Published online Sep 26, 2023. doi: 10.12998/wjcc.v11.i27.6344
Peer-review started: June 24, 2023
First decision: August 9, 2023
Revised: August 18, 2023
Accepted: August 31, 2023
Article in press: August 31, 2023
Published online: September 26, 2023
Processing time: 88 Days and 4.2 Hours
Breast cancer is regarded as a highly malignant neoplasm in the female popu
To identify diagnostic and prognostic biomarkers for breast cancer.
Overlapping differentially expressed genes were screened based on Gene Expression Omnibus (GSE36765, GSE10810, and GSE20086) and The Cancer Genome Atlas datasets. A protein-protein interaction network was applied to excavate the hub genes among these differentially expressed genes. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analyses, as well as gene set enrichment analyses, were conducted to examine the functions of these genes and their potential mechanisms in the development of breast cancer. For clarification of the diagnostic and prognostic roles of these genes, Kaplan–Mei
This study demonstrated that calreticulin, heat shock protein family B member 1, insulin-like growth Factor 1, interleukin-1 receptor 1, Krüppel-like factor 4, suppressor of cytokine signaling 3, and triosephosphate isomerase 1 are potential diagnostic biomarkers of breast cancer as well as potential treatment targets with clinical implications.
The screening of biomarkers is of guiding significance for the diagnosis and prognosis of the diseases.
Core Tip: Breast cancer is one of the most common malignant tumors in women, according to statistics, the incidence of this disease accounts for 7%-10% of all kinds of malignant tumors in the whole body. However, the treatment of breast cancer is still not optimistic, and it is crucial to reveal the pathogenesis and biomarkers. Therefore, this study used bioinformatics statistics to mine the characteristic markers of breast cancer based on the database, in order to provide a more solid foundation for the treatment of the disease.
- Citation: Zhang X, Mi ZH. Identification of potential diagnostic and prognostic biomarkers for breast cancer based on gene expression omnibus. World J Clin Cases 2023; 11(27): 6344-6362
- URL: https://www.wjgnet.com/2307-8960/full/v11/i27/6344.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i27.6344
The global incidence of breast cancer worldwide has exceeded that of lung cancer, with 2.26 million new cases reported, making it the most prevalent cancer worldwide[1]. Invasive breast cancer, known for its high malignancy and unfavorable prognosis, represents the predominant form of breast cancer[2].
According to the latest report on breast cancer in China in 2020, the incidence and death rates of breast cancer in Chinese women accounted for 11.2% and 9.2% of the global incidence and death rates, respectively, ranking among the top in the world. The diagnosis and management of breast cancer are currently experiencing a paradigm shift, transitioning from a standardized approach to personalized medicine. This shift encompasses a range of treatment options, including resection surgery, radiotherapy and targeted adjuvant therapy[3]. However, progress in breast cancer treatment remains limited, and the prognosis is not promising. Various factors, such as genetics, lifestyle choices, obesity, and environmental influences, may all contribute to the onset and progression of breast cancer[4]. The study has documented that engaging in physical activity among individuals with breast cancer patients not only enhance their quality of life but also exerts an impact on their immune system[5]. In order to gain a deeper comprehension of the pathogenesis of breast cancer and enhance the precision of treatment, it is imperative to direct attention towards genetic research, tumor signaling pathways, and targeted therapies, which are progressively being implemented in clinic work. Furthermore, the adoption of molecular stratification of therapies and the utilization of biomarkers to inform prognosis and treatment choices are on the rise[4].
The occurrence, development, overall survival, recurrence, and non-recurrence of tumors are influenced by both the pathological type and clinical stage of tumors, as well as the expression and pathways of tumor genes. Extensive research has demonstrated a significant elevation in abnormal gene expression in breast cancer compared to normal tissues, and these genes playing a crucial role in proliferation, invasion, apoptosis, and overall survival[6-8]. The analysis of these abnormally expressed genes holds great clinical significance in terms of targeted therapy, prognosis, and predicting the risk of recurrence in breast cancer. Presently, numerous clinical investigations are being conducted on genes associated with tumor recurrence genes and signaling pathways. As a result, a predictive model has been developed to elucidate the role of genes in enhancing the conventional tumor classification and recurrence prediction. This model provides a greater wealth of genetic information and more precise predictive data[9-11]. For instance, Rodrigues-Ferreira et al[12] identified potential implications of EB1 and ATIP3 in the diagnosis and prognosis of breast cancer. Additionally, Ki67 has proven to be an efficient prognostic indicator in the Chinese population following neoadjuvant chemotherapy[13].
