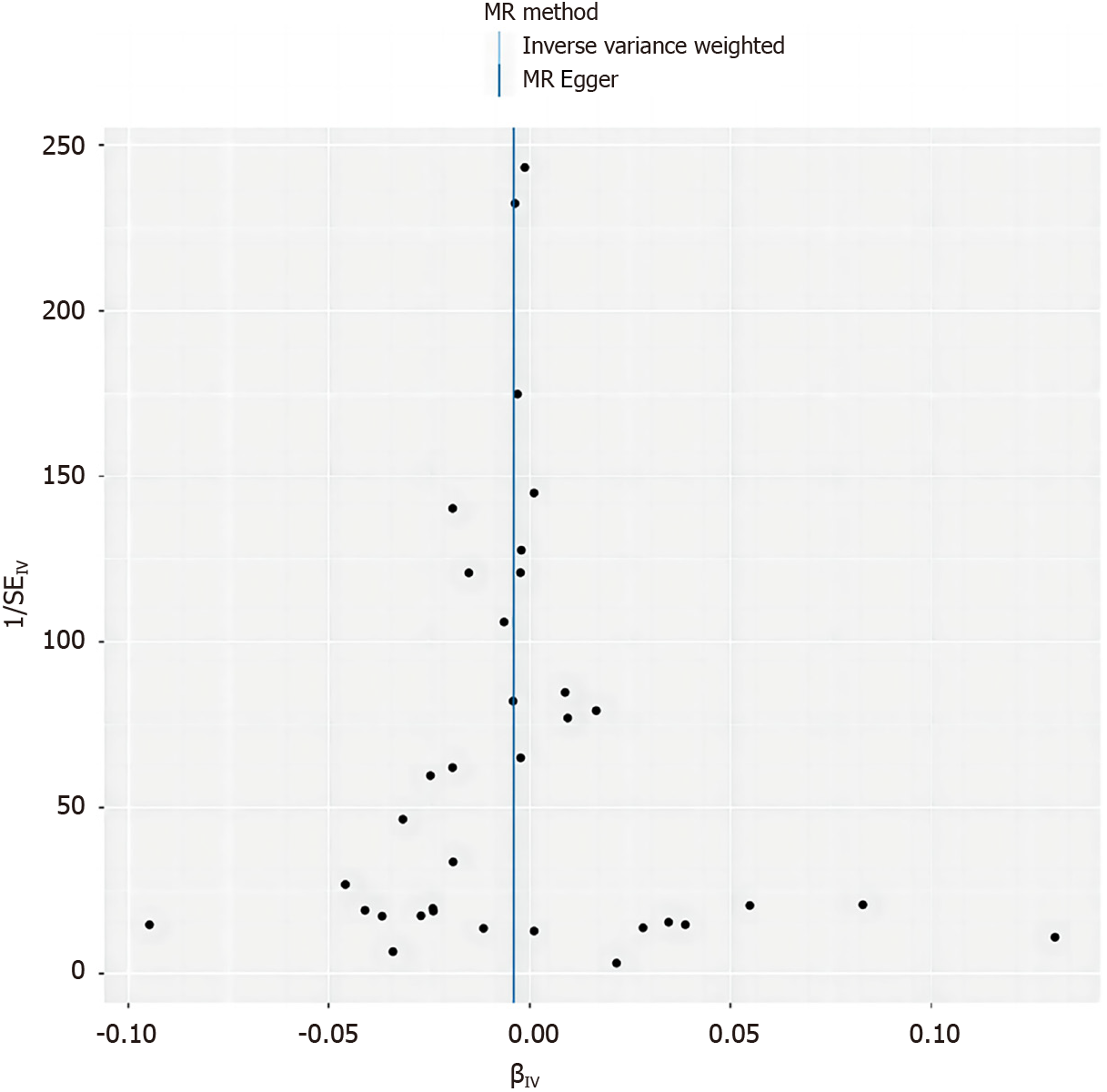
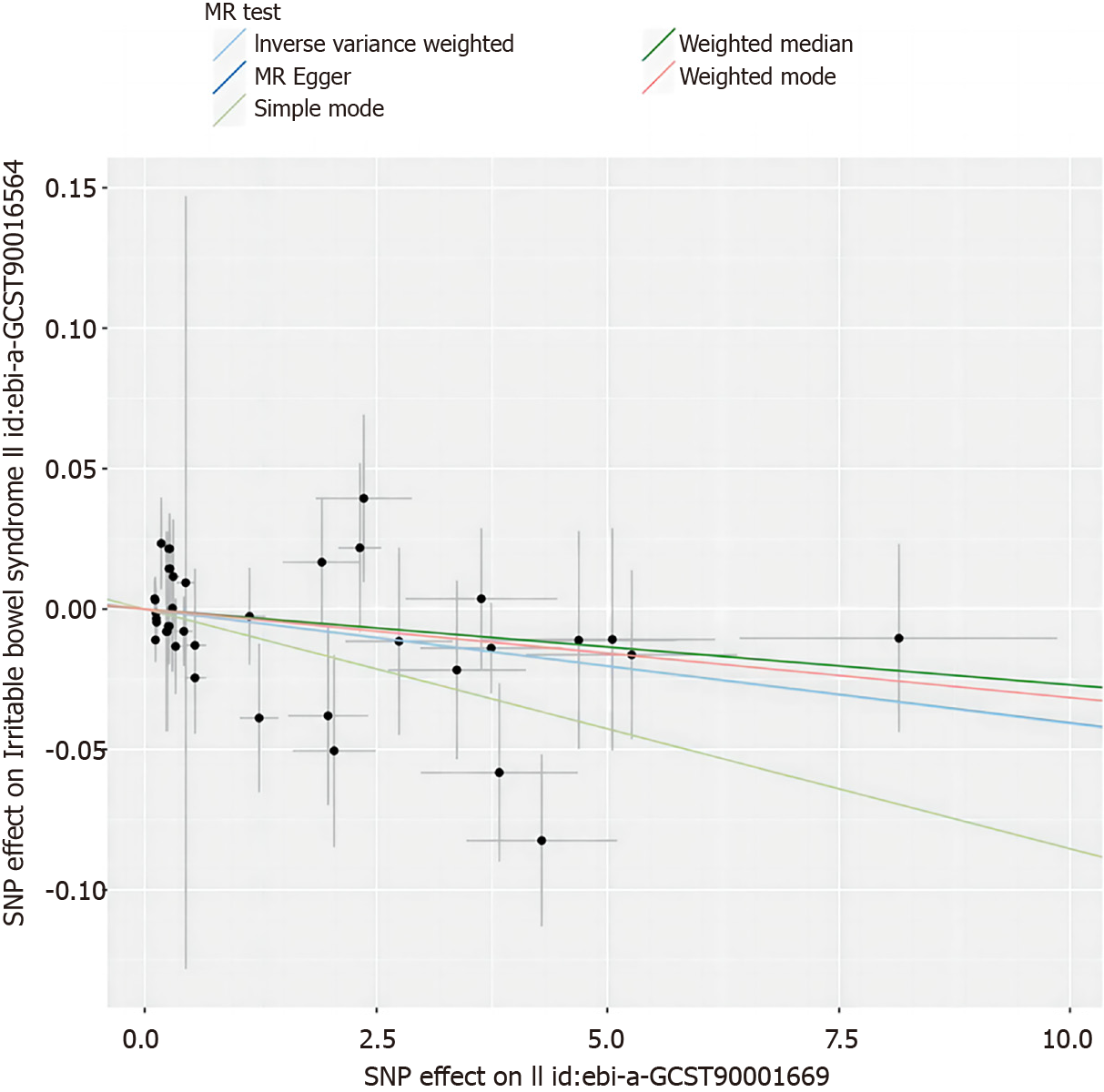
We also employed scatter plots and funnel plots for data visualization. Scatter plots assist in identifying any outliers that may be affecting the results, while funnel plots provide insight into the consistency of the correlations among data points. These graphical tools are instrumental in ensuring the precision and reliability of our research findings.
