Published online May 6, 2024. doi: 10.12998/wjcc.v12.i13.2160
Peer-review started: December 14, 2023
First decision: February 12, 2024
Revised: February 21, 2024
Accepted: March 28, 2024
Article in press: March 28, 2024
Published online: May 6, 2024
Processing time: 133 Days and 0.6 Hours
Analyzing the variations in serum bile acid (BA) profile can provide a certain biological basis for early warning and prevention of various diseases. There is currently no comprehensive study on the relationship between the serum BA pro
To study the serum BA profile detection results of patients with colonic polyps, and analyze the correlation between BA and colonic polyps.
From January 1, 2022, to June 1, 2023, 204 patients with colonic polyps who were diagnosed and treated at Zhongda Hospital Southeast University were chosen as the study subjects, and 135 non-polyp people who underwent physical examina
Compared with the control group, the serum levels of taurocholic acid, glycocho
The serum BA profile showed significant changes in patients with colonic polyps, with a significant increase in primary conjugated BA content and a decrease in secondary free bile acid DCA content. There is a certain correlation between primary free BA and pathological parameters of polyps.
Core Tip: This study shows that the serum primary conjugated bile acid (BA) levels in the colonic polyp group were significantly higher than those in the control group (P < 0.05), while the secondary free BA, deoxycholic acid content was lower than that in the control group. Patients with various polyp sizes, locations, morphologies, and pathological types had variable serum BA profile, according to subgroup study of colonic polyps. Therefore, analyzing the changes in serum BA profile may provide new ideas for finding new targets for the treatment of colonic tumors.
- Citation: Ji X, Chen H. Detection and analysis of serum bile acid profile in patients with colonic polyps. World J Clin Cases 2024; 12(13): 2160-2172
- URL: https://www.wjgnet.com/2307-8960/full/v12/i13/2160.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i13.2160
Colonic polyps are lesions that protrude from the mucosal surface into the large intestine lumen, and they can be further classified into adenomatous polyps and non-adenomatous polyps based on their pathology[1]. The second-highest death rate of all malignancies is associated with colon cancer, which is the third most frequent malignancy worldwide[2]. Colonic polyps are precancerous lesions of colonic cancer, especially adenomatous polyps. Over 50% of colonic cancer is derived from adenomas, which make up about two-thirds of colonic polyps[3]. Early-stage colon cancer is typically found via a colonoscopy and does not typically present with any overt clinical symptoms. The incidence of colonic cancer can be decreased and the survival rate increased by early detection of precancerous lesions, early diagnosis, and early treatment. At present, the initial diagnosis of the disease mainly relies on endoscopic examination, further diagnosis requires pa
Bile acid (BA) is a major component of bile, synthesized by cholesterol in the liver and stored in the gallbladder. It is secreted into the small intestine after eating to promote the digestion and absorption of lipids and lipophilic vitamins[5]. Meanwhile, as a cellular signaling molecule, BA also regulates biological processes by stimulating various signaling pathways, participating in the regulation of glucose metabolism, energy homeostasis, and immune response in the body. Analyzing the variations in serum BA profile can provide a certain biological