In this study, we obtained breast cancer gene profiles (GSE36765, GSE20086, and GSE10810) from the Gene Expression Omnibus (GEO). By performing GEO2R online analysis, differentially expressed genes (DEGs) were identified in breast cancer tissues compared to non-cancerous tissues. Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment, co-expression, and protein-protein interaction (PPI) analyses were conducted on the DEGs. We then performed overall survival analyses with respect to the potential target genes. In addition, we employed the GEPIA and UALCAN web-based platforms to identify potential genes linked to pathological stage using The Cancer Genome Atlas (TCGA) data. Our findings indicate that calreticulin (CALR), heat shock protein family B member 1 (HSPB1), insulin-like growth Factor 1 (IGF1), interleukin-1 receptor 1 (IL1R1), Krüppel-like factor 4 (KLF4), suppressor of cytokine signaling 3 (SOCS3), and triosephosphate isomerase 1 (TPI1) exhibit potential as therapeutic targets with significant clinical implications.
GEO datasets were selected based on the following criteria: studies with invasive breast cancer tissue samples, a description of the technology and platforms utilized, and the inclusion of adjacent normal tissues as controls. Three datasets (GSE36765[14], GSE20086[15], GSE10810[16]) were downloaded from the GEO database, meeting these criteria. All three studies utilized the Affymetrix Human Genome U133 Plus 2.0 Array [HG-U133 plus 2] platform. GSE 36765 consisted of 30 breast cancer tumor tissue samples and 4 normal tissue samples; GSE20086 consisted of 6 breast cancer tumor tissue samples and 6 normal samples; GSE10810 consisted of 31 breast cancer samples and 27 normal samples. A total of 67 breast cancer tumor tissues samples and 37 normal tissue samples were included in this study. The detailed analysis process is shown in Figure 1.
The identification of DEGs in selected datasets was conducted using the GEO2R tool, which integrates the GEO query and limma R software packages from the Bioconductor project. Through this analysis, the three GEO datasets were examined, and genes with a P value < 0.05 and a fold change > 1.2 were determined as DEGs. Subsequently, a Venn diagram was constructed using the online tool Venny (v2.1.0) to identify the DEGs that were consistently present across all three datasets.
The GO terms (http://www.geneontology.org) and KEGG pathways (http://www.genome.jp) were identified and analyzed using the DAVID Functional Annotation Tool (v6.8) with the identifier parameter set as “official_gene_symbol” and the species parameter set as “Homo sapiens”. A significant enrichment was defined as a P value < 0.05. The results obtained from DAVID were visualized using the ggplot2 package in R language (v3.6.3).
STRING (v11.0), which incorporates known and predicted interactions among over 932000000 proteins from various organisms, encompassing 24584628 proteins from 5090 organisms[17]. The DEGs were uploaded for modeling of multiple proteins, and the organism was set as “Homo Sapiens”. The statistical significance of the network interaction relationship was determined based on an interaction score > 0.4, and disconnected nodes in the network were excluded. The high-confidence interaction relationships were imported into the Cytoscape software (v3.6.0) for visualization of gene interactions[18]. The cytohubba plugin was utilized to identify the hub genes in the PPI network, which were subsequently chosen as candidate DEGs for subsequent experiments.
GEPIA (http://gepia.cancer-pku.cn/) is an online database that encompasses comprehensive RNA sequencing expression data derived from 9736 tumors and 8537 normal samples sourced from the TCGA and GTEx projects[19]. GEPIA is structured around three primary functional modules: namely single gene analysis, cancer type analysis, and multiple gene analysis. These modules enable the identification of differential gene expression between tumor and normal tissues, survival analysis, gene correlations, and other related analyses. In this study, we conducted Kaplan–Meier survival analysis to examine the association between the relative expression of candidate DEGs in patients with breast cancer and the overall survival time. Hazard ratios and corresponding 95% confidence intervals were calculated to elucidate the relationship between patient survival and high/Low gene expression, thereby providing insights into the roles of genes in disease development. Multiple gene comparisons and principal component analysis were employed to visually assess the discriminatory ability of candidate genes in distinguishing between tumor and normal models.
The UALCAN (http://ualcan.path.uab.edu/index.html) is an online database that specializes in transcriptome and clinical data derived from TCGA data. It facilitates the examination of differential expression patterns between tumor and normal tissues, as well as the exploration of tumor stage, lymph node metastasis, and other pertinent related clinical parameters[20]. In this study, the UALCAN database was utilized to validate the expression of DEGs in both breast cancer and normal tissues.
This study incorporated three gene sets, namely (GSE36765, GSE20086, and GSE10810), of which GSE36765 comprised of 30 tumor samples and 4 normal samples, GSE20086 consisted of 6 tumor samples and 6 normal samples, and GSE10810 encompassed 31 tumor samples and 27 normal samples. In comparison to the normal samples, a total of significant DEGs were identified across all datasets (Figure 2), including 87 upregulated genes, 120 downregulated genes, and 24 genes exhibiting both upregulation and downregulation.