DISCUSSION
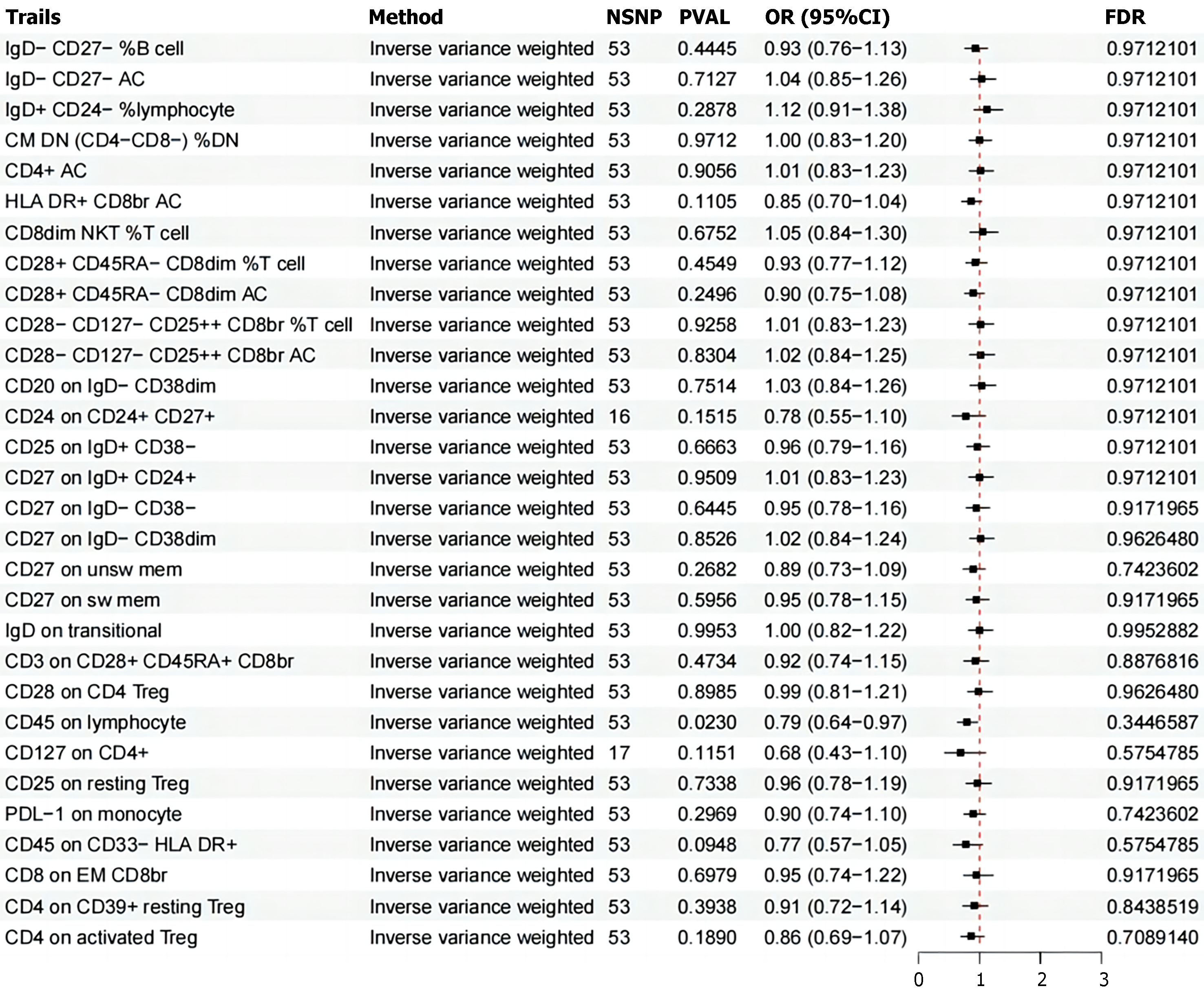
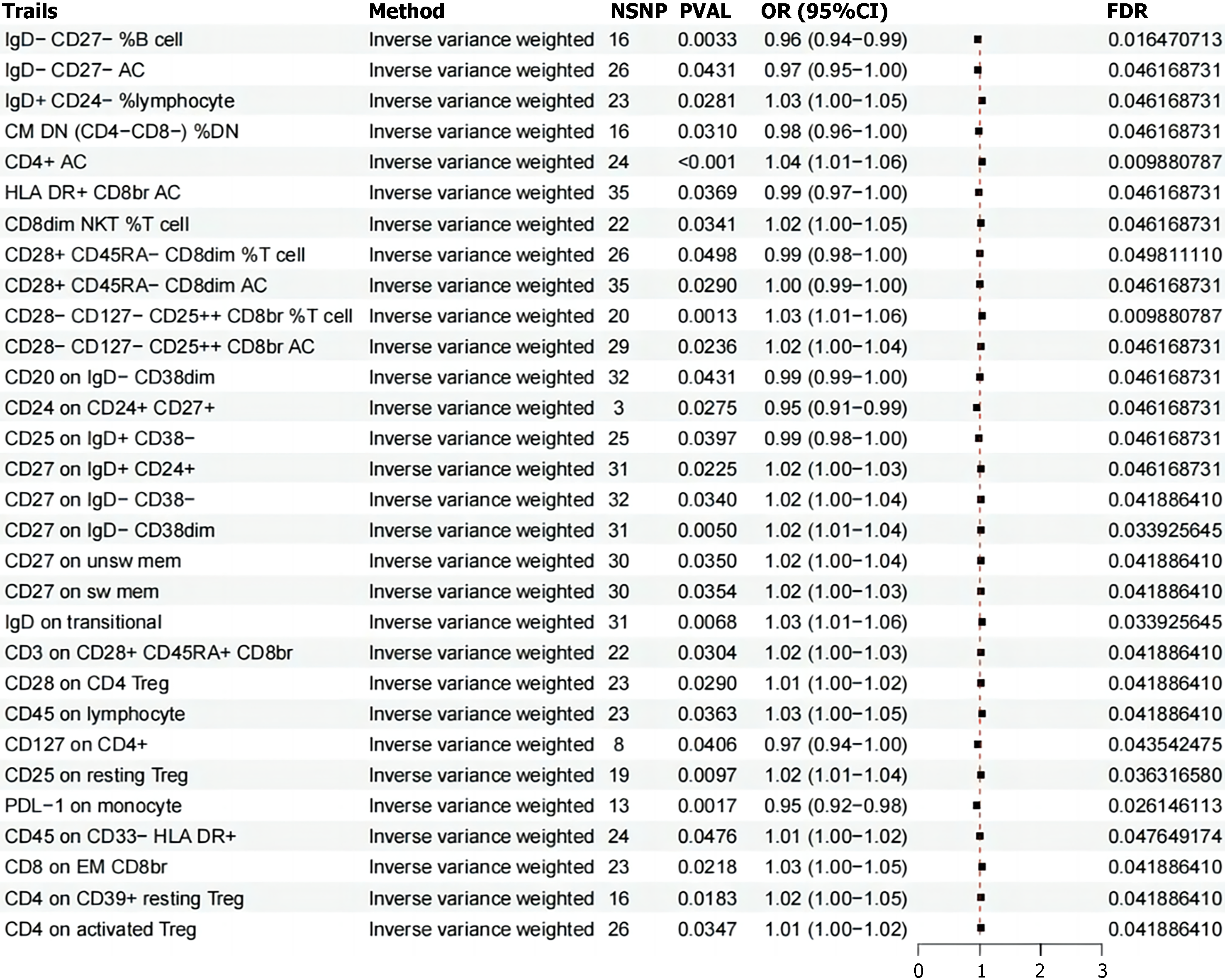
Leveraging extensive public genetic datasets, our study explored the causal associations between 731 immune cell traits and IBS. To our knowledge, this is the first MR analysis to investigate the causal relationships between a multitude of immunophenotypes and IBS. While the immune system's role in inflammatory bowel disease (IBD) is well-established, its specific contributions to IBS development are less clear and more complex. This research represents a pioneering effort to examine potential causal relationships between diverse immune-related traits and IBS. Our findings indicate the presence of significant immune traits across various immune cell types, with notable subsets of T cells and B cells standing out. These results collectively suggest a more extensive role for immune biomarkers in IBS pathophysiology than previously recognized.
Human B cells are classified into four traditional subsets based on the expression of CD27 and immunoglobulin (Ig) D[34]. Among these, the CD27−IgD− B cells, termed double-negative (DN) B cells[35], are less studied compared to the other three subsets. DN B cells have been observed to be elevated in autoimmune diseases, including systemic lupus erythematosus, myasthenia gravis, multiple sclerosis, and rheumatoid arthritis (RA). The research by Pararasa et al[36] aimed to elucidate the distinct subsets of peripheral B cells in IBD and how these subsets differ between periods of active disease and remission. The study consistently reported a reduced proportion of CD27−IgD− B cells that produce IgM, IgA, and IgG in the bloodstream of IBD patients. Furthermore, an increase in the CD27−IgD− B cell subset was noted within mucosal-associated lymphoid tissues in individuals with IBD, indicating a recruitment of CD27−IgD− B cells from the circulation to the gut during the course of IBD[37]. These findings contribute to the growing understanding of the immune cell dynamics in IBD and may have implications for disease monitoring and therapeutic strategies.