basis for early warning and prevention of various diseases. There is currently no comprehensive study on the relationship between the serum bile acid profile and colonic polyps, despite the fact that numerous studies have demonstrated that high levels of total bile acid (TBA) are a risk factor for colonic cancer[6]. In this study, the levels of 15 serum BA components were compared between patients with colonic polyps and healthy people. Additionally, alterations in the serum BA profile of patients with colonic polyps were initially explored, and the relationship between BA components and colonic polyps was analyzed.
204 individuals who were hospitalized and diagnosed with colonic polyps at Zhongda Hospital Southeast University between January 1, 2022, and June 1, 2023 were chosen as the colonic polyp group by reviewing the electronic medical record system. There were 114 men and 90 women in this group, with an average age of (57.19 ± 9.43) years. Inclusion criteria: (1) Patients with pathological diagnosis of colonic polyps through colonoscopy, aged between 30 and 75 years old; and (2) Routine biochemical tests and serum BA profile have been completed before undergoing colonoscopy. Exclusion criteria: (1) Previous history of inflammatory bowel disease; (2) Previous intestinal surgery (excluding appen
Gathering demographic data and clinical test results about the research subjects, such as age, gender, body mass index (BMI), alanine transaminase (ALT), aspartate transaminase (AST), total cholesterol (TC), and serum BA profile. Ad
Statistical analysis was conducted using SPSS 26.0 software. The normality test of the data was conducted using the Kolmogorov-Smirnon test. The measurement data of normal distribution was expressed by mean ± SD, and the com
In this retrospective analysis, 204 people made up the colonic polyp group and 135 people made up the control group. Age, BMI, gender, ALT, AST, and TC did not statistically differ between the two groups (P > 0.05), demonstrating com
| General information | Colonic polyp group (n = 204) | Control group (n = 135) | P value |
| Age (year) | 57.19 ± 9.43 | 55.35 ± 8.79 | 0.072 |
| BMI (kg/m2) | 23.82 ± 2.28 | 23.52 ± 2.37 | 0.244 |
| Gender | 0.054 | ||
| Males | 114 (55.88) | 61 (45.19) | |
| Females | 90 (44.12) | 74 (54.81) | |
| ALT (U/L) | 18.00 (13.00, 26.00) | 17.00 (13.00, 24.00) | 0.277 |
| AST (U/L) | 20.00 (17.00, 24.00) | 19.00 (16.00, 23.00) | 0.155 |
| TC (mmol/L) | 4.49 ± 0.84 | 4.56 ± 0.91 | 0.463 |
The TBA content did not differ statistically significantly between the colonic polyp group and the control group, ac
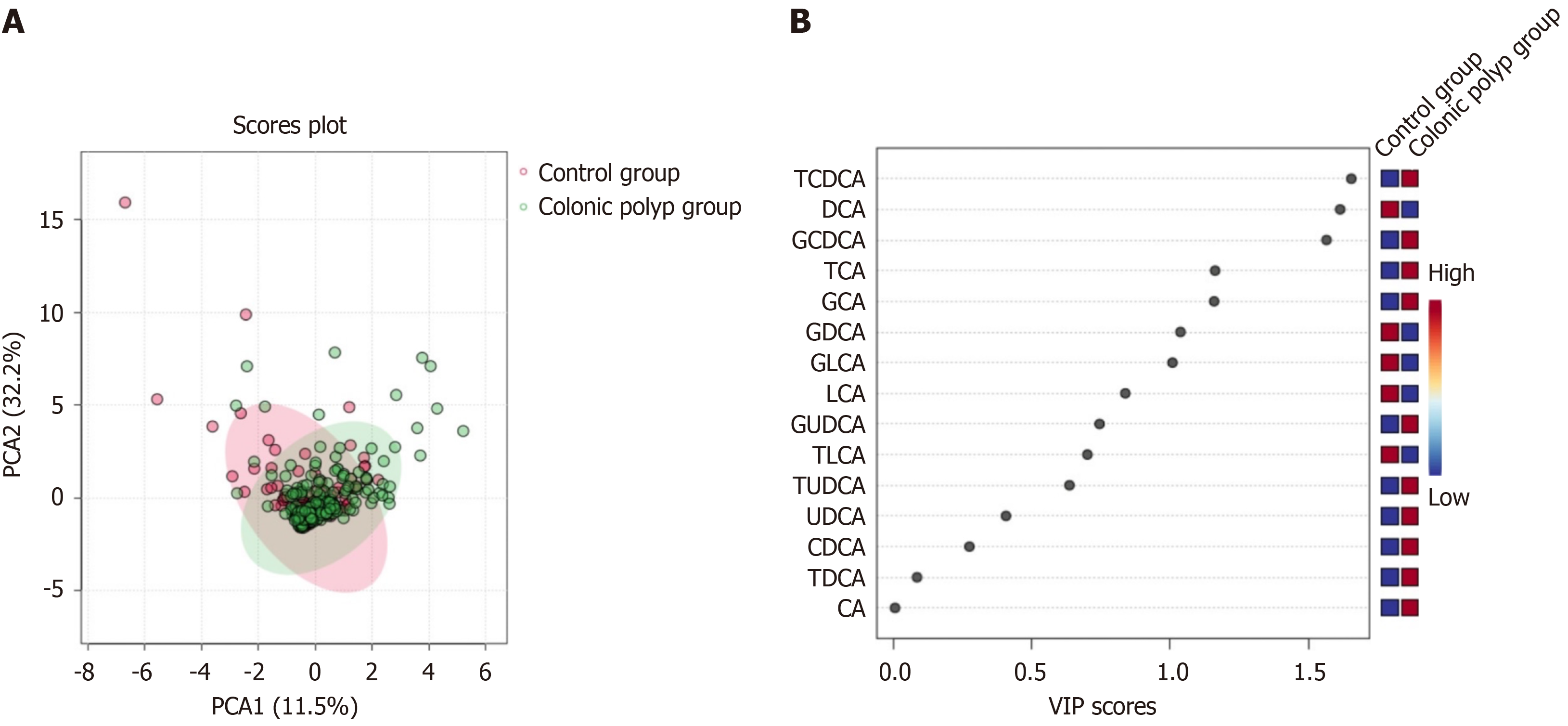
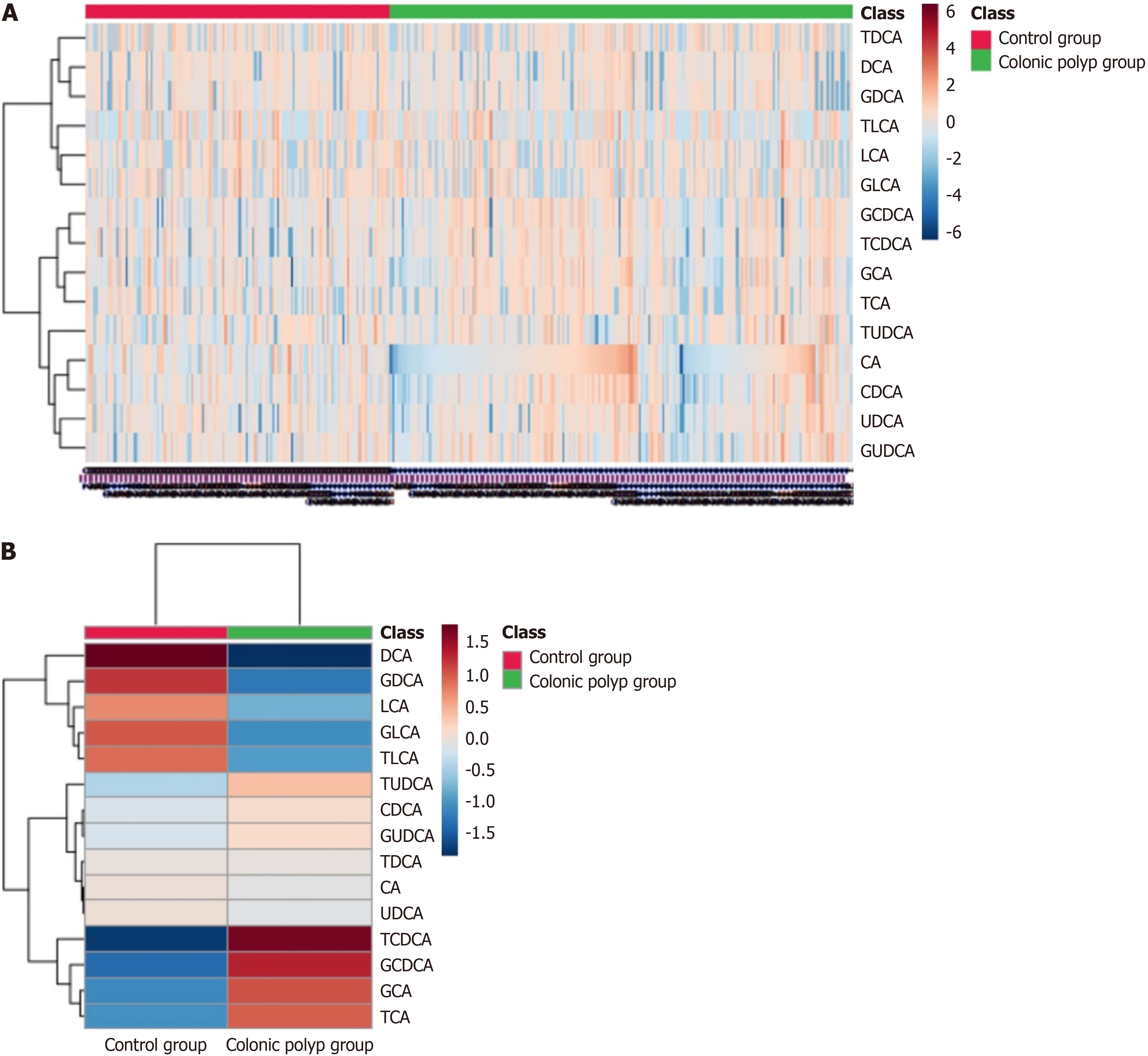
The results of two sets of BA profile detection are shown in Table 2. Using the OPLS-DA model to search for differential metabolites between the colonic polyp group and the control group, it can be observed from the score chart (Figure 1A) that the sample points of the two groups are relatively concentrated, and the differences between the data groups are not significant. To further screen for BA with discrepancies, use VIP values (Figure 1B). It is evident that the two groups' BAs differ in the following ways: GDCA, DCA, GCA, GCDCA, TCA, TCDCA (VIP > 1). DCA, GCA, GCDCA, TCA, and TCDCA were all statistically different (P < 0.05) between the two groups, according to SPSS software analysis. While the concentration of DCA was lower than that of the control group, it was significantly greater than that of GCDCA, GCA, TCA, and TCDCA in the colonic polyp group. The other BA components (Table 2) showed no statistically significant change (P > 0.05). Differential BA components GCA, GCDCA, TCA, TCDCA, and DCA were screened under the con