Functional enrichment analysis of the obtained DEGs was performed using DAVID. The GO enrichment analysis primarily aimed to predict the functions of the target genes based on biological processes, cell components, and molecular functions. Utilizing DAVID, the enrichment analysis revealed that several biological processes, including signal transduction, negative regulation of apoptotic process, negative regulation of cell proliferation, protein stabilization, positive regulation of cell migration, positive regulation of fibroblast proliferation, and positive regulation of mitogen-activated protein kinases (MAPK) cascade, were significantly enriched among the DEGs (Figure 3A). In the category of cell components, the DEGs were found to be enriched in terms such as extracellular exosome, cell–cell junction, vesicle, plasma membrane, and secretory granule membrane, as depicted in (Figure 3B). Furthermore, the DEGs were associated with molecular protein binding, cadherin binding, MHC class II protein complex binding, signaling adaptor activity, and protein binding involved in heterotypic cell–cell adhesion, as shown in Figure 3D.
The following 16 KEGG pathways were enriched among the 231 DEGs: Inflammatory bowel disease, central carbon metabolism in cancer, melanoma, prostate cancer, glioma, proteoglycans in cancer, viral carcinogenesis, transcriptional deregulation in cancer, Epsrein-Barr virus infection, human T-cell leukemia virus 1 infection, leishmaniasis, osteoclast differentiation, protein processing in endoplasmic reticulum, HIF-1 signaling pathway, EGFR tyrosine kinase inhibitor resistance, and inositol phosphate metabolism (Figure 3C).
A PPI network was constructed based on the 231 significant DEGs, and the hub genes were identified using the STRING database and Cytoscape software. The PPI network (Figure 4A) encompassed a total of 169 nodes and 290 edges with any disconnected nodes being concealed. An interaction score exceeding 0.4 was deemed indicative of a high-confidence interaction relationship. The cytohubba module was employed to identify the genes with the highest degree of connectivity, as illustrated in Figure 4B. The set of genes that exhibited higher connectivity included CALR, CHD4, EEF1A1, EGFR, HSPB1, IGF1, IL1R1, KLF4, MANF, PKM, PTPRC, SEC11C, SOCS3, PIK3R1, and TPI1. Gene expression profiles of the 15 central genes in breast cancer tumor vs normal samples identified using GEPIA are shown in Figure 5. Significantly differential was observed for CALR, EGFR, HSPB1, IGF1, IL1R1, KLF4, SOCS3, and TPI1 between the breast cancer tumor and normal groups. Thus, analyses of the correlations between these eight genes and the pathological stage of breast cancer were performed using GEPIA. The expression levels of CALR, HSPB1, IGF1, IL1R1, KLF4, SOCS3, and TPI1 were found to be significantly associated with the pathological stage of breast cancer (P value < 0.05), whereas EGFR exhibited no significant correlation (P value > 0.05) (Figure 6). Therefore, this study focused on the seven genes CALR, HSPB1, IGF1, IL1R1, KLF4, SOCS3, and TPI1.
In order to enhance the understanding of the functions of the 231 DEGs derived from the three datasets, functional and pathway enrichment analyses were conducted using DAVID. Within the core biological process (BPs), particular attention was given to those that are closely associated with the development and advancement of cancer, including signal transduction, inactivation of MAPK activity, positive regulation of tumor necrosis factor production, regulation of cell migration, and regulation of fibroblast proliferation (Figure 7), the diagrammatic figure based on the enrichment pathway analysis was provided in Supplementary Figure 1.