Tregs are an essential part of the immune system's regulatory network[38], critically contributing to the maintenance of intestinal homeostasis and the modulation of overly vigorous immune responses. In the context of IBS, our research has underscored the importance of specific Treg subsets, identifiable by their unique surface marker profiles. These include Tregs characterized by CD28+, CD45RA-dim, CD8dim, and AC; CD28−, CD127−, CD25++, CD8br%; CD3+, CD28+, CD45RA+, CD8br; CD28+ on CD4 Tregs; CD25 on resting Tregs; CD4 on CD39+, resting Tregs; and CD4 on activated Treg molecules. These markers are indicative of Tregs' role in regulating intestinal immune responses and their potential impact on the pathogenesis of IBS. Moreover, the activity of Tregs is closely associated with the human leukocyte antigen (HLA) gene[39], which may provide insights into the etiology of IBS. CD4+CD25highCD127Low/−FOXP3+ regulatory T cells are pivotal in preserving immune tolerance and curbing excessive immune reactions[40]. Some studies have investigated the effects of retinoic acid on a specific human Treg cell subset (CD4+CD25+CD127Low/−CD45RA+), showing that these cells, when expanded, form a uniform and epigenetically stable population that does not produce pro-inflammatory cytokines. When transplanted into SCID mice with xenografts of human small intestine, these cells selectively home to the lamina propria[41,42]. The exploration of Tregs as a therapeutic intervention and the development of strategies to enhance their therapeutic potential are active areas of research[43].
The TBNK cell subset, comprising T lymphocytes, B lymphocytes, and natural killer (NK) cells[44], is a cornerstone of the immune system's functionality. These cells are essential in mounting the human immune response, and any deviation in their numbers or functions can serve as a biomarker for changes in immune health. Our latest research has shed light on the significant role that TBNK cells play in mitigating the onset and progression of IBS. This is achieved primarily through the regulation of specific cell populations, including CD4+AC, CD8dim NKT% T cells, and those expressing the CD45 marker. TBNK cells act as a vital bridge, mediating the complex interactions between a range of immune features and the pathological mechanisms underlying IBS. The directed movement of activated T lymphocytes to the gut, particularly the small intestine, is governed by two main pathways[45,46]: The first involves the α4β7-mucosal vascular addressin cell-adhesion molecule 1 (MAdCAM-1), and the second is the chemokine ligand 25 and CCR9 pathway. This migration is meticulously orchestrated by dendritic cells present in the gut-associated lymphoid tissues, such as Peyer's patches and mesenteric lymph nodes, which enhance the expression of key integrins like α4β7 or CCR9. The α4β7 integrin engages with its ligand, MAdCAM-1, predominantly located on the luminal surface of post-capillary venules in lymphoid organs. There is a growing body of clinical and experimental data suggesting that the targeted disruption of B cell function may offer therapeutic benefits in certain contexts of human IBD. The therapeutic agent rituximab, which targets CD20—a cell surface molecule predominantly expressed on mature B lymphocytes—has demonstrated substantial efficacy in treating a variety of hematological and immune-mediated conditions[47]. Although the application of such a targeted approach to IBS is still under investigation, it represents an exciting and promising area of research that could lead to novel treatment paradigms for IBS. As we delve deeper into the intricate relationship between the immune system and IBS, we are presented with the opportunity to refine our understanding of the disease's etiology and to develop more precise therapeutic interventions. The potential of harnessing the knowledge of immune cell dynamics, such as those of the TBNK cell subset, could revolutionize the way we approach the management and treatment of IBS, ultimately leading to improved patient outcomes and quality of life.