| BA components | Colonic polyp group (n = 204) | Control group (n = 135) | P value |
| Primary free BAs | |||
| CA | 62.75 (24.33, 232.00) | 53.80 (27.60, 149.00) | 0.571 |
| CDCA | 382.50 (105.50, 851.50) | 294.00 (130.00, 625.00) | 0.164 |
| Primary conjugated BAs | |||
| TCA | 21.85 (5.50, 50.50) | 12.70 (1.50, 32.30) | 0.015a |
| GCA | 166.50 (76.60, 330.00) | 126.00 (52.90, 234.00) | 0.025a |
| GCDCA | 935.50 (430.50, 1967.50) | 708.00 (298.00, 1250.00) | 0.005a |
| TCDCA | 74.35 (27.20, 164.50) | 41.60 (18.30, 119.00) | 0.006a |
| Secondary free BAs | |||
| DCA | 142.00 (30.90, 424.25) | 234.00 (82.60, 502.00) | 0.011a |
| LCA | 6.20 (0.13, 17.10) | 6.40 (0.60, 21.00) | 0.539 |
| UDCA | 73.70 (23.03, 221.50) | 70.70 (19.00, 199.00) | 0.545 |
| Secondary conjugated BAs | |||
| TDCA | 8.50 (0.05, 32.65) | 7.70 (0.00, 35.30) | 0.615 |
| GDCA | 113.50 (12.08, 248.50) | 125.00 (34.60, 335.00) | 0.274 |
| TLCA | 0.00 (0.00, 2.40) | 0.10 (0.00, 4.00) | 0.255 |
| GLCA | 4.60 (0.00, 16.35) | 5.40 (0.00, 18.10) | 0.399 |
| TUDCA | 7.65 (3.15, 15.00) | 8.20 (2.50, 15.00) | 0.369 |
| GUDCA | 137.50 (47.25, 343.00) | 122.00 (63.80, 283.00) | 0.604 |
Colonic polyps can be classified using subgroup analysis in accordance with different pathological types, numbers, sizes, locations, and shapes (Table 3). Through subgroup analysis, we found that: (1) In terms of CA, CDCA, UDCA, and TUDCA, there was a statistically difference (P < 0.05) between the adenomatous colonic group and the non-adenomatous polyp group. In comparison to the non-adenomatous polyp group, the CA, CDCA, UDCA, and TUDCA content in the adenomatous polyp group was lower (Table 4); (2) There is no statistical difference in the composition of BA between the single and multiple groups (P > 0.05) (Table 5); (3) There was a statistical difference (P < 0.05) between the two groups with polyp diameter < 1 cm and ≥ 1 cm in CA, CDCA, UDCA, GUDCA, and TUDCA, and the content of CA, CDCA, UDCA, GUDCA, and TUDCA in the group with polyp diameter ≥ 1cm was higher than that in the group with polyp diameter < 1 cm (Table 6); (4) There were statistical differences (P < 0.05) among CA, CDCA, GCA, and GCDCA in the left colon group, right colon group, and total colon group (Table 7). Through pairwise analysis, it was found that there was a significant statistical difference in GCDCA between the left and right colon groups (P = 0.008), and the GCDCA content in the right colon group was significantly higher than that in the left colon group; There was a significant statistical di
| Group | Cases, n (%) |
| Pathological type | |
| Adenomatous polyp | |
| Tubular adenoma | 30 (14.71) |
| Villous tubular adenoma | 109 (53.43) |
| High grade intraepithelial neoplasia | 6 (2.94) |
| Non adenomatous polyp | |
| Hyperplastic polyp | 59 (28.92) |
| Number of polyps | |
| Single polyp | 73 (35.78) |
| Multiple polyps | 131 (64.22) |
| Size of polyp | |
| Diameter < 1 cm | 169 (82.84) |
| Diameter ≥ 1 cm | 35 (17.16) |
| Location of polyp | |
| Left colon | 114 (55.88) |
| Right colon | 48 (23.53) |
| Total colon | 42 (20.59) |
| The polyp is pedicled or not | |
| Pedicled polyp | 22 (10.78) |
| Sessile polyp | 182 (89.22) |
| Non adenomatous polyp group | Adenomatous polyp group | P value | |
| Primary free BAs | |||
| CA | 107.00 (39.50, 357.00) | 53.20 (20.35, 185.50) | 0.003a |
| CDCA | 408.00 (191.00, 1130.00) | 373.00 (80.50, 785.00) | 0.034a |
| Primary conjugated BAs | |||
| TCA | 24.80 (8.90, 71.50) | 20.10 (5.45, 45.80) | 0.189 |