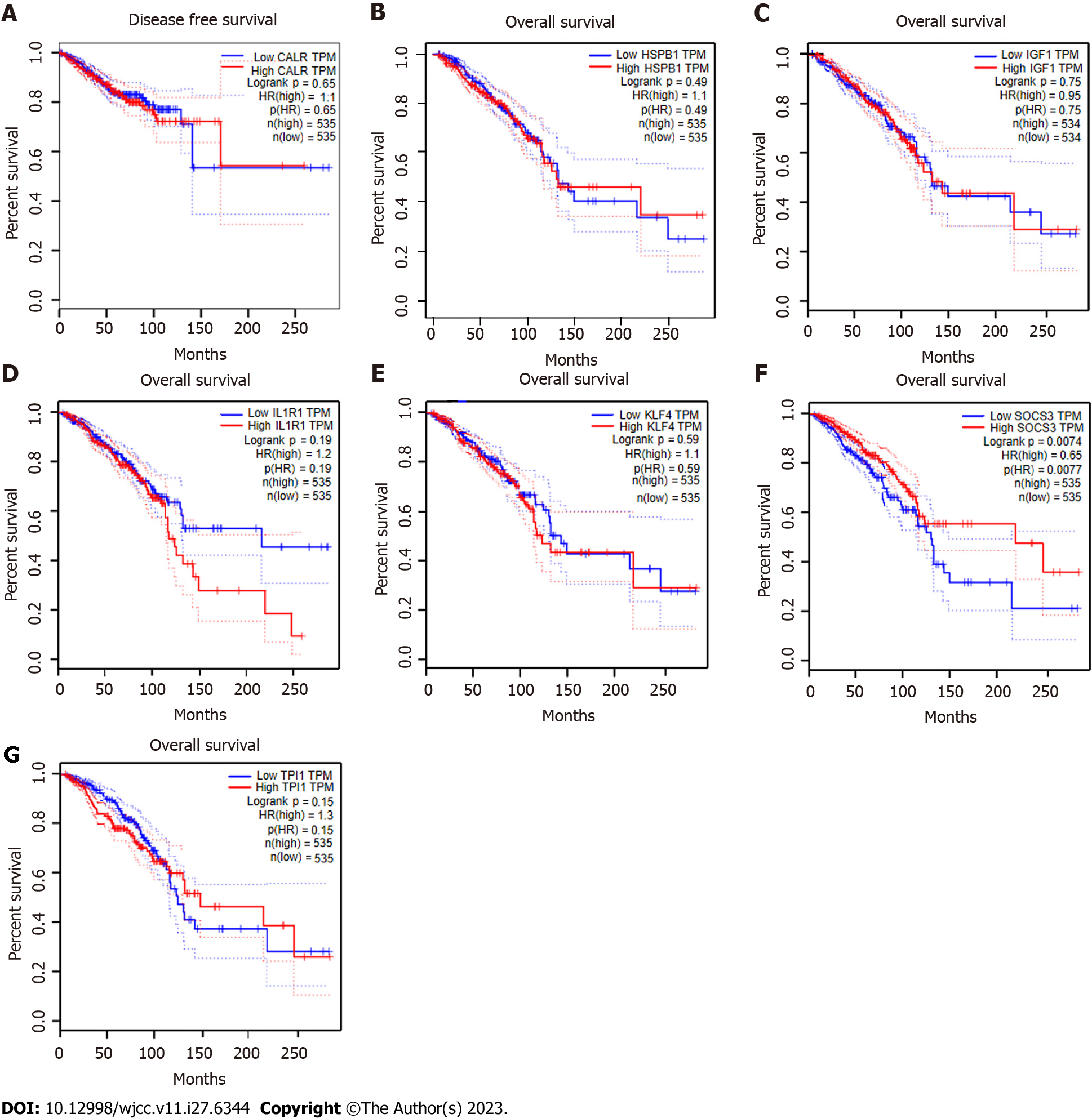
The overall survival and disease-free survival of patients with breast cancer were analyzed using the GEPIA database, with a focus on the expression of seven hub genes.
The results indicated that there was no association between the expression of CALR, HSPB1, IGF1, IL1R1, KLF4, SOCS3, or TPI1 and either overall survival or disease-free survival (Figure 8). Only the expression of the reserve gene CDH4 was found to be significantly associated with overall survival in patients with breast cancer (P value < 0.05) (Supplementary Figure 2).
Correlation analysis was conducted on the expression levels of seven candidate genes in patients with breast cancer. Moderate positive correlations, with a threshold of R < 0.5, were observed between CALR and TPI1, IGF1 and IL1R1, IGF1 and KLF4, IGF1 and SOCS3, and KLF4 and SOCS3 (Figure 9).
UALCAN analysis revealed significant differences in the expression of these seven candidate genes between the normal and tumor groups, confirming their potential as marker genes (Figure 10). Based on the reference group of normal samples, all seven genes exhibited significant deregulation in expression when considering age group, race, and lymph node metastasis status. In terms of age (Supplementary Figure 3), CALR, HSPB1, and TPI1 demonstrated high expression across all age groups. Notably, there were significant variations in CALR, HSPB1, and IGF1 expression between the age groups of 41–60 and 81–100 years, as well as in IGF1 and TPI1 expression between the age groups of 41–60 and 61–80 years. Additionally, IL1R1 expression exhibited significant differences between the age groups of 21–40 and 61–80 years. Regarding race (Supplementary Figure 4), our study specifically examined gene expression differences between Asian and non-Asian countries, given our focus on China. Notably, the expression levels of IGF1, SOCS3, and IL1R1 were found to be significantly reduced in Asian countries compared to non-Asian countries, whereas KLF4 expression was observed to be increased. Additionally, when considering the node metastasis status, significant variations in the expression of these genes were observed across the N0, N1, N2, and N3 stages (Supplementary Figure 5).