NK cells and monocytes are increasingly recognized for their role in the pathophysiology of IBS, contributing to its sustained effects on affected individuals[48,49]. A multitude of studies have uncovered elevated levels of inflammatory cytokines within the bloodstream of those suffering from IBS, underscoring the intricate relationship between inflammation and this complex disorder. NK cells, with their versatile immunoregulatory capabilities, can exert a range of effects within the immune system. Notably, their expression of a high-affinity receptor for interleukin (IL)-2 positions them to compete with other immune cells for this critical cytokine, potentially influencing the overall immune response. The Fc receptor CD16a (FcγRIIIa), a feature of CD56Dim NK cells, interacts with the constant region of Immunoglobulin G (IgG) antibodies. This interaction triggers Antibody-Dependent Cell-Mediated Cytotoxicity, a process that is crucial for the NK cells' ability to eliminate target cells[50]. In the gastrointestinal tract, NK cells may exert a more pronounced influence on gut homeostasis compared to their circulating counterparts, owing to their localized cytokine production. This local effect is particularly significant given the gut's role as a key site of immune activity and its intimate connection to IBS symptoms. Dominik Aschenbrenner's seminal study[51] has shed light on the role of monocytes in inflammatory bowel disease through transcriptomic analysis. His findings reveal an IL-1 cytokine network that regulates the production of IL-23, particularly in the context of both genetic and acquired resistance to IL-10. This discovery not only enhances our understanding of the immune mechanisms at play but also points to a potential therapeutic avenue for clinical application. Moreover, research has indicated that signaling through the IL-6 receptor (IL6R) is implicated in the development of inflammatory bowel disease[52]. This pathway's involvement suggests that it may also play a role in the pathogenesis of IBS, opening up new avenues for potential therapeutic interventions. In summary, the contributions of NK cells and monocytes to the immune response in IBS are multifaceted and warrant further investigation. Understanding the precise mechanisms by which these immune cells and the cytokines they produce influence IBS pathophysiology is crucial for developing targeted therapies. As our knowledge of the immune system's role in IBS continues to grow, so too does the potential for more effective treatment strategies that can alleviate the burden of this prevalent condition.
In our investigation, we conducted a rigorous two-sample MR analysis, utilizing data from extensive GWAS cohorts that have been made publicly available[24]. By incorporating a large cohort of approximately 486601 individuals, we significantly bolstered the statistical power of our analysis. Our results are anchored in genetic instrumental variables, and we employed a diverse suite of MR techniques to infer causal relationships. The consistency of our findings was maintained, and they appeared to be robust against horizontal pleiotropy and other potential confounders. However, our study does have its limitations. A significant one is the potential incomplete assessment of horizontal pleiotropy, an issue that persists despite our execution of multiple sensitivity analyses. Moreover, the lack of individual-level data precludes us from conducting more nuanced stratified analyses, which could account for variables such as the average age, health status, and socio-environmental factors of the participants. This limitation may introduce bias into our MR estimates, affecting the precision of our conclusions. Additionally, our research was conducted using data from a European database, which raises questions about the generalizability of our findings to other ethnic groups. This could limit the broader applicability of our results to diverse populations. Lastly, while our study provides preliminary evidence of a relationship between immune cells and IBS, it is essential to note that further research is required. Additional basic experimental research and clinical trials are necessary to validate our hypotheses and to expand upon the implications of our findings for the understanding and treatment of IBS. In summary, our MR analysis has contributed valuable insights into the potential causal links between genetic factors, immune cell activity, and IBS. While our results are robust and consistent, they must be interpreted within the context of the study's limitations. Future research should aim to address these limitations and to build upon the foundation laid by our investigation to advance the field and improve the lives of those affected by IBS.
CONCLUSION
In summary, our research has undertaken an extensive examination of four key immune-related traits—MFI, RC, AC, and MP—employing bidirectional MR to elucidate the complex interplay between these factors and IBS. Our findings have established a causal nexus between IBS and a spectrum of 30 immune phenotypes, with nine demonstrating a protective effect against the condition and 21 being correlated with an elevated susceptibility.
This study has not only unveiled the intricate relationship between the immune system and IBS but has also significantly mitigated the impact of confounding variables, reverse causation, and other potential biases. These insights may herald a paradigm shift in our comprehension of the biological underpinnings of IBS, potentially leading to the formulation of more efficacious preventative measures and therapeutic strategies.
While our work has shed light on the genetic interplay between the immune system and IBS, the specific pathways and mechanisms that underlie these observed causal associations warrant further exploration. By delving deeper into these connections, we aim to refine our understanding and potentially illuminate targeted avenues for the clinical management and prevention of IBS.
The implications of our findings are far-reaching, promising to reshape the landscape of IBS research and practice. By harnessing this newfound knowledge, we can envision a future where the diagnosis, treatment, and day-to-day management of IBS are informed by a more nuanced appreciation of the immune system's role in the condition's etiology. This represents a significant step forward in our ongoing quest to alleviate the burden of IBS for patients and to enhance their quality of life.