| GCA | 160.00 (81.70, 423.00) | 174.00 (72.60, 326.00) | 0.676 |
| GCDCA | 866.00 (458.00, 2190.00) | 961.00 (397.00, 1785.00) | 0.465 |
| TCDCA | 113.00(40.70, 185.00) | 64.50 (24.60, 152.50) | 0.060 |
| Secondary free BAs | |||
| DCA | 182.00 (38.50, 448.00) | 118.00 (21.25, 401.00) | 0.226 |
| LCA | 5.30 (0.00, 16.80) | 6.70 (0.50, 17.20) | 0.588 |
| UDCA | 107.00 (49.00, 311.00) | 64.00 (16.15, 190.00) | 0.003a |
| Secondary conjugated BAs | |||
| TDCA | 17.90 (2.50, 39.10) | 6.80 (0.00, 24.35) | 0.078 |
| GDCA | 135.00 (7.40, 398.00) | 107.00 (19.25, 229.00) | 0.593 |
| TLCA | 0.00 (0.00, 2.10) | 0.00 (0.00, 2.55) | 0.566 |
| GLCA | 4.40 (0.00, 20.30) | 4.60 (0.00, 15.50) | 0.646 |
| TUDCA | 11.70 (4.50, 20.10) | 6.50 (2.95, 15.00) | 0.023a |
| GUDCA | 220.00 (58.10, 543.00) | 114.00 (43.55, 303.00) | 0.067 |
| Single polyp group | Multiple polyps group | P value | |
| Primary free BAs | |||
| CA | 92.40 (35.40, 275.50) | 59.70 (23.50, 170.00) | 0.067 |
| CDCA | 492.00 (193.50, 905.50) | 357.00 (84.80, 836.00) | 0.185 |
| Primary conjugated BAs | |||
| TCA | 23.50 (8.70, 47.35) | 20.20 (5.00, 52.10) | 0.809 |
| GCA | 203.00 (93.35, 330.00) | 157.00 (60.90, 342.00) | 0.363 |
| GCDCA | 1050.00 (506.50, 1945.00) | 881.00 (354.00, 2030.00) | 0.346 |
| TCDCA | 80.70 (29.70, 170.50) | 69.10 (25.10, 165.00) | 0.707 |
| Secondary free BAs | |||
| DCA | 119.00 (10.90, 401.00) | 145.00 (32.30, 437.00) | 0.588 |
| LCA | 5.50 (0.25, 15.50) | 6.70 (0.00, 17.60) | 0.715 |
| UDCA | 74.90 (33.15, 210.00) | 73.70 (17.20, 253.00) | 0.540 |
| Secondary conjugated BAs | |||
| TDCA | 7.60 (0.00, 24.70) | 11.20 (0.70, 36.10) | 0.371 |
| GDCA | 116.00 (10.60, 239.00) | 113.00 (12.70, 253.00) | 0.991 |
| TLCA | 0.00 (0.00, 2.10) | 0.00 (0.00, 2.80) | 0.520 |
| GLCA | 2.70 (0.00, 16.70) | 5.50 (0.00, 16.20) | 0.340 |
| TUDCA | 9.10 (3.05, 15.00) | 6.90 (3.30, 15.00) | 0.865 |
| GUDCA | 169.00 (47.20, 399.50) | 115.00 (47.10, 340.00) | 0.482 |
| Diameter < 1cm group | Diameter ≥ 1 cm group | P value | |
| Primary free BAs | |||
| CA | 55.80 (22.70, 200.00) | 155.00 (32.30, 343.00) | 0.005a |
| CDCA | 365.00 (85.20, 835.00) | 586.00 (278.00, 1130.00) | 0.015a |
| Primary conjugated BAs | |||
| TCA | 21.70 (5.50, 46.85) | 22.00 (11.50, 75.50) | 0.391 |
| GCA | 167.00 (84.05, 329.50) | 166.00 (57.50, 416.00) | 0.927 |
| GCDCA | 961.00 (389.00, 1845.00) | 900.00 (556.00, 2410.00) | 0.333 |
| TCDCA | 69.10 (24.05, 152.50) | 90.20 (40.70, 262.00) | 0.060 |
| Secondary free BAs | |||
| DCA | 127.00 (21.90, 389.00) | 274.00 (77.30, 525.00) | 0.063 |
| LCA | 5.50 (0.00, 16.15) | 8.50 (2.10, 21.60) | 0.163 |
| UDCA | 64.50 (16.75, 182.50) | 196.00 (52.70, 421.00) | 0.003a |
| Secondary conjugated BAs | |||
| TDCA | 8.20 (0.00, 29.80) | 13.30 (3.10, 37.10) | 0.317 |
| GDCA | 108.00 (12.65, 235.00) | 144.00 (10.80, 324.00) | 0.610 |
| TLCA | 0.00 (0.00, 2.30) | 0.80 (0.00, 3.20) | 0.189 |
| GLCA | 4.60 (0.00, 15.80) | 2.50 (0.00, 21.20) | 0.736 |
| TUDCA | 7.00 (3.00, 15.00) | 14.10 (3.90, 34.60) | 0.034a |
| GUDCA | 122.00 (42.95, 315.50) | 234.00 (59.00, 556.00) | 0.030a |
| Left colon group | Right colon group | Total colon group | P value | |
| Primary free BAs | ||||
| CA | 108.00 (24.53, 334.00) | 65.60 (48.23, 126.00) | 23.20 (11.05, 157.48) | 0.000a |
| CDCA | 447.00 (130.25, 1000.00) | 448.00 (191.00, 813.00) | 135.50 (41.43, 678.00) | 0.047a |
| Primary conjugated BAs | ||||
| TCA | 20.75 (4.88, 53.78) | 34.70 (12.73, 58.85) | 15.00 (4.15, 39.43) | 0.148 |
| GCA | 156.50 (65.53, 342.25) | 257.00 (136.00, 373.00) | 134.00 (42.95, 204.00) | 0.006a |