Comparative analyses were conducted using GEPIA to evaluate seven potential biomarkers for breast cancer, based solely on tumor data. Among the seven genes, CALR exhibited the highest expression level among the seven genes, followed by HSPB1, TPI1, IL1R1, KLF4, SOCS3, and IGF1 (Figure 11). Principle component analysis was performed on the seven genes using both breast cancer tumor data and normal tissue data. The results demonstrated that the seven genes cohort effectively distinguished breast cancer samples from normal samples (Figure 12), thereby supporting the utilization of this seven-gene biomarker for the diagnosis of breast cancer. To further validate the rationality and predictive capacity of biomarkers, we conducted an assessment of the expression of a gene set consisting of CALR, HSPB1, IGF1, IL1R1, KLF4, SOCS3, and TPI1. This evaluation aimed to differentiate between patients with breast cancer and controls using the receiver operating characteristic curve. The area under the curve value of 1 provided evidence that the selected biomarker could effectively distinguish the model.
Breast cancer exhibits the highest global incidence among all cancers and is the primary contributor to cancer-related morbidity and mortality among females[21]. A growing body of evidence suggests that various factors, including genetic and environmental factors, may contribute to the onset and progression of breast cancer. Some lifestyle factors, such as excessive nutrition, obesity, a high-fat diet, and excessive alcohol consumption, have been found to impact the occurrence of breast cancer[17,22]. The symptoms of early-stage breast cancer may not be readily apparent, making it easy to overlook breast lumps, abnormalities in breast skin, and other symptoms[23]. Detecting symptoms during the mid- or advanced stages of breast cancer poses significant challenges for treatment. The recommended multidisciplinary treatment approach encompasses surgical intervention, radiotherapy, neoadjuvant therapy, and adjuvant therapy[24]. Targeted therapy, as an emerging modality, offers the advantages of specificity, notable efficacy, and reduced incidence of side effects. Currently, the molecules targeted in the treatment of breast cancer primarily consist of HER-2, VEGF, EGFR, PARP, PI3K/Akt/mTOR, and CDK4/6[25-28]. In patients with HER2-positive early-stage breast cancer who experienced invasive cancer post-neoadjuvant therapy, the T-DM1 adjuvant group exhibited a 50% lower risk of recurrence of death from invasive breast cancer compared to the trastuzumab group[29]. Nevertheless, the early detection, reduction of recurrence, and improvement of overall survival continue to pose challenges in the clinical management treatment of breast cancer. Hence, the identification of novel biomarkers capable of predicting the recurrence of breast cancer and overall survival assumes significance, as it enables the classification of individuals into high and low risk groups based on these markers, thereby enhancing the effectiveness of subsequent treatment interventions. Numerous clinical studies pertaining to tumor recurrence are available in public databases. In this particular investigation, our attention was directed towards studies encompassing both tumor and adjacent normal tissue samples, with the aim of discerning genes linked to overall survival in patients afflicted with breast cancer.
The analysis incorporated three datasets (GSE36765, GSE20086, and GSE10810) sourced from the GEO database. Among these datasets, a total of 231 DEGs were identified as common, comprising 87 upregulated DEGs, 120 downregulated DEGs, and 24 DEGs exhibiting both up and downregulation. The chosen database exhibited an appropriate sample size, and the distribution of DEGs were deemed reasonable.
The present study reveals a significant association between the gene expression of CALR, HSPB1, IGF1, IL1R1, KLF4, SOCS3, CHD4, and TPI1 and the prognosis of breast cancer. Specifically, only CHD4 expression demonstrated a correlation with overall survival in breast cancer patients, whereas the expression of CALR, HSPB1, IGF1, IL1R1, KLF4, SOCS3 and TPI1 did not exhibit any significant relationship with overall survival. Notably, a previous investigation documented the overexpression of CALR in breast cancer tissue compared to normal tissue, and further established its association with morality and the stemness index. Furthermore, the depletion of CALR results in the impairment of breast cancer stem cells, thereby impacting tumor initiation and metastasis, and augmenting the sensitivity to chemotherapy[30]. Our investigation demonstrated a significant association between aberrant CALR expression and lymph node metastasis. Previous research has also indicated that CALR exhibits potential as a biomarker of tumor status, exhibiting high expression in oral squamous cell carcinoma, pancreatic cancer, gastric cancer, and esophageal squamous cell carcinoma[31-33]. Clinical studies have demonstrated that the overexpression of HSPB1 and IGF1 in breast cancer has an impact on disease outcome and the responsiveness of tumors to chemotherapy and radiotherapy[34]. Another study reported that the interference of the IL6 pathway through SOCS3 or IL6R inhibits tumor growth and metastasis in mouse xenograft models[35]. Increased TPI1 expression has been associated with a poor prognosis in patients with lung adenocarcinoma and has influenced the infiltration of immune cell[36]. In this study, we have discovered that these abnormally expressed genes not only contribute to the initiation and progression of tumors but also have a significant relationship with the pathological stage of tumors and lymph node metastasis. Our study revealed that the expression of reserve CDH4 was solely linked to the overall survival of breast cancer. Furthermore, the expression of CALR, HSPB1, IGF1, IL1R1, KLF4, SOCS3, and TPI1 exhibited significant associations with the pathological and lymph node metastasis stages of breast cancer.
These gene serve as crucial indicators for evaluating the prognosis and guiding the treatment of breast cancer. Notably, the expression levels of these seven genes effectively differentiated breast cancer samples from normal samples in a principal component analysis. The integration of these seven genes into a biomarker panel holds potential for enhancing the accuracy of breast cancer diagnoses. By discerning the specific locations where aberrant gene expression triggers tumorigenesis, a theoretical foundation can be established for the advancement of gene-level targeted therapies with greater precision.
Breast cancer is widely recognized as a highly malignant neoplasm in women, posing a significant risk to their overall health. The diagnostic challenges of breast cancer arise from the heterogeneity of samples and the limitations of conventional techniques. Consequently, the identification of more stable biomarkers assumes paramount importance in facilitating early breast cancer screening. In this context, bioinformatics methods have been employed to detect differentially expressed genes associated with proliferation, invasion, apoptosis, and overall survival in breast cancer.
To ascertain fundamental prognostic biomarkers in breast cancer, three databases were queried for genes associated with breast cancer as tumor markers.
Bioinformatics analysis of the molecular mechanism involved in breast cancer revealed that seven differentially expressed genes (DEGs) [calreticulin (CALR), heat shock protein family B member 1 (HSPB1), insulin-like growth Factor 1 (IGF1), interleukin-1 receptor 1 (IL1R1), Krüppel-like factor 4 (KLF4), suppressor of cytokine signaling 3 (SOCS3), and triosephosphate isomerase 1 (TPI1)] play critical roles in the progression of breast cancer. Bioinformatics were used to identify hub genes and enrichment pathways in breast cancer, illustrating a biological relationship between the pathways and gene expression in breast cancer.
Microarray data information, data processing of differentially expressed genes, protein-protein interaction network and module analysis, and survival analysis were used to mine potential biomarkers. In addition, pathway enrichment analysis was also conducted to elaborate the pathogenesis of disease.
Three Gene Expression Omnibus datasets that included breast cancer tissues and normal tissues were analyzed; 231 DEGs were identified. 7 potential biomarkers (CALR, HSPB1, IGF1, IL1R1, KLF4, SOCS3, and TPI1) were closely related to the occurrence and progression of breast cancer. The discrimination of potential markers for the model is also relatively perfect, and the discrimination for cancer group and normal group is 100%.
Through the utilization of bioinformatics analysis, the molecular mechanism of breast cancer was investigated, revealing the significant involvement of seven differentially expressed genes (CALR, HSPB1, IGF1, IL1R1, KLF4, SOCS3, and TPI1) in the advancement of breast cancer. These findings hold promise in enhancing our understanding of breast cancer pathogenesis, as well as in the identification of novel biomarkers and potential drug targets, thereby facilitating advancements in breast cancer diagnosis and therapeutics.