| GCDCA | 812.50 (334.00, 1822.50) | 1420.00 (764.00, 2387.50) | 655.50 (290.50, 1622.50) | 0.005a |
| TCDCA | 73.95 (23.68, 154.25) | 110.00 (45.63, 257.00) | 64.20 (28.18, 125.50) | 0.060 |
| Secondary free BAs | ||||
| DCA | 185.00 (48.85, 466.25) | 105.50 (3.18, 383.75) | 110.50 (18.08, 280.75) | 0.098 |
| LCA | 6.20 (0.68, 15.08) | 8.85 (0.00, 17.20) | 5.50 (0.00, 28.83) | 0.963 |
| UDCA | 73.55 (27.00, 209.50) | 102.50 (29.80, 212.75) | 46.45 (9.78, 252.75) | 0.314 |
| Secondary conjugated BAs | ||||
| TDCA | 8.50 (1.25, 34.63) | 8.50 (0.00, 22.825) | 7.55 (0.00, 30.33) | 0.635 |
| GDCA | 122.50 (20.03, 278.50) | 118.00 (6.48, 242.00) | 86.70 (7.18, 213.75) | 0.464 |
| TLCA | 0.00 (0.00, 2.30) | 0.45 (0.00, 3.00) | 0.65 (0.00, 2.48) | 0.478 |
| GLCA | 4.60 (0.00, 15.90) | 3.35 (0.00, 18.25) | 5.85 (0.38, 19.28) | 0.555 |
| TUDCA | 9.10 (2.20, 15.00) | 6.40 (4.13, 15.00) | 6.75 (3.83, 15.00) | 0.933 |
| GUDCA | 113.00 (34.43, 341.00) | 228.50 (83.25, 514.50) | 112.00 (47.55, 308.75) | 0.064 |
| Pedicled polyp group | Sessile polyp group | P value | |
| Primary free BAs | |||
| CA | 420.00 (32.48, 791.00) | 59.80 (24.08, 173.00) | 0.006a |
| CDCA | 711.00 (214.75, 2845.00) | 373.50 (94.68, 834.50) | 0.016a |
| Primary conjugated BAs | |||
| TCA | 17.90 (11.00, 94.98) | 22.10 (5.30, 49.90) | 0.635 |
| GCA | 166.50 (92.35, 491.25) | 168.50 (73.75, 330.00) | 0.709 |
| GCDCA | 1230.00 (625.25, 2775.00) | 900.00 (413.25, 1860.00) | 0.096 |
| TCDCA | 111.50 (44.68, 307.50) | 68.15 (25.35, 162.00) | 0.075 |
| Secondary free BAs | |||
| DCA | 155.50 (0.68, 806.00) | 142.00 (34.03, 418.25) | 0.976 |
| LCA | 3.50 (0.00, 22.13) | 6.45 (0.50, 15.93) | 0.662 |
| UDCA | 228.00 (64.38, 454.75) | 64.90 (19.63, 196.50) | 0.003a |
| Secondary conjugated BAs | |||
| TDCA | 13.90 (0.00, 56.58) | 8.15 (0.50, 30.33) | 0.539 |
| GDCA | 118.15 (0.00, 494.50) | 113.50 (18.50, 238.00) | 0.595 |
| TLCA | 0.25 (0.00, 3.78) | 0.00 (0.00, 2.30) | 0.540 |
| GLCA | 3.40 (0.00, 30.45) | 4.65 (0.00, 16.05) | 0.723 |
| TUDCA | 7.20 (4.15, 24.43) | 7.65 (3.08, 15.00) | 0.472 |
| GUDCA | 330.50 (133.75, 573.00) | 114.50 (44.40, 314.75) | 0.008a |
A univariate logistic regression analysis using the presence or absence of colonic polyps as the dependent variable and other indicators as the independent variables was carried out to evaluate the risk factors for colonic polyps. The results showed that CDCA (B = 0.000, OR = 1.000), GCDCA (B = 0.000, OR = 1.000), and primary BA (B = 0.000, OR = 1.000) were associated with the risk of colonic polyps and were risk factors for colonic polyps (P < 0.05), as shown in Table 9. The results of multivariate logistic regression analysis using the statistically differences in the aforementioned univariate ana
| Variable | Univariate analysis | Multivariate analysis | ||
| OR (95%CI) | P value | OR (95%CI) | P value | |
| TBA | 1.000 (1.000, 1.000) | 0.104 | ||
| CA | 1.000 (1.000, 1.001) | 0.181 | ||
| CDCA | 1.000 (1.000, 1.000) | 0.046 | 1.001 (1.000, 1.001) | 0.073 |
| DCA | 1.000 (1.000, 1.000) | 0.799 | ||
| LCA | 1.000 (0.999, 1.001) | 0.636 | ||
| UDCA | 1.000 (1.000, 1.001) | 0.329 | ||
| GCA | 1.000 (1.000, 1.001) | 0.512 | ||
| GCDCA | 1.000 (1.000, 1.000) | 0.027 | 1.001 (1.000, 1.001) | 0.074 |
| GDCA | 1.000 (0.999, 1.000) | 0.080 | ||
| GLCA | 1.000 (0.999, 1.001) | 0.394 | ||
| GUDCA | 1.000 (1.000, 1.001) | 0.154 | ||
| TCA | 1.000 (0.998, 1.002) | 0.927 | ||
| TCDCA | 1.00 1(0.999, 1.002) | 0.328 | ||
| TDCA | 0.998 (0.995, 1.002) | 0.310 | ||
| TLCA | 0.999 (0.989, 1.009) | 0.900 | ||
| TUDCA | 1.005 (0.993, 1.018) | 0.413 | ||
| primary BA | 1.000 (1.000, 1.000) | 0.018 | 1.000 (0.999, 1.000) | 0.182 |
| primary free BA | 1.000 (1.000, 1.000) | 0.053 | ||
| primary conjugated BA | 1.000 (1.000, 1.000) | 0.071 | ||
| secondary BA | 1.000 (1.000, 1.000) | 0.720 | ||
| secondary free BA | 1.000 (1.000, 1,000) | 0.710 | ||
| secondary conjugated BA | 1.000 (1.000, 1.000) | 0.363 | ||
This study found that compared with the control group, the serum primary conjugated BAs, TCA, GCA, GCDCA, and TCDCA levels in the colonic polyp group were significantly higher than those in the control group (P < 0.05), while the secondary free BAs, DCA content was lower than that in the control group. Kühn et al[7] included 581 cases of primary colonic cancer diagnosed between 1993 and 2008, found that five primary conjugated BAs, GCA, TCA, GCDCA, TCDCA, and GHCA, as well as two secondary conjugated BAs, GDCA and TDCA were positively correlated with colonic cancer risk. Experts believed that an increase in primary conjugated BAs can promote the occurrence of colonic cancer, and the outcomes of this investigation supported those of our study. The concentration of fecal BA is the main subject of several relevant investigations. Sun et al[8] demonstrated that CDCA, DCA, and LCA increased in the feces of colon cancer patients whereas GCDCA decreased. By comparing the Alaskan aboriginals (AN) with the highest incidence rate of colonic cancer and the African rural people (RA) with the lowest incidence rate, Ocvirk et al[9] discovered that the de
Previous studies have analyzed the role and mechanism of BA profile in the occurrence and development of colonic tumors. The commonly accepted theory holds that while increasing the concentration of UDCA may restrict the onset and development of cancers, increasing the concentration of DCA in the BA profile may promote the emergence of colonic malignancies[11,12]. In 1940, DCA was first proven to be a carcinogen capable of causing mouse colonic cancer[11]. It can induce excessive proliferation of colonic epithelium, disrupt cell membranes, promote excessive production of reactive oxygen species and reactive nitrogen species, cause oxidative stress, damage DNA, induce gene mutations, and nuclear factor kappa B (NF-κB) activation by activating epidermal growth factor receptor and protein kinase C leads to patho
This study went on to conduct grouping analysis based on a comparison of the BA profile detection results between the colonic polyp group and the control group. The results showed that the CA, CDCA, UDCA, and TUDCA contents of the adenomatous polyp group were lower than those of the non adenomatous polyp group. The content of CA, CDCA, UDCA, GUDCA, TUDCA in the group with polyp diameter ≥ 1 cm was higher than that in the group with polyp dia
In summary, the serum BA profile showed significant changes in patients with colonic polyps. The etiology of colon cancers may be intimately associated with secondary bile acid DCA, one of them. At present, the widely recognized view on the role of serum BA metabolism in the occurrence and development of colon polyps is that BA can induce changes in the colon environment by activating various signaling pathways in the body, thereby promoting the occurrence of colonic polyps and even colonic cancer. Among them, a large number of studies have shown that DCA can induce NF-κB ac
This study shows that the serum BA profile of patients with colonic polyps has changed compared to normal individuals. The serum GCA, GCDCA, TCA, and TCDCA levels in the colonic polyp group are significantly higher than those in the control group (P < 0.05), while the DCA content is lower than that in the control group. Patients with various polyp sizes, locations, morphologies, and pathological types had variable serum BA profile, according to subgroup study of colonic polyps. Therefore, analyzing the changes in serum BA profile may provide new ideas for finding new targets for the treatment of colonic tumors.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Wu YH, Taiwan S-Editor: Liu JH L-Editor: A P-Editor: Xu ZH
| 1. | Sullivan BA, Noujaim M, Roper J. Cause, Epidemiology, and Histology of Polyps and Pathways to Colorectal Cancer. Gastrointest Endosc Clin N Am. 2022;32:177-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 68] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 2. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64583] [Article Influence: 16145.8] [Reference Citation Analysis (176)] |
| 3. | Cao PX, Shen YZ, Huang YQ, Jiang CX, Ma HQ, Wang ZY. Clinical characteristics and pathological types of 7408 cases of intestinal lesions found in colorectal cancer screening. Zhonghua Xiaohua Neijing Zazhi. 2018;35:630-633. |
| 4. | Shaukat A, Levin TR. Current and future colorectal cancer screening strategies. Nat Rev Gastroenterol Hepatol. 2022;19:521-531. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 239] [Article Influence: 79.7] [Reference Citation Analysis (1)] |
| 5. | Marin JJ, Macias RI, Briz O, Banales JM, Monte MJ. Bile Acids in Physiology, Pathology and Pharmacology. Curr Drug Metab. 2015;17:4-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 127] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 6. | Qi L, Chen Y. Circulating Bile Acids as Biomarkers for Disease Diagnosis and Prevention. J Clin Endocrinol Metab. 2023;108:251-270. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 7. | Kühn T, Stepien M, López-Nogueroles M, Damms-Machado A, Sookthai D, Johnson T, Roca M, Hüsing A, Maldonado SG, Cross AJ, Murphy N, Freisling H, Rinaldi S, Scalbert A, Fedirko V, Severi G, Boutron-Ruault MC, Mancini FR, Sowah SA, Boeing H, Jakszyn P, Sánchez MJ, Merino S, Colorado-Yohar S, Barricarte A, Khaw KT, Schmidt JA, Perez-Cornago A, Trichopoulou A, Karakatsani A, Thriskos P, Palli D, Agnoli C, Tumino R, Sacerdote C, Panico S, Bueno-de-Mesquita B, van Gils CH, Heath AK, Gunter MJ, Riboli E, Lahoz A, Jenab M, Kaaks R. Prediagnostic Plasma Bile Acid Levels and Colon Cancer Risk: A Prospective Study. J Natl Cancer Inst. 2020;112:516-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 89] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 8. | Sun L, Zhang Y, Cai J, Rimal B, Rocha ER, Coleman JP, Zhang C, Nichols RG, Luo Y, Kim B, Chen Y, Krausz KW, Harris CC, Patterson AD, Zhang Z, Takahashi S, Gonzalez FJ. Bile salt hydrolase in non-enterotoxigenic Bacteroides potentiates colorectal cancer. Nat Commun. 2023;14:755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 61] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 9. | Ocvirk S, Wilson AS, Posma JM, Li JV, Koller KR, Day GM, Flanagan CA, Otto JE, Sacco PE, Sacco FD, Sapp FR, Newton K, Brouard F, DeLany JP, Behnning M, Appolonia CN, Soni D, Bhatti F, Methé B, Fitch A, Morris A, Gaskins HR, Kinross J, Nicholson JK, Thomas TK, O'Keefe SJD. A prospective cohort analysis of gut microbial co-metabolism in Alaska Native and rural African people at high and low risk of colorectal cancer. Am J Clin Nutr. 2020;111:406-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 59] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 10. | Kawano A, Ishikawa H, Kamano T, Kanoh M, Sakamoto K, Nakamura T, Otani T, Sakai T, Kono K. Significance of fecal deoxycholic acid concentration for colorectal tumor enlargement. Asian Pac J Cancer Prev. 2010;11:1541-1546. [PubMed] |
| 11. | Cook J, Kennaway E, Kennaway N. Production of Tumours in Mice by Deoxycholic Acid. Nature. 1940;145:627. [RCA] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 92] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 12. | Fang Y, Yan C, Zhao Q, Xu J, Liu Z, Gao J, Zhu H, Dai Z, Wang D, Tang D. The roles of microbial products in the development of colorectal cancer: a review. Bioengineered. 2021;12:720-735. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 78] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 13. | Di Ciaula A, Wang DQ, Molina-Molina E, Lunardi Baccetto R, Calamita G, Palmieri VO, Portincasa P. Bile Acids and Cancer: Direct and Environmental-Dependent Effects. Ann Hepatol. 2017;16:s87-s105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 84] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 14. | Wang S, Liu Z, Wang L, Zhang X. NF-kappaB signaling pathway, inflammation and colorectal cancer. Cell Mol Immunol. 2009;6:327-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 330] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 15. | Jia W, Xie G, Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol. 2018;15:111-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1355] [Cited by in RCA: 1256] [Article Influence: 179.4] [Reference Citation Analysis (0)] |
| 16. | Ocvirk S, O'Keefe SJD. Dietary fat, bile acid metabolism and colorectal cancer. Semin Cancer Biol. 2021;73:347-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 155] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 17. | Liu L, Dong W, Wang S, Zhang Y, Liu T, Xie R, Wang B, Cao H. Deoxycholic acid disrupts the intestinal mucosal barrier and promotes intestinal tumorigenesis. Food Funct. 2018;9:5588-5597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 110] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 18. | Serfaty L, De Leusse A, Rosmorduc O, Desaint B, Flejou JF, Chazouilleres O, Poupon RE, Poupon R. Ursodeoxycholic acid therapy and the risk of colorectal adenoma in patients with primary biliary cirrhosis: an observational study. Hepatology. 2003;38:203-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 67] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 19. | Khare S, Mustafi R, Cerda S, Yuan W, Jagadeeswaran S, Dougherty U, Tretiakova M, Samarel A, Cohen G, Wang J, Moore C, Wali R, Holgren C, Joseph L, Fichera A, Li YC, Bissonnette M. Ursodeoxycholic acid suppresses Cox-2 expression in colon cancer: roles of Ras, p38, and CCAAT/enhancer-binding protein. Nutr Cancer. 2008;60:389-400. [PubMed] |
| 20. | Shah SA, Volkov Y, Arfin Q, Abdel-Latif MM, Kelleher D. Ursodeoxycholic acid inhibits interleukin 1 beta [corrected] and deoxycholic acid-induced activation of NF-kappaB and AP-1 in human colon cancer cells. Int J Cancer. 2006;118:532-539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 65] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 21. | Zhang H, Xu H, Zhang C, Tang Q, Bi F. Ursodeoxycholic acid suppresses the malignant progression of colorectal cancer through TGR5-YAP axis. Cell Death Discov. 2021;7:207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 51] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 22. | Cai Y, Shen X, Lu L, Yan H, Huang H, Gaule P, Muca E, Theriot CM, Rattray Z, Rattray NJW, Lu J, Ahuja N, Zhang Y, Paty PB, Khan SA, Johnson CH. Bile acid distributions, sex-specificity, and prognosis in colorectal cancer. Biol Sex Differ. 2022;13:61. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 7.3] [Reference Citation Analysis (0)] |