Bioinformatics was employed to identify hub genes and significant pathways in breast cancer, thereby establishing a biological association between the pathways and gene expression potentially implicated in the advancement of breast cancer. The utilization of bioinformatics analysis revealed the relevant genes and cellular pathways implicated in the genesis and progression of breast cancer.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Biology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: El-Arabey AA, Egypt S-Editor: Liu JH L-Editor: A P-Editor: Wu RR
| 1. | Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J (Engl). 2021;134:783-791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1624] [Cited by in RCA: 1799] [Article Influence: 449.8] [Reference Citation Analysis (1)] |
| 2. | Thomas M, Kelly ED, Abraham J, Kruse M. Invasive lobular breast cancer: A review of pathogenesis, diagnosis, management, and future directions of early stage disease. Semin Oncol. 2019;46:121-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 3. | Bertozzi N, Pesce M, Santi PL, Raposio E. Oncoplastic breast surgery: comprehensive review. Eur Rev Med Pharmacol Sci. 2017;21:2572-2585. [PubMed] |
| 4. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55843] [Article Influence: 7977.6] [Reference Citation Analysis (132)] |
| 5. | Schmidt T, van Mackelenbergh M, Wesch D, Mundhenke C. Physical activity influences the immune system of breast cancer patients. J Cancer Res Ther. 2017;13:392-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 28] [Reference Citation Analysis (0)] |
| 6. | Juríková M, Danihel Ľ, Polák Š, Varga I. Ki67, PCNA, and MCM proteins: Markers of proliferation in the diagnosis of breast cancer. Acta Histochem. 2016;118:544-552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 436] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 7. | Qin J, Zhou Z, Chen W, Wang C, Zhang H, Ge G, Shao M, You D, Fan Z, Xia H, Liu R, Chen C. BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5. Nat Commun. 2015;6:8471. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 155] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 8. | Zhang R, Zhang S, Xing R, Zhang Q. High expression of EZR (ezrin) gene is correlated with the poor overall survival of breast cancer patients. Thorac Cancer. 2019;10:1953-1961. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 9. | Filipits M, Rudas M, Jakesz R, Dubsky P, Fitzal F, Singer CF, Dietze O, Greil R, Jelen A, Sevelda P, Freibauer C, Müller V, Jänicke F, Schmidt M, Kölbl H, Rody A, Kaufmann M, Schroth W, Brauch H, Schwab M, Fritz P, Weber KE, Feder IS, Hennig G, Kronenwett R, Gehrmann M, Gnant M; EP Investigators. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin Cancer Res. 2011;17:6012-6020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 466] [Cited by in RCA: 508] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 10. | Lin H, Wu Y, Liang G, Chen L. Establishing a predicted model to evaluate prognosis for initially diagnosed metastatic Her2-positive breast cancer patients and exploring the benefit from local surgery. PLoS One. 2020;15:e0242155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 11. | Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817-2826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4524] [Cited by in RCA: 4437] [Article Influence: 211.3] [Reference Citation Analysis (0)] |
| 12. | Rodrigues-Ferreira S, Nehlig A, Monchecourt C, Nasr S, Fuhrmann L, Lacroix-Triki M, Garberis I, Scott V, Delaloge S, Pistilli B, Vielh P, Dubois T, Vincent-Salomon A, André F, Nahmias C. Combinatorial expression of microtubule-associated EB1 and ATIP3 biomarkers improves breast cancer prognosis. Breast Cancer Res Treat. 2019;173:573-583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 13. | Wang J, Sang D, Xu B, Yuan P, Ma F, Luo Y, Li Q, Zhang P, Cai R, Fan Y, Chen S. Value of Breast Cancer Molecular Subtypes and Ki67 Expression for the Prediction of Efficacy and Prognosis of Neoadjuvant Chemotherapy in a Chinese Population. Medicine (Baltimore). 2016;95:e3518. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 14. | Gu-Trantien C, Loi S, Garaud S, Equeter C, Libin M, de Wind A, Ravoet M, Le Buanec H, Sibille C, Manfouo-Foutsop G, Veys I, Haibe-Kains B, Singhal SK, Michiels S, Rothé F, Salgado R, Duvillier H, Ignatiadis M, Desmedt C, Bron D, Larsimont D, Piccart M, Sotiriou C, Willard-Gallo K. CD4⁺ follicular helper T cell infiltration predicts breast cancer survival. J Clin Invest. 2013;123:2873-2892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 585] [Cited by in RCA: 812] [Article Influence: 67.7] [Reference Citation Analysis (0)] |
| 15. | Bauer M, Su G, Casper C, He R, Rehrauer W, Friedl A. Heterogeneity of gene expression in stromal fibroblasts of human breast carcinomas and normal breast. Oncogene. 2010;29:1732-1740. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 139] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 16. | Pedraza V, Gomez-Capilla JA, Escaramis G, Gomez C, Torné P, Rivera JM, Gil A, Araque P, Olea N, Estivill X, Fárez-Vidal ME. Gene expression signatures in breast cancer distinguish phenotype characteristics, histologic subtypes, and tumor invasiveness. Cancer. 2010;116:486-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 17. | Khushalani JS, Qin J, Ekwueme DU, White A. Awareness of breast cancer risk related to a positive family history and alcohol consumption among women aged 15-44 years in United States. Prev Med Rep. 2020;17:101029. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 18. | Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J Proteome Res. 2019;18:623-632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 637] [Cited by in RCA: 1398] [Article Influence: 199.7] [Reference Citation Analysis (0)] |
| 19. | Tang Z, Li C, Kang B, Gao G, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5550] [Cited by in RCA: 7089] [Article Influence: 886.1] [Reference Citation Analysis (0)] |
| 20. | Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649-658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2365] [Cited by in RCA: 4228] [Article Influence: 528.5] [Reference Citation Analysis (0)] |
| 21. | Katsura C, Ogunmwonyi I, Kankam HK, Saha S. Breast cancer: presentation, investigation and management. Br J Hosp Med (Lond). 2022;83:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 22. | Barone I, Giordano C, Bonofiglio D, Andò S, Catalano S. The weight of obesity in breast cancer progression and metastasis: Clinical and molecular perspectives. Semin Cancer Biol. 2020;60:274-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 80] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 23. | Alkabban FM, Ferguson T. Breast Cancer. 2022 Sep 26. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2023 Jan-. [PubMed] |
| 24. | Fisusi FA, Akala EO. Drug Combinations in Breast Cancer Therapy. Pharm Nanotechnol. 2019;7:3-23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 254] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 25. | Cortesi L, Rugo HS, Jackisch C. An Overview of PARP Inhibitors for the Treatment of Breast Cancer. Target Oncol. 2021;16:255-282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 135] [Cited by in RCA: 237] [Article Influence: 59.3] [Reference Citation Analysis (0)] |
| 26. | Neesham D, Richards A, McGauran M. Advances in epithelial ovarian cancer. Aust J Gen Pract. 2020;49:665-669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 27. | Miricescu D, Totan A, Stanescu-Spinu II, Badoiu SC, Stefani C, Greabu M. PI3K/AKT/mTOR Signaling Pathway in Breast Cancer: From Molecular Landscape to Clinical Aspects. Int J Mol Sci. 2020;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 291] [Cited by in RCA: 481] [Article Influence: 96.2] [Reference Citation Analysis (0)] |
| 28. | Pandey K, An HJ, Kim SK, Lee SA, Kim S, Lim SM, Kim GM, Sohn J, Moon YW. Molecular mechanisms of resistance to CDK4/6 inhibitors in breast cancer: A review. Int J Cancer. 2019;145:1179-1188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 221] [Cited by in RCA: 249] [Article Influence: 41.5] [Reference Citation Analysis (0)] |
| 29. | von Minckwitz G, Huang CS, Mano MS, Loibl S, Mamounas EP, Untch M, Wolmark N, Rastogi P, Schneeweiss A, Redondo A, Fischer HH, Jacot W, Conlin AK, Arce-Salinas C, Wapnir IL, Jackisch C, DiGiovanna MP, Fasching PA, Crown JP, Wülfing P, Shao Z, Rota Caremoli E, Wu H, Lam LH, Tesarowski D, Smitt M, Douthwaite H, Singel SM, Geyer CE Jr; KATHERINE Investigators. Trastuzumab Emtansine for Residual Invasive HER2-Positive Breast Cancer. N Engl J Med. 2019;380:617-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1122] [Cited by in RCA: 1689] [Article Influence: 281.5] [Reference Citation Analysis (0)] |
| 30. | Liu X, Xie P, Hao N, Zhang M, Liu Y, Liu P, Semenza GL, He J, Zhang H. HIF-1-regulated expression of calreticulin promotes breast tumorigenesis and progression through Wnt/β-catenin pathway activation. Proc Natl Acad Sci U S A. 2021;118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 62] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 31. | Chiang WF, Hwang TZ, Hour TC, Wang LH, Chiu CC, Chen HR, Wu YJ, Wang CC, Wang LF, Chien CY, Chen JH, Hsu CT, Chen JY. Calreticulin, an endoplasmic reticulum-resident protein, is highly expressed and essential for cell proliferation and migration in oral squamous cell carcinoma. Oral Oncol. 2013;49:534-541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 54] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 32. | Li Z, Huang Y, Xu Y, Wang X, Wang H, Zhao S, Liu H, Yu G, Che X. Targeting ADAR1 suppresses progression and peritoneal metastasis of gastric cancer through Wnt / β-catenin pathway. J Cancer. 2021;12:7334-7348. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 33. | Yoneda A, Minomi K, Tamura Y. Heat shock protein 47 confers chemoresistance on pancreatic cancer cells by interacting with calreticulin and IRE1α. Cancer Sci. 2021;112:2803-2820. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 34. | Ciocca DR, Calderwood SK. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones. 2005;10:86-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 962] [Cited by in RCA: 1026] [Article Influence: 54.0] [Reference Citation Analysis (0)] |
| 35. | Kim G, Ouzounova M, Quraishi AA, Davis A, Tawakkol N, Clouthier SG, Malik F, Paulson AK, D'Angelo RC, Korkaya S, Baker TL, Esen ES, Prat A, Liu S, Kleer CG, Thomas DG, Wicha MS, Korkaya H. SOCS3-mediated regulation of inflammatory cytokines in PTEN and p53 inactivated triple negative breast cancer model. Oncogene. 2015;34:671-680. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 71] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 36. | Yang X, Ye C, Zheng H, Dai C, Zhu Y. Systemic Analyses of the Expression of TPI1 and Its Associations with Tumor Microenvironment in Lung Adenocarcinoma and Squamous Cell Carcinoma. Dis Markers. 2022;2022:6258268. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |