Published online Sep 26, 2023. doi: 10.12998/wjcc.v11.i27.6327
Peer-review started: April 21, 2023
First decision: May 19, 2023
Revised: June 24, 2023
Accepted: July 28, 2023
Article in press: July 28, 2023
Published online: September 26, 2023
Processing time: 152 Days and 9.3 Hours
Wuzhuyu decoction, a traditional Chinese medicinal formula, is effective in treating hepatocellular carcinoma (HCC).
To explore the potential mechanism of action of Wuzhuyu decoction against HCC.
The active components of each Chinese herbal medicinal ingredient in Wuzhuyu decoction and their targets were obtained from the Traditional Chinese Medicine Database and Analysis Platform. HCC was used as a search query in GeneCards, Online Mendelian Inheritance in Man, Malacards, DisGeNET, Therapeutic Target Database, and Comparative Toxicogenomics Database. The overlapping targets of the Wuzhuyu decoction and HCC were defined, and then protein-protein interaction, Gene Ontology, and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses were performed. CytoHubba was used to select hub genes, and their binding activities and key active components were verified using molecular docking.
A total of 764 compounds, 77 active compounds, and 204 potential target genes were identified in Wuzhuyu decoction. For HCC, 9468 potential therapeutic target genes were identified by combining the results from the six databases and removing duplicates. A total of 179 overlapping targets of Wuzhuyu decoction and HCC were defined, including 10 hub genes (tumor necrosis factor, interleukin-6, AKT1, TP53, caspase-3, mitogen-activated protein kinase 1, epidermal growth factor receptor, MYC, mitogen-activated protein kinase 8, and JUN). There were six main active components (quercetin, kaempferol, ginsenoside Rh2, rutaecarpine, β-carotene, and β-sitosterol) that may act on hub genes to treat HCC in Wuzhuyu decoction. Kyoto Encyclopedia of Genes and Genomes enrichment analysis mainly involved the mitogen-activated protein kinase, p53, phosphatidylinositol-4,5-bisphosphate 3-kinase-Akt, Janus kinase-signal transducer of activators of transcription, and Hippo signaling pathways. Further verification based on molecular docking results showed that the small molecule compounds (quercetin, kaempferol, ginsenoside Rh2, rutaecarpine, β-carotene, and β-sitosterol) contained in Wuzhuyu decoction generally have excellent binding affinity to the macromolecular target proteins encoded by the top 10 genes.
This study revealed that Wuzhuyu decoction may be a latent multicomponent, multitarget, and multipathway treatment for HCC. It provided novel insights for verifying the mechanism of Wuzhuyu decoction in the treatment of HCC.
Core Tip: Six main active components (quercetin, kaempferol, ginsenoside Rh2, rutaecarpine, β-carotene, and β-sitosterol) and ten core genes (tumor necrosis factor, interleukin-6, AKT1, TP53, caspase-3, mitogen-activated protein kinase 1, epidermal growth factor receptor, MYC, mitogen-activated protein kinase 8, and JUN) were obtained from Wuzhuyu decoction. In addition, 179 pathways were obtained from Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis, which indicated that Wuzhuyu decoction is a multicomponent, multitarget, and multipathway treatment for hepatocellular carcinoma. The active components of Wuzhuyu decoction may participate in these pathways via the top 10 hub genes, which achieves the therapeutic effect on hepatocellular carcinoma.
- Citation: Ouyang JY, Lin WJ, Dong JM, Yang Y, Yang HK, Zhou ZL, Wang RQ. Exploring the pharmacological mechanism of Wuzhuyu decoction on hepatocellular carcinoma using network pharmacology. World J Clin Cases 2023; 11(27): 6327-6343
- URL: https://www.wjgnet.com/2307-8960/full/v11/i27/6327.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i27.6327
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors in the world, ranking sixth in the global mortality rate of malignant tumors. It is the most common pathological type of primary liver carcinoma, accounting for approximately 75%-85% of all cases[1]. The etiology and pathogenesis of HCC have not been defined; however, it is generally believed to be related to cirrhosis, viral hepatitis[2], excessive alcohol consumption[3], and chemical carcinogens such as aflatoxins[4]. Presently, the treatment method for HCC is relatively simple. If patients are diagnosed and treated early, they have a good prognosis through surgery and chemotherapy. However, the invasion of HCC is inconspicuous. The majority of the patients are diagnosed late, losing the best opportunity to undergo surgery and therefore only receive drug maintenance therapy[5]. Clinical first-line drugs for treating HCC include sorafenib and lenvatinib[6]. These drugs are expensive and impose heavy family and socioeconomic burdens. In addition, adverse drug reactions and tolerance often occur in clinical practice. Therefore, it is necessary to search for other effective, safe, and inexpensive drugs to treat HCC.
Traditional Chinese medicine has been practiced for thousands of years to treat various diseases[7]. For some patients with HCC who are unable to receive surgical treatment, whose bodies develop resistance to targeted drugs, or whose families cannot afford the relatively high cost of clinical treatment, traditional Chinese medicine treatment is selected to improve their quality of life and prolong life. Wuzhuyu decoction, as a warming interior formula, can warm the middle jiao (stomach, liver, spleen, and gallbladder) and tonify deficiencies from the Treatise on Febrile Diseases. Wuzhuyu decoction usually consists of four herbs: Wuzhuyu (Evodiae Fructus), Renshen (Panax ginseng C. A. Mey.), Shengjiang (Zingiber officinale Roscoe), and Dazhao (Jujubae Fructus). According to traditional Chinese medicine, Wuzhuyu plays a role in dispersing cold and relieving pain, stopping diarrhea, and relieving vomiting[8].
Recent studies have reported that it also exhibits anti-inflammatory and antitumor activities, induces cell cycle arrest, and suppresses tumor cell migration[8-10]. Renshen and Dazhao can boost immune function[11,12]. Ginsenoside, a major active component of Renshen, has strong pharmacological activities, such as immune function stimulation, cardiovascular system protection, antitumor proliferation, antitumor invasion, and antitumor metastasis[13]. Shengjiang, with a long clinical history, has various biological activities, including anti-inflammatory, antimicrobial, and antiemetic activities[14]. Modern preclinical studies have also shown that Shengjiang is effective in ameliorating the side effects of doxorubicin and cisplatin and exerts chemosensitization effects in certain tumor cells[15].
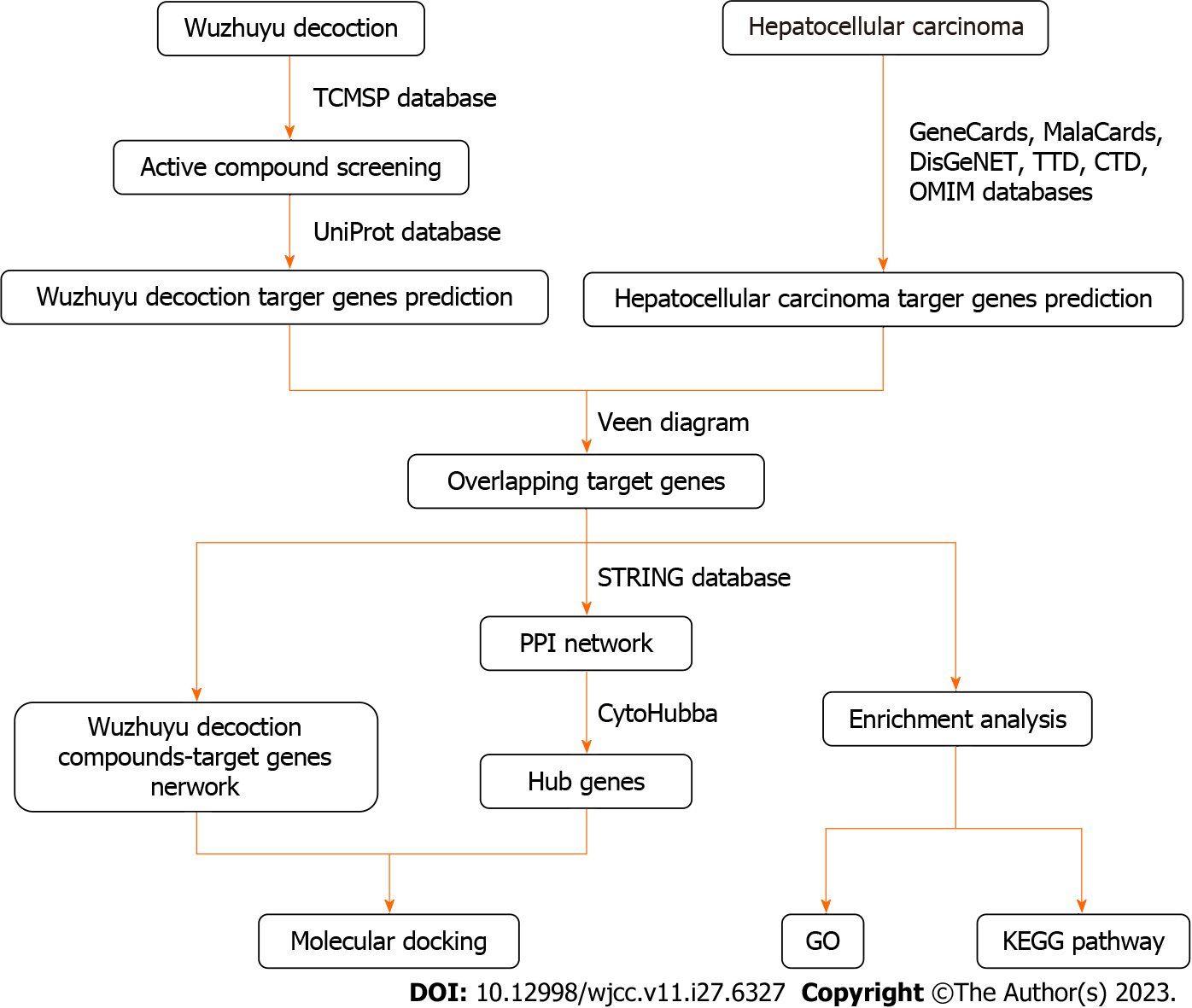
The relationships among drugs, active components, and diseases can be studied using the network pharmacology of traditional Chinese medicine, which conforms to the characteristics of traditional Chinese medicine with multiple components, targets, and pathways[16,17]. They can overcome the difficulty of elucidating the mechanism of action of prescriptions against diseases because prescriptions used in traditional Chinese medicine contain a variety of medicinal materials with complex chemical compositions. Traditional Chinese medicine network pharmacology provides a novel strategy for developing traditional Chinese medicine and elucidating its underlying mechanisms. This study utilized network pharmacology to explore the mechanism of Wuzhuyu decoction against HCC and reveal active compounds, key genes, and potential pathways of Wuzhuyu decoction treatment of HCC, which would be useful for further investigation (Figure 1).
The relationships among drugs, targets, and diseases were determined using the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, version 2.3, https://tcmsp-e.com/), a systematic pharmacological platform for Chinese herbal medicines[18]. All herbal compounds used in Wuzhuyu decoction were retrieved by searching “Wuzhuyu,” “Renshen,” “Shengjiang,” and “Dazhao” as the keywords. Based on the absorption, distribution, metabolism, and excretion protocols, the active compounds were screened out with oral bioavailability ≥ 30 and drug-likeness ≥ 0.18 as the criteria[19]. Potential target proteins of selected active compounds were identified in TCMSP. Each of the potential target proteins were then used as keywords searched using the UniProt database (https://www.uniprot.org/)[20] with the “Homo sapiens” species. Finally, the potential target genes of Wuzhuyu decoction and the corresponding UniProt ID of the potential targets were obtained.
The GeneCards (https://www.genecards.org/)[21], Online Mendelian Inheritance in Man (https://omim.org/)[22], MalaCards (https://www.malacards.org/)[23], DisGeNET (https://www.disgenet.org/)[24], Therapeutic Target Database (http://db.idrblab.net/ttd/)[25], and Comparative Toxicogenomics Database (CTD) (http://ctdbase.org/)[26] were searched using the keyword “hepatocellular carcinoma” to identify targets relevant to HCC. In the search results of GeneCards, the targets with relevance score ≥ 10 were screened. In the search results of DisGeNET, the targets with score ≥ 0.1 were screened. In the search results of CTD, the targets with inference score ≥ 50 were screened. After integrating and removing duplicates, a set of potential targets relevant to HCC were obtained and screened from the six databases mentioned above.
According to the potential target genes of Wuzhuyu decoction and targets relevant to HCC, a Venn online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/) was used to identify common targets that were potential target genes of Wuzhuyu decoction against HCC. Cytoscape software (version 3.8.2)[27] was used to construct the interaction between Wuzhuyu decoction-active compounds and target genes.
The STRING database (http://string-db.org/; version 11.0) is a database of known and predicted protein-protein interactions (PPIs). The PPI analysis was obtained from this database with the species limited to “Homo sapiens” and confidence score > 0.4[28,29]. The PPI analysis data were then imported into the Cytoscape software (version 3.8.2) to construct the network. Subsequently, the first 10 nodes, as hub genes, were identified via the following plug-in 12 CytoHubba algorithms of the Cytoscape software: betweenness; bottleneck; closeness; clustering coefficient; degree; density of maximum neighborhood component; EcCentricity; Edge Percolated Component; Maximal Clique Centrality; Maximum Neighborhood Component; radiality; and stress[30]. A Sankey diagram (https://www.omicstudio.cn/tool/) was constructed according to the hub genes, active compounds, and herbs of Wuzhuyu decoction.
The Gene Ontology (GO)[31] and Kyoto Encyclopedia of Genes and Genomes (KEGG)[32] pathway enrichment analyses were performed by the clusterProfiler package in R (R 4.1.0 for Windows) to confirm the biological processes (BPs) and signaling pathway of Wuzhuyu decoction active compounds involved in the treatment of HCC. An adjusted P value of < 0.05 was considered statistically significant, which validated the enriched terms.
AutoDock Vina[33] was used to conduct molecular docking among the top 10 hub genes screened using the plug-in CytoHubba of the Cytoscape software and the key active components obtained in the Sankey diagram. The SDF format of ligands and PDB format of proteins were downloaded from the PubChem (https://pubchem.ncbi.nlm.nih.gov/) and RCSB PDB databases (https://www.rcsb.org/). The ligands were then imported into ChemBio3D Ultra (ChemBio3D Ultra 14.0.0.117 for Windows) to transfer the format to MOL2. Proteins were dehydrated and separated using the PyMol software (https://pymol.org/2/; version 2.5). Ligands and proteins were then introduced into AutoDock Tools for molecular docking. Vina scores between hub genes and key active components were obtained. When the Vina score was lower than 0, molecular docking was considered successful. A lower Vina score indicated a higher binding affinity between the protein and the ligand. PyMol software was used to visualize the complexes of proteins and ligands.
The TCMSP database was used to screen for potential active compounds contained in Wuzhuyu decoction. In the TCMSP database, 764 compounds were identified, including 176 from Wuzhuyu, 190 from Renshen, 265 from Shengjiang, and 133 from Dazhao. Based on the standard of oral bioavailability ≥ 30 and drug-likeness ≥ 0.18, there were 86 active compounds of Wuzhuyu decoction that were searched, including 30 from Wuzhuyu, 22 form Renshen, 5 from Shengjiang, and 29 from Dazhao. After removing duplicates, there were 77 active compounds. They were used for subsequent analysis. Supplementary Table 1 shows essential information on some active compounds in the Wuzhuyu decoction.
Using the retrieval and screening function of the TCMSP database, it was confirmed that 56 of the 77 active compounds in Wuzhuyu decoction have target proteins, and 187 proteins were targeted. Subsequently, the UniProt database was then utilized to transform the target proteins predicted by the active compounds of the Wuzhuyu decoction into the corresponding gene names. In total, 204 possible target genes were identified.
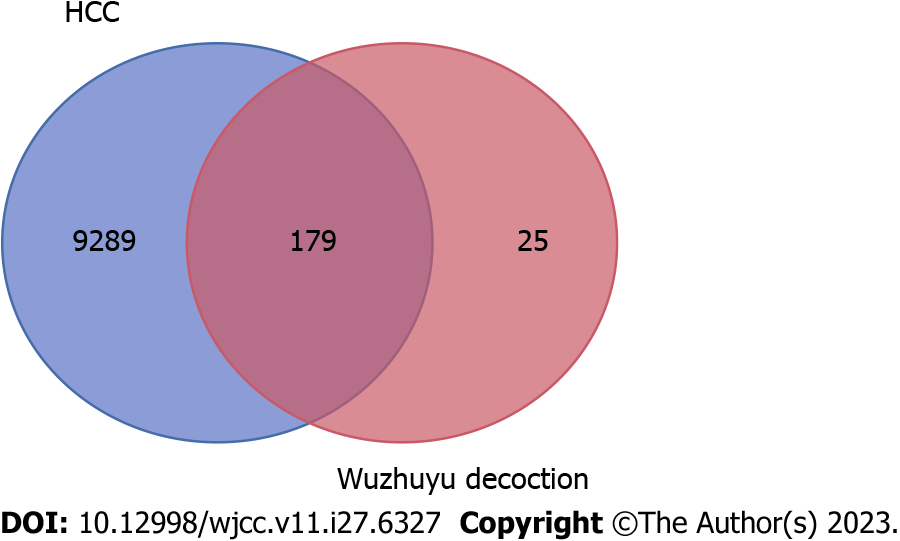
A total of 9468 potential therapeutic target genes for HCC were identified after searching the GeneCard, Online Mendelian Inheritance in Man, MalaCard, DisGeNET, Therapeutic Target Database, and CTD databases. Based on the 204 possible target genes of the active compounds of Wuzhuyu decoction and potential therapeutic target genes related to HCC, the Venn online platform was used to draw a Venn diagram. As shown in Figure 2, 197 core target were obtained, which were potential target genes of Wuzhuyu decoction against HCC. Supplementary Table 2 shows the basic information of the 179 potential target genes of Wuzhuyu decoction against HCC.
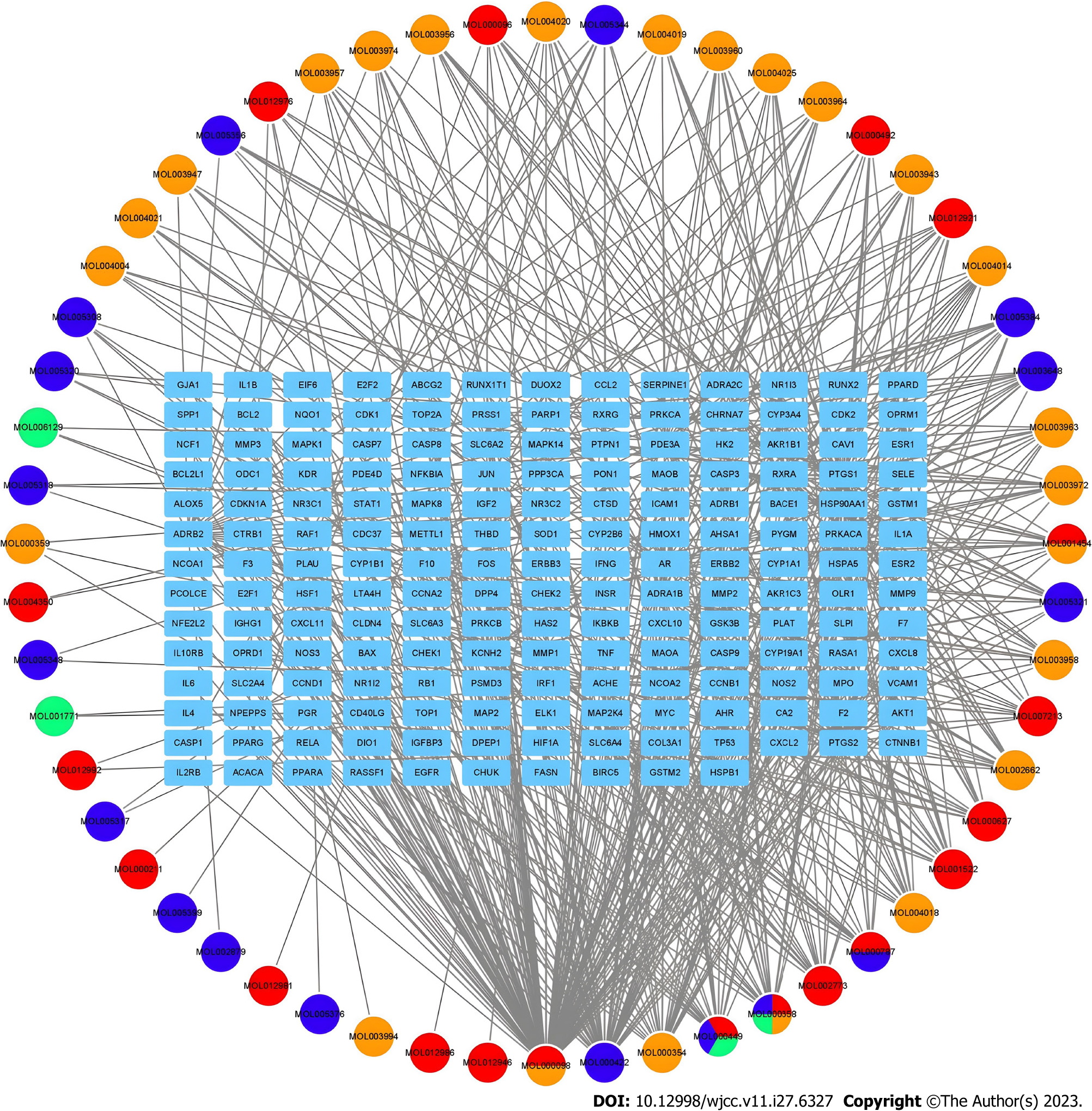
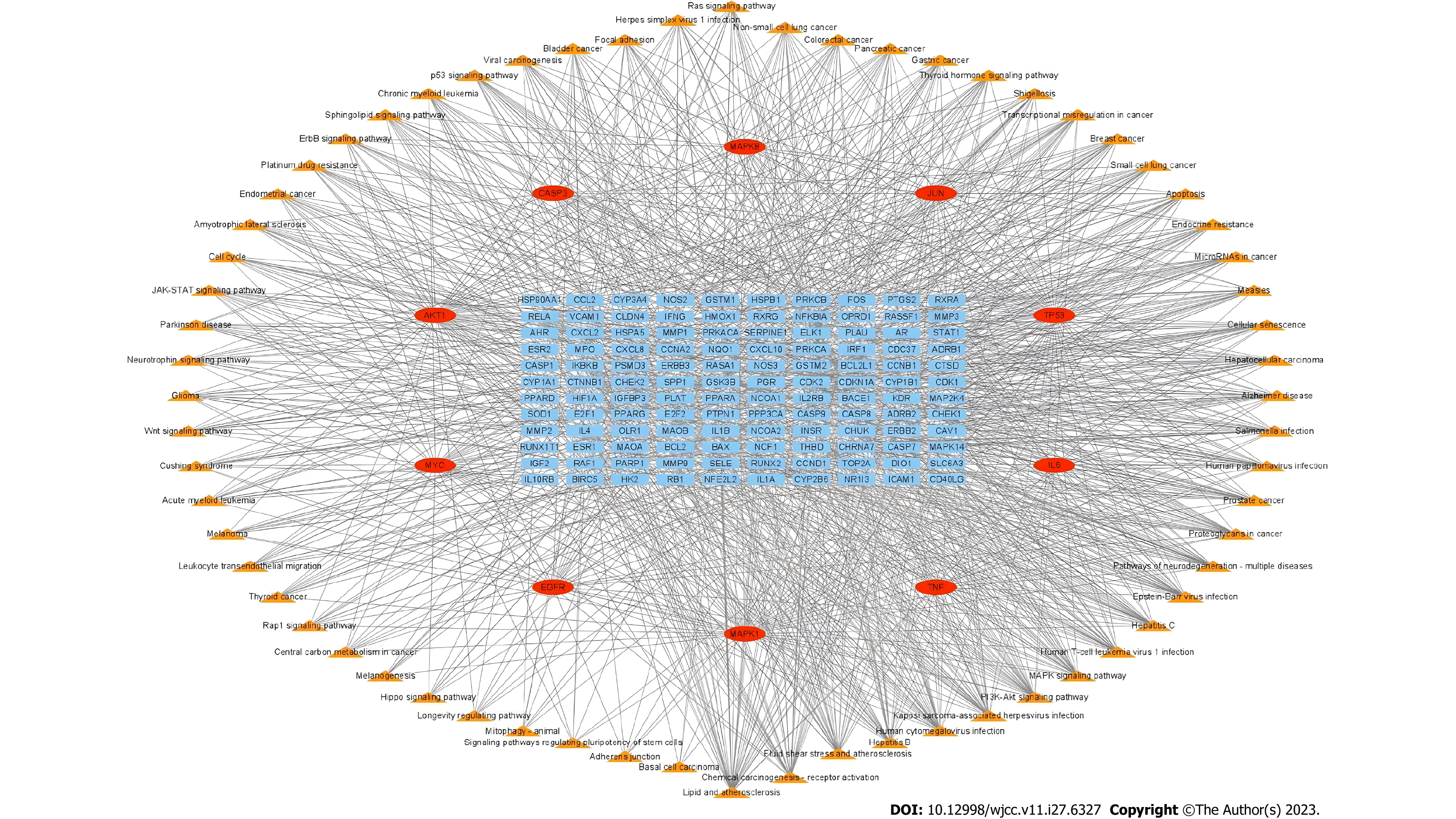
Traditional Chinese medicine prescriptions with multiple targets have shown various pharmacological activities. Studying the possible mechanisms of traditional Chinese medicine prescriptions is highly significant in the treatment of diseases through network analysis. Figure 3 shows the Wuzhuyu decoction active compounds and target genes network of Wuzhuyu decoction treating in HCC, which was structured using Cytoscape software. It can visually show the association between active compounds and target genes.
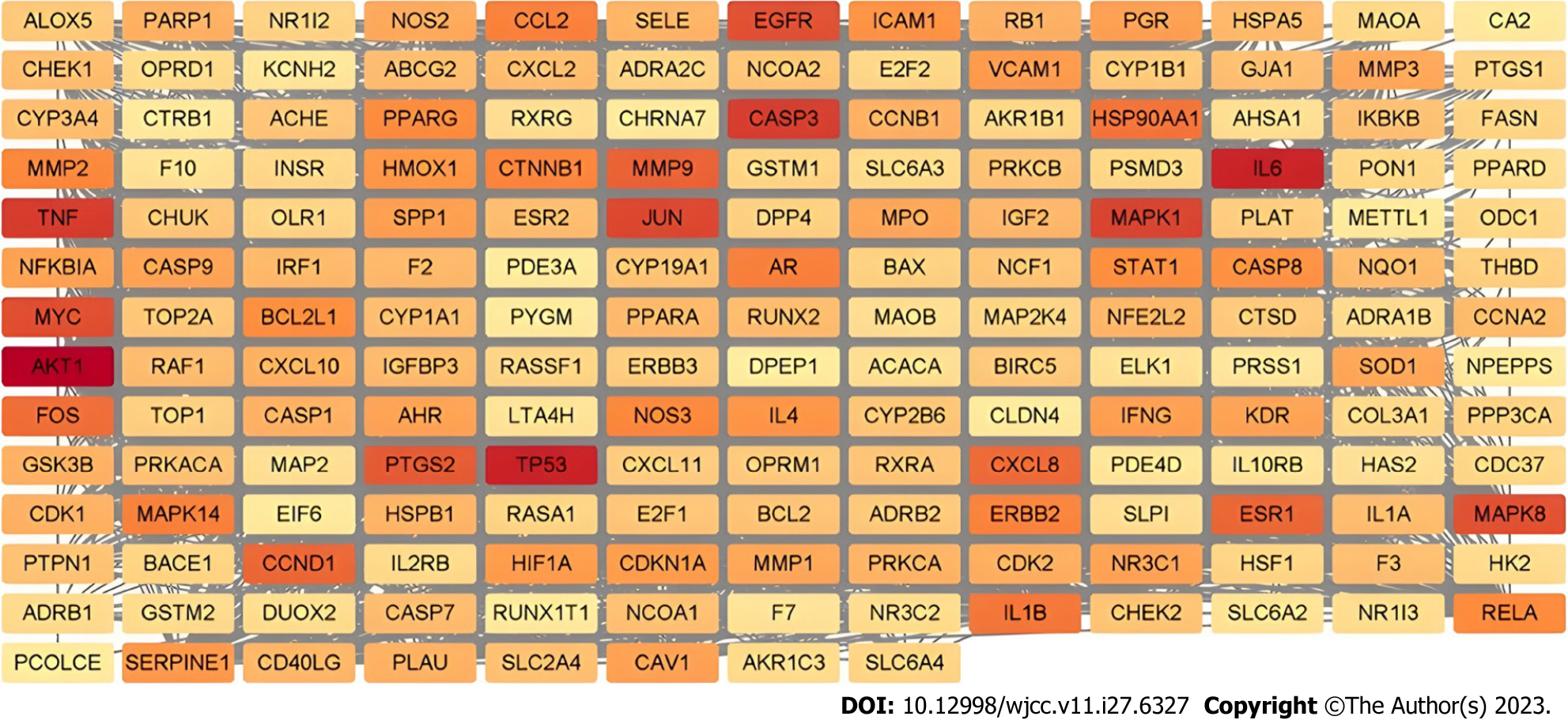
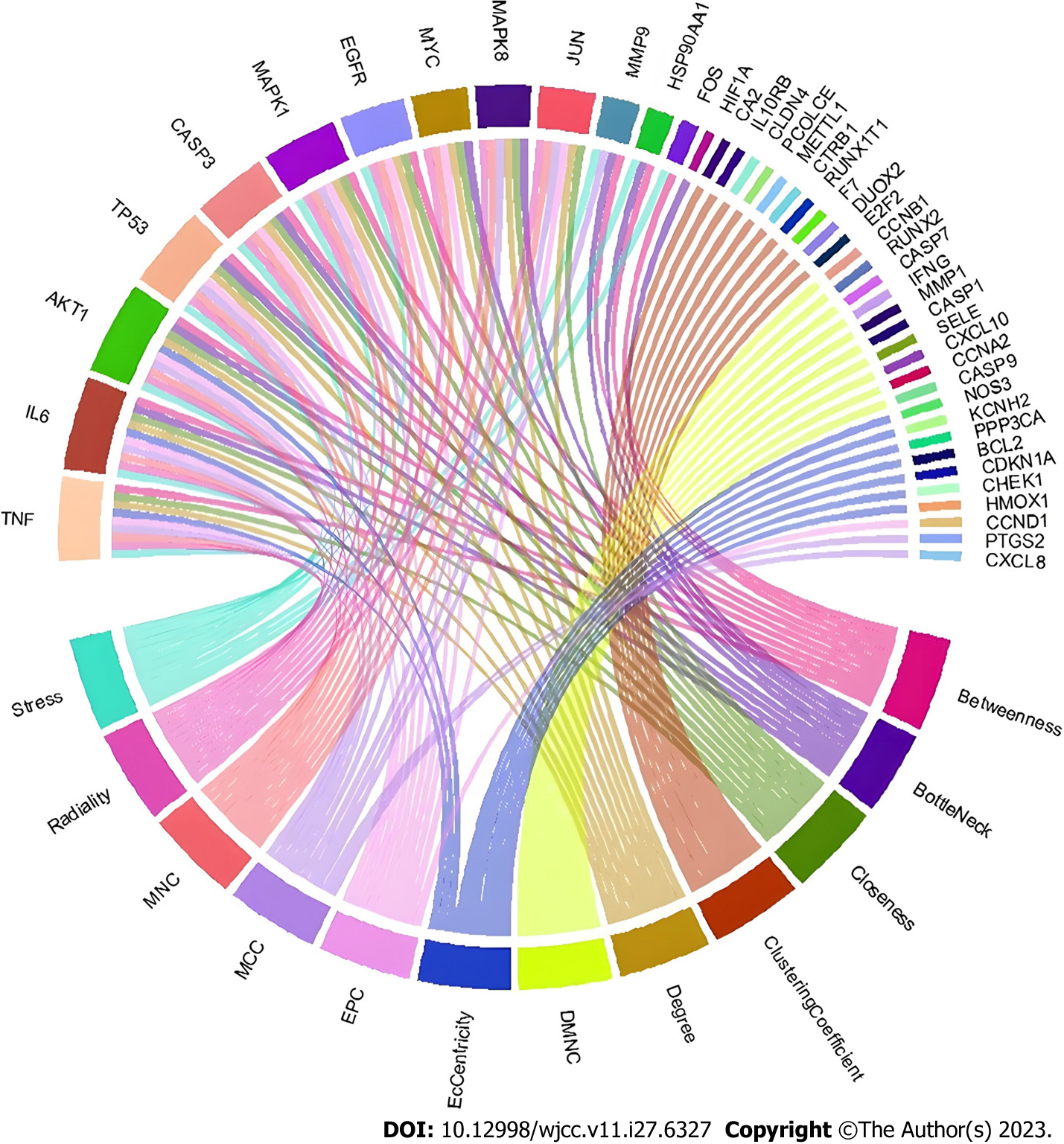
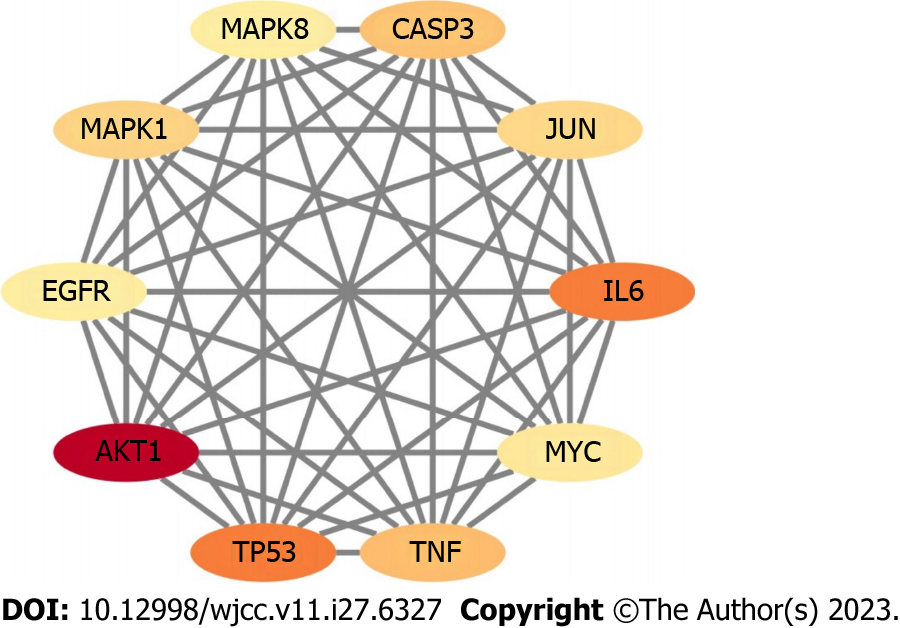
The PPI network can be used to identify potential targets of Wuzhuyu decoction in HCC treatment. Based on the 179 potential target genes identified, the PPI network was obtained using the STRING database, which involved 178 nodes and 1040 edges. The obtained data were further visualized using Cytoscape software (Figure 4). According to the 12 CytoHubba algorithms (see “PPI analysis”) of Cytoscape software, the top 10 hub genes were selected based on each algorithm. According to the results, the top 10 hub genes of the 12 algorithms contained 44 different genes. In addition, the chord diagram (Figure 5) showed the relationship between the 44 genes and the corresponding algorithms. Based on the number of algorithms corresponding to genes, those 44 genes were sorted, and the top 10 hub genes were identified as tumor necrosis factor (TNF), interleukin (IL) 6, AKT1, TP53, caspase-3 (CASP3), mitogen-activated protein kinase 1 (MAPK1), epidermal growth factor receptor (EGFR), MYC, MAPK8, and JUN. These results were close to closeness, degree, Maximum Neighborhood Component, and radiality algorithms, which contain the same genes and are only slightly different in sequence, and are the closest to the radiality result (Figure 6).
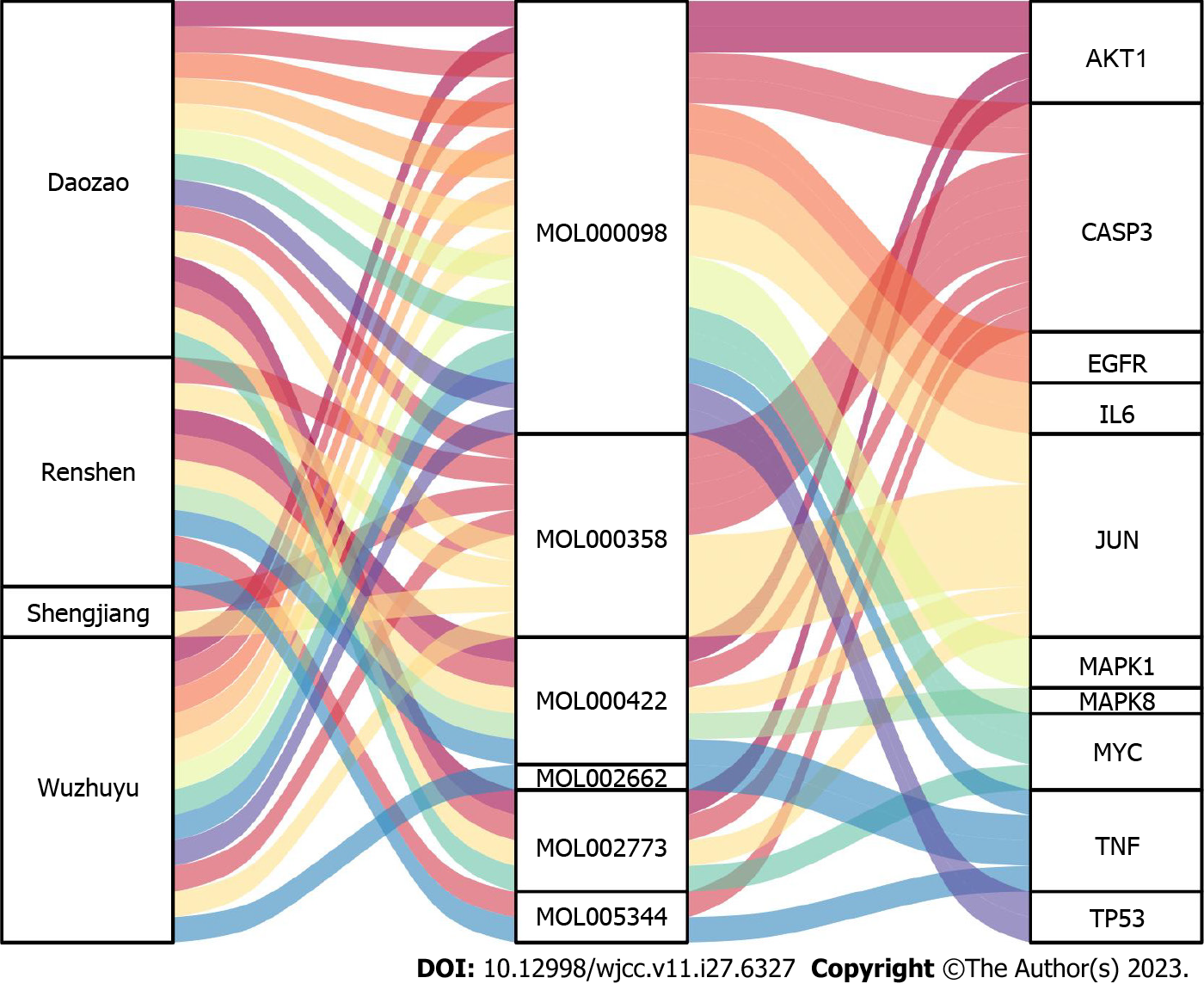
Additionally, a Sankey diagram (Figure 7) was constructed based on the top 10 hub genes, active components, and traditional Chinese medicines of the Wuzhuyu decoction, in which MOL000098 (quercetin) targeted most of the top 10 hub genes. MOL000422 (kaempferol), MOL005344 (ginsenoside Rh2), MOL002662 (rutaecarpine), MOL002773 (β-carotene), and MOL000358 (β-sitosterol) were the main components of the Wuzhuyu decoction against HCC.
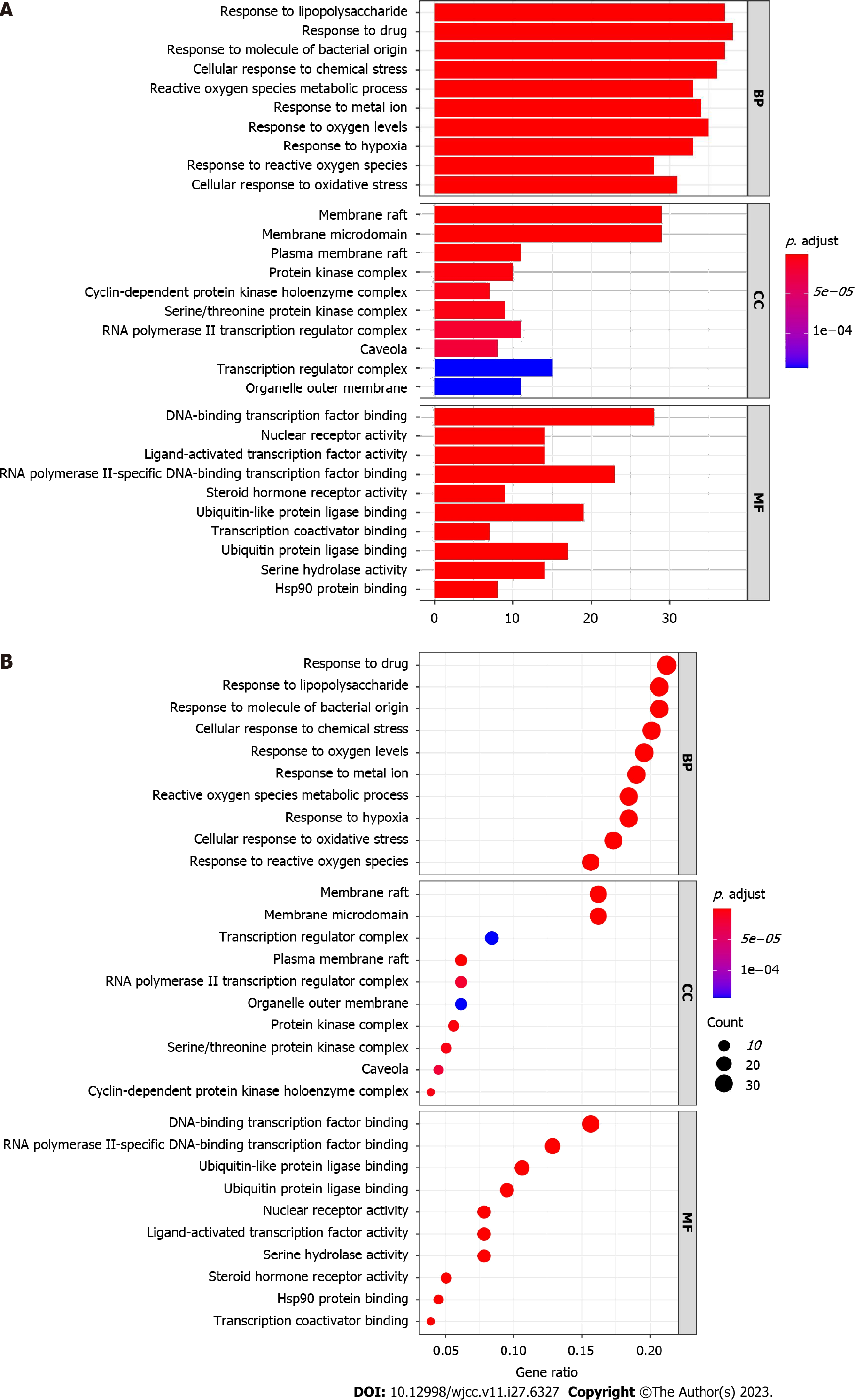
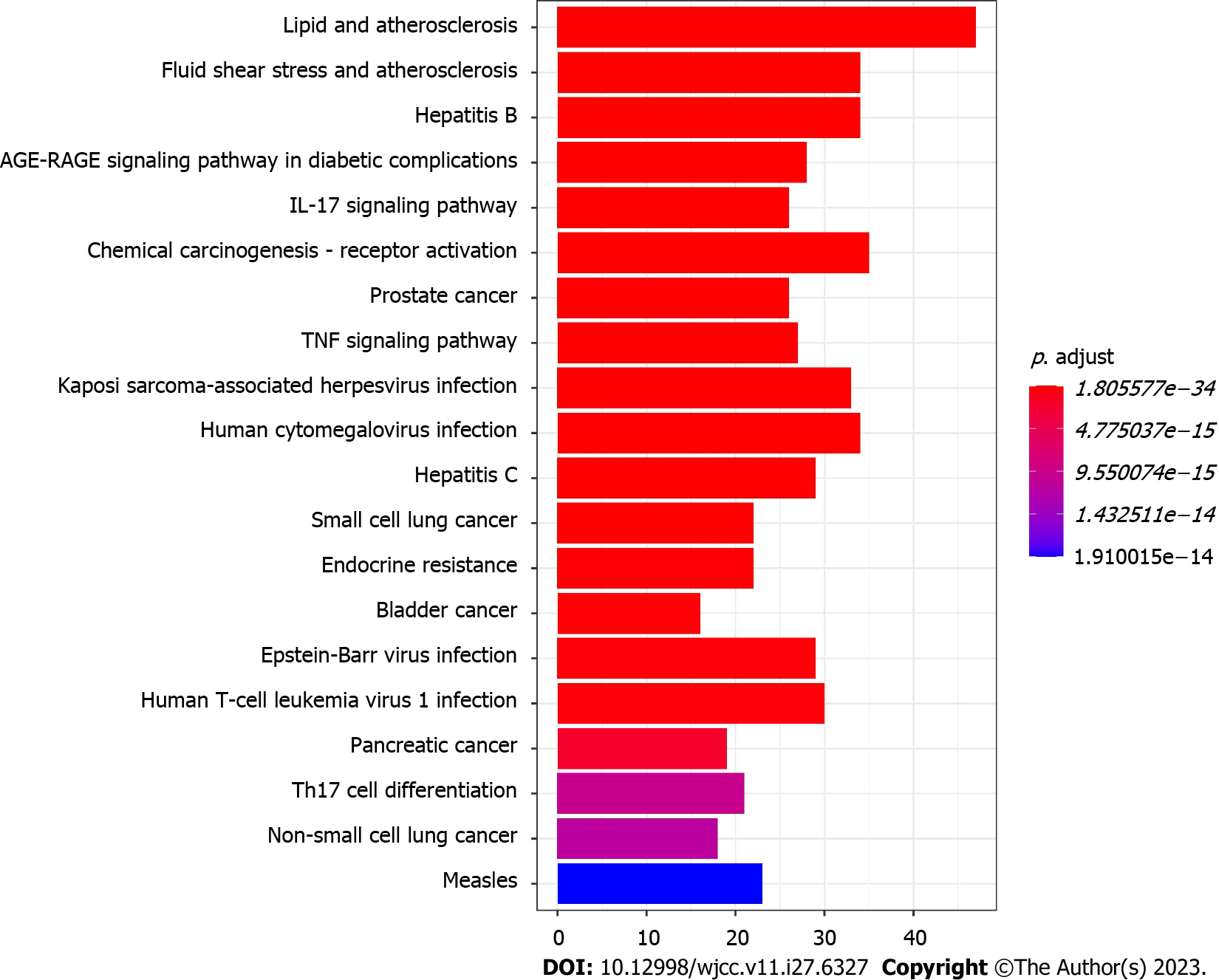
GO and KEGG pathway enrichment analyses were conducted in clusterProfiler of R to elucidate the biological mechanism of Wuzhuyu decoction against HCC. The 179 potential targets were entered into R, and 2513 GO terms were obtained (adjusted, P < 0.05), including 2257 BP, 199 molecular function (MF), and 57 cellular component terms. The top 10 of GO-BP terms, GO-molecular function terms, and GO-cellular component terms (Table 1) were sorted according to the adjusted P values from small to large. The top 10 GO enrichment items were selected for further visualization using bar (Figure 8A) and bubble diagrams (Figure 8B).
| ID | Description | P value | Adjust P value | Gene number | GO items |
| GO:0032496 | Response to lipopolysaccharide | 2.70E-29 | 1.11E-25 | 37 | BP |
| GO:0042493 | Response to drug | 5.99E-29 | 1.23E-25 | 38 | BP |
| GO:0002237 | Response to molecule of bacterial origin | 2.40E-28 | 3.27E-25 | 37 | BP |
| GO:0062197 | Cellular response to chemical stress | 4.08E-27 | 4.17E-24 | 36 | BP |
| GO:0072593 | Reactive oxygen species metabolic process | 1.25E-26 | 1.02E-23 | 33 | BP |
| GO:0010038 | Response to metal ion | 1.37E-24 | 9.36E-22 | 34 | BP |
| GO:0070482 | Response to oxygen levels | 2.05E-24 | 1.20E-21 | 35 | BP |
| GO:0001666 | Response to hypoxia | 1.28E-23 | 6.56E-21 | 33 | BP |
| GO:0000302 | Response to reactive oxygen species | 1.99E-23 | 9.03E-21 | 28 | BP |
| GO:0034599 | Cellular response to oxidative stress | 2.21E-23 | 9.04E-21 | 31 | BP |
| GO:0045121 | Membrane raft | 1.49E-20 | 2.36E-18 | 29 | CC |
| GO:0098857 | Membrane microdomain | 1.49E-20 | 2.36E-18 | 29 | CC |
| GO:0044853 | Plasma membrane raft | 6.06E-09 | 6.38E-07 | 11 | CC |
| GO:1902911 | Protein kinase complex | 4.05E-08 | 3.20E-06 | 10 | CC |
| GO:0000307 | Cyclin-dependent protein kinase holoenzyme complex | 1.18E-07 | 6.68E-06 | 7 | CC |
| GO:1902554 | Serine/threonine protein kinase complex | 1.27E-07 | 6.68E-06 | 9 | CC |
| GO:0090575 | RNA polymerase II transcription regulator complex | 4.95E-07 | 2.24E-05 | 11 | CC |
| GO:0005901 | Caveola | 7.06E-07 | 2.79E-05 | 8 | CC |
| GO:0005667 | Transcription regulator complex | 5.82E-06 | 0.000192173 | 15 | CC |
| GO:0031968 | Organelle outer membrane | 6.14E-06 | 0.000192173 | 11 | CC |
| GO:0140297 | DNA-binding transcription factor binding | 5.06E-17 | 9.61E-15 | 28 | MF |
| GO:0004879 | Nuclear receptor activity | 5.46E-17 | 9.61E-15 | 14 | MF |
| GO:0098531 | Ligand-activated transcription factor activity | 5.46E-17 | 9.61E-15 | 14 | MF |
| GO:0061629 | RNA polymerase II-specific DNA-binding transcription factor binding | 2.32E-15 | 3.06E-13 | 23 | MF |
| GO:0003707 | Steroid hormone receptor activity | 1.78E-12 | 1.88E-10 | 9 | MF |
| GO:0044389 | Ubiquitin-like protein ligase binding | 2.35E-10 | 2.07E-08 | 19 | MF |
| GO:0001223 | Transcription coactivator binding | 4.22E-09 | 2.70E-07 | 7 | MF |
| GO:0031625 | Ubiquitin protein ligase binding | 4.53E-09 | 2.70E-07 | 17 | MF |
| GO:0017171 | Serine hydrolase activity | 4.71E-09 | 2.70E-07 | 14 | MF |
| GO:0051879 | Hsp90 protein binding | 5.12E-09 | 2.70E-07 | 8 | MF |
The 179 potential targets of Wuzhuyu decoction against HCC were put into R, and 177 enriched KEGG pathways were obtained (adjusted, P < 0.05). Based on the adjusted P values ranging from small to large, the top 20 KEGG pathway enrichment terms were selected and further visualized using a bar diagram (Figure 9). “Hepatocellular carcinoma,” as the keyword, was searched in the KEGG database to obtain potential pathways related to HCC. From the 177 results of enriched KEGG pathways, 64 were selected, which were possibly associated with HCC. They included the MAPK, p53, phosphatidylinositol 3-kinase (PI3K)-Akt, Janus kinase (JAK)-signal transducer of activators of transcription (STAT), and Hippo signaling pathways, among others. Details of the 64 pathways can be found in Supplementary Table 3. Finally, a network of target genes and pathways was constructed using the Cytoscape software (Figure 10).
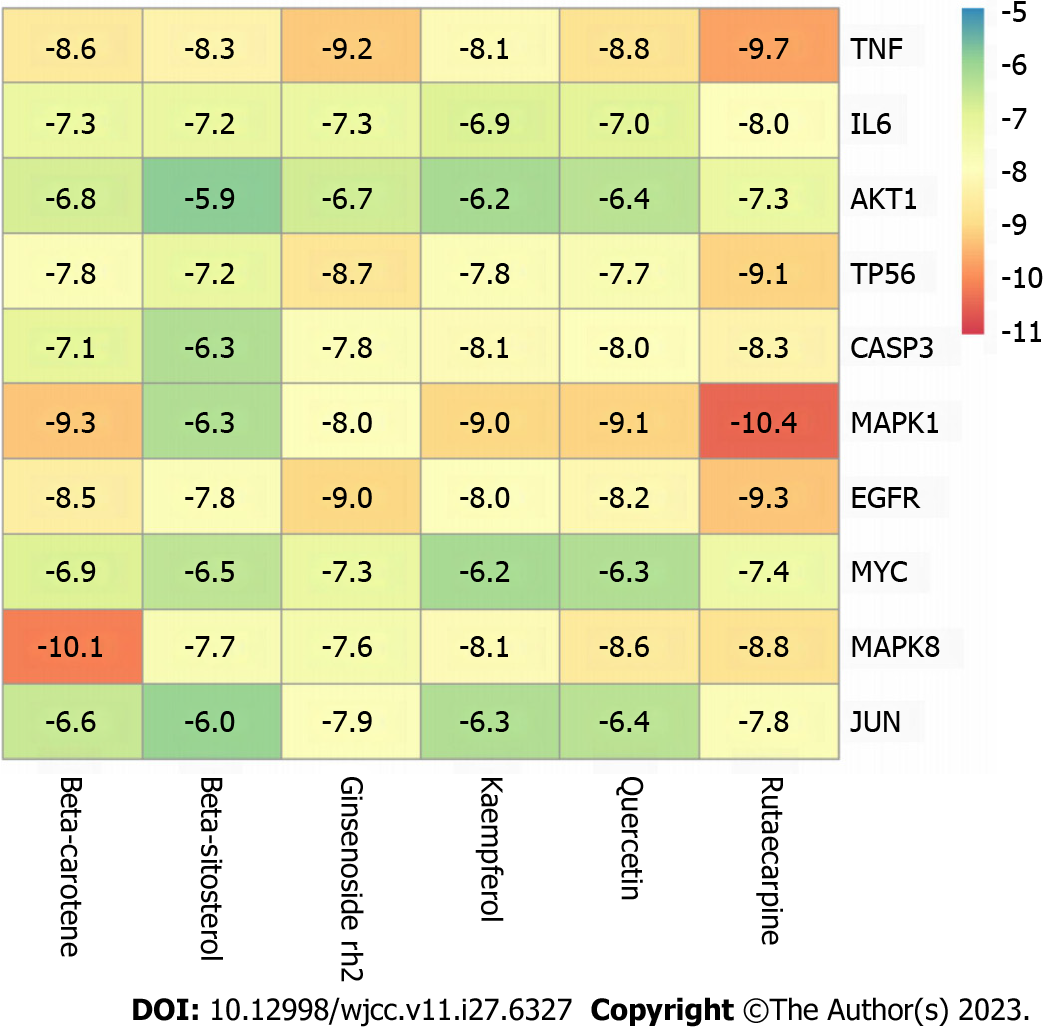
Based on the CytoHubba algorithm analysis results of the Cytoscape software, the top 10 hub genes were TNF, IL6, AKT1, TP53, CASP3, MAPK1, EGFR, MYC, MAPK8, and JUN, which were core target proteins. According to the Sankey diagram (Figure 7), the key active components were MOL000098 (quercetin), MOL000422 (kaempferol), MOL005344 (ginsenoside Rh2), MOL002662 (rutaecarpine), MOL002773 (β-carotene), and MOL000358 (β-sitosterol). Based on these results, molecular docking of the core target proteins and active compounds was performed.
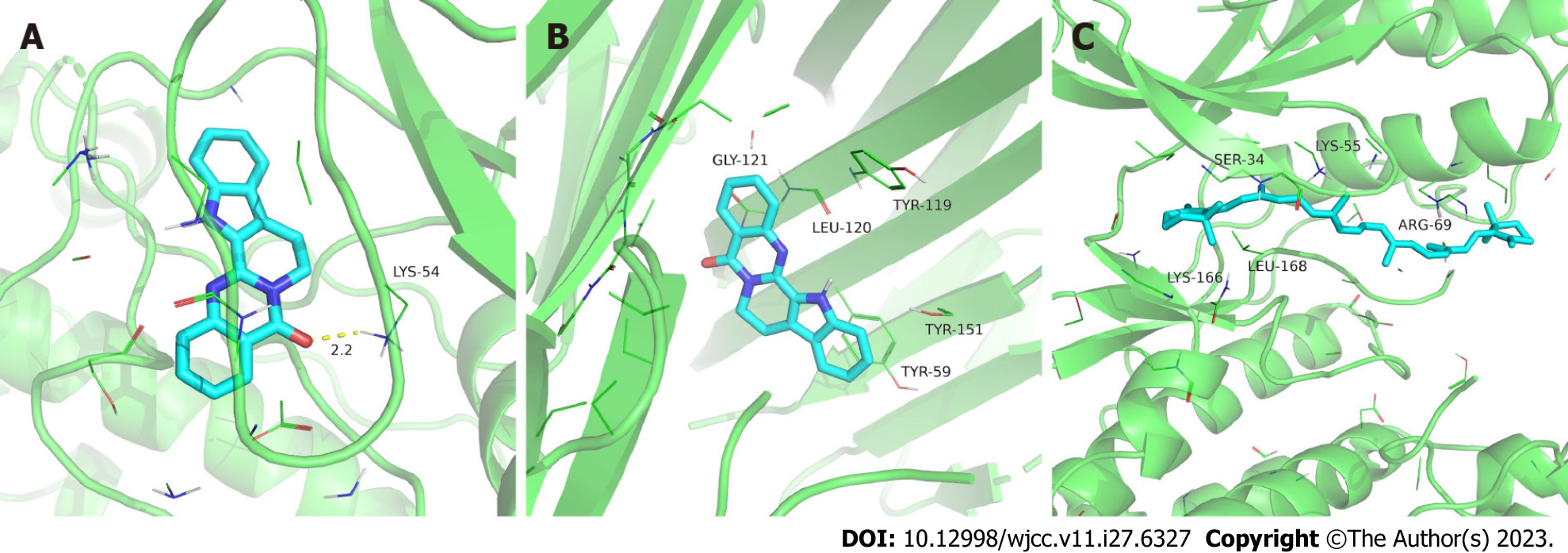
The 10 core target proteins were molecularly docked with the six key active components using AutoDock Vine. A heat map was used to visualize the results of the docking score with the highest affinity, as shown in Figure 11. The binding energies between core target proteins and the key active components were between -5.9 and -10.4 kcal/mol, which indicated that the six key active components of Wuzhuyu decoction all bound well with the 10 core target proteins. In addition, TNF had a high affinity for all key active components. Finally, the top three core target proteins and key active component molecules with the best docking affinities were selected for visualization using PyMOL (Figure 12).
HCC is one of the most common cancers worldwide. As medical treatment and care continue to improve, morbidity and mortality rates for most tumors are declining. However, the mortality rate of HCC continues to increase[34]. The majority of the patients diagnosed with HCC progress to advanced stages of the disease quite soon and become ineligible for curative treatments. However, recurrence rates 5 years after liver resection have been reported to exceed 70%[34]. Presently, drug therapy has become an important method to prolong the overall survival and improve the quality of life of patients.
Traditional Chinese medicine has a long history and is widely used for treating various diseases. However, the choice of adjuvant therapies for HCC remains limited. In the guidelines of the Chinese Society of Clinical Oncology, modern traditional Chinese medicine preparations (Huai Er granules, elemene, compound cantharidin capsule, etc) and traditional Chinese medicine syndrome differentiation treatments have been included in the guidelines and are recommended as level I treatments. Traditional Chinese medicine prescriptions comprise multiple herbs that can act on multiple targets and pathways simultaneously and have a wide range of pharmacological activities. The occurrence and development of HCC are complex processes involving multiple etiologies, genes, and steps, including gene mutations, changes in protein expression, and pathway activation. Traditional Chinese medicine may be helpful in HCC treatment. Studies have revealed that when used as an adjuvant therapy it may not only prolong the overall survival time of HCC patients but also enhance the anti-HCC efficacy of sorafenib by promoting tumor apoptosis and regulating inflammation in the tumor microenvironment[35,36]. Traditional Chinese medicine is characterized by several active ingredients, targets, and complex pathways of action, which make the mechanism of its treatment unclear and difficult to study.
Network pharmacology emphasizes a holistic approach to treatment. It analyzes the relationship between drugs and diseases by predicting potential targets, constructing protein networks, and performing molecular target docking, which is a research concept consistent with the holism of traditional Chinese medicine. Network pharmacology has been widely used to explain the mechanisms underlying Chinese medicine. The specific distribution of the active ingredients in the body is unclear. The results of network pharmacological analysis are predicted based on the network database. The sources of information, theory, and experimental results for each database may be inconsistent, and the analysis results must be verified experimentally.
Nevertheless, network pharmacology breaks the “one disease-one target-one drug” approach of clinicians. It focuses on understanding diseases from the perspective of systems and biological networks, analyzing the molecular association between drugs and their therapeutic targets and revealing the systemic pharmacological mechanisms of drugs. This can guide new drug development and clinical treatment. Network pharmacology provides a new method for scientifically interpreting the principles of traditional Chinese medicine and solves problems through well-informed development.
In the past 3 years, several studies have analyzed the mechanism of action of traditional Chinese medicine through network pharmacology and verified its accuracy through experiments. Network pharmacology technology can elucidate the chemical composition, action targets, pharmacological mechanisms, and formulation laws of traditional Chinese medicine prescriptions, and the recognition and acceptance of traditional Chinese medicine will continue to improve. This study adopted network pharmacology to preliminarily reveal the potential therapeutic mechanism of Wuzhuyu decoction in HCC.
Wuzhuyu decoction has been used to treat digestive diseases. Studies have shown that alkaloids extracted from Evodia have anti-liver cancer activity in vitro and in vivo through targets such as NOD1, AKT, and WWOX[37-39]. This study analyzed the active components of Wuzhuyu decoction and constructed an interaction network diagram of active compounds and target genes. It was observed that the six main active components (quercetin, kaempferol, ginsenoside Rh2, rutaecarpine, β-carotene, and β-sitosterol) may be the main components of Wuzhuyu decoction in adjuvant therapy of HCC.
Quercetin is a flavonoid with antioxidant, anti-inflammatory, and antitumor activities. It has notable antiproliferative and antiapoptotic effects in treating HCC and has a regulatory effect on the cell cycle of tumors[40]. Furthermore, quercetin has been shown to not only regulate apoptosis, migration, and invasion of cells by blocking the JAK2/STAT3 signaling pathway but also inhibit glycolysis by reducing hexokinase 2 and inhibiting the AKT/mammalian target of rapamycin signaling pathway, ultimately hindering the progression of HCC[41,42].
Kaempferol, also a member of the flavonoids, can inhibit both MAPK and hypoxia-inducible factor-1 activity at physiologically relevant concentrations (5-10 mM), reduce liver fibrosis by targeted binding to the ATP-binding pocket of ALK5, and effectively suppress the survival of HCC cells under hypoxia[43,44]. In addition, a natural compound extracted from Abies georgei, which is structurally related to kaempferol, has been shown to enhance the tumor immunosuppressive microenvironment and strengthen the therapeutic effects of low-dose sorafenib without obvious toxicity[45]. It also enhanced the efficacy of sorafenib in HCC in vitro[46].
Recently, ginsenosides have been shown to affect a variety of tumors. Ginsenoside Rh2 has been shown to decrease EZH2, downregulate p14, p15, and p16 mRNA levels, inhibit HCC cell proliferation[47], and inhibit HCC cell growth, b-catenin, and autophagy[48].
β-sitosterol is a phytosterol with antioxidant activity that has ameliorative effects on HCC, and its structural optimization can improve the efficacy of HCC treatment[49]. Rutaecarpine alleviates weight loss, liver injury, and inflammation[50]. Long-term β-carotene treatment can increase blood natural killer, IL2, TNF-α, white blood cells, total protein, albumin, and A/G levels and decrease blood alanine aminotransferase, aspartate aminotransferase, and alkaline phosphatase activities in HCC in rats[51]. Although rutaecarpine and β-carotene have no direct antitumor activity, they can slow the development of tumors, which is beneficial for inhibiting the progression of HCC. These results support the predicted results of network pharmacology. This also proves the feasibility of network pharmacology in identifying and analyzing the mechanisms of action of traditional Chinese medicine. This study preliminarily revealed the potential active substance of Wuzhuyu decoction in treating HCC.
According to the results of comprehensive drug target prediction and pathway analysis, Wuzhuyu decoction may exert an antitumor effect on HCC by inhibiting cell proliferation and angiogenesis and by inducing cell apoptosis. The occurrence and development of HCC are accompanied by the dysregulation of multiple signaling pathways, including cell proliferation, invasion, migration, and angiogenesis. From a treatment perspective, dysregulated signaling pathways are also important targets for treating HCC.
In this study, network pharmacological methods were used to analyze and predict HCC, and the results showed that Wuzhuyu decoction had two main effects on HCC. First, it regulated HCC cell proliferation and apoptosis through the TP53, Ras-MAPK, PI3K-Akt, and JAK-STAT signaling pathways. TP53, a negative regulator of the cell cycle, is one of the most common protein variants expressed in cancer cells and plays important roles in cell cycle regulation, DNA damage repair, cell senescence, and apoptosis[52]. In addition, TP53 has been shown to influence the microenvironment and immune response, which can affect the rate of tumor expansion and aggressiveness[53]. The Ras-MAPK and PI3K-Akt signaling pathways are classical tumor signaling pathways that play crucial roles in regulating the proliferation, apoptosis, and metastasis of tumor cells. They are often considered key therapeutic targets for HCC, and many related small-molecule targeted inhibitors have entered clinical trials for HCC[54,55].
Second, Wuzhuyu decoction regulated HCC angiogenesis through the Hippo signaling pathway. Increasing evidence suggests that the Hippo signaling pathway plays a role in regulating vascular germination, vascular barrier formation, and vascular remodeling[56]. In addition, AKT has biological functions such as regulating cell survival, proliferation, and metabolism. Studies have revealed that AKT not only participates in the migration and invasion of cancer cells but also promotes tumor angiogenesis and enhances vascular permeability[57]. Finally, the KEGG pathway analysis results also included signaling pathways related to lipids and atherosclerosis, hepatitis B, hepatitis C, and the AGE-RAGE signaling pathway in diabetic complications, which were related to the risk factors for HCC.
The results of this study indicate that Wuzhuyu decoction may inhibit HCC cell proliferation and angiogenesis by targeting P53, Ras-MAPK, PI3K-Akt, Hippo, and other signaling pathways via quercetin, kaempferol, ginsenoside Rh2, and other active compounds. However, network pharmacological analysis methods have some limitations. Regarding the detailed pharmacological mechanism of Wuzhuyu decoction in the treatment of HCC, we aim to further conduct accurate target verification and clinical effectiveness verification in subsequent studies.
In this study, network pharmacology was used to integrate potential targets, construct networks, and perform enrichment analysis. It was preliminarily predicted that quercetin, kaempferol, ginsenoside Rh2, rutaecarpine, β-carotene, and β-sitosterol were identified as key components of Wuzhuyu decoction in the treatment of HCC. TNF, IL6, AKT1, TP53, CASP3, MAPK1, EGFR, MYC, MAPK8, and JUN may be important target genes in the pharmacological network. Furthermore, P53, Ras-MAPK, PI3K-Akt and Hippo signaling pathways may play an important role in the treatment of HCC by Wuzhuyu decoction. These results provide preliminary information and a basis for further exploration of the potential mechanism of action of Wuzhuyu decoction in HCC treatment.
The occurrence and development of hepatocellular carcinoma (HCC) are complex processes involving multiple etiologies, genes, and steps, including gene mutations, changes in protein expression, and pathway activation. Traditional Chinese medicine prescriptions comprise multiple herbs that can act on multiple targets and pathways simultaneously and have a wide range of pharmacological activities. In the guidelines of the Chinese Society of Clinical Oncology, modern traditional Chinese medicine preparations and traditional Chinese medicine syndrome differentiation treatments have been included in the guidelines. Wuzhuyu decoction, as a warming interior formula, can warm the middle jiao (stomach, liver, spleen, and gallbladder). Recent studies have reported that it also exhibits anti-inflammatory and antitumor activities, induces cell cycle arrest, and suppresses tumor cell migration.
This study utilized network pharmacology to explore the mechanism of Wuzhuyu decoction against HCC and reveal active compounds, key genes, and potential pathways of Wuzhuyu decoction treatment of HCC, which would be useful for further investigation.
This study aimed to explore the potential mechanism of action of Wuzhuyu decoction against HCC.
This study utilized network pharmacology to explore the mechanism of Wuzhuyu decoction against HCC. Network pharmacology emphasizes a holistic approach to treatment. It analyzes the relationship between drugs and diseases by predicting potential targets, constructing protein networks, and performing molecular target docking, which is a research concept consistent with the holism of traditional Chinese medicine. Network pharmacology has been widely used to explain the mechanisms underlying Chinese medicine.
This study analyzed the active components of Wuzhuyu decoction and constructed an interaction network diagram of active compounds and target genes. According to the analysis results, Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses were conducted. The results of this study indicate that quercetin, kaempferol, ginsenoside Rh2, and other active compounds in the Wuzhuyu decoction may be involved in the regulation of P53, Ras-MAPK, phosphatidylinositol 3-kinase (PI3K)-Akt, Hippo, and other signaling pathways by acting on the first 10 hub genes to achieve therapeutic effects on HCC.
In this study, network pharmacology was used to integrate potential targets, construct networks, and perform enrichment analysis. Wuzhuyu decoction may inhibit HCC cell proliferation and angiogenesis by targeting P53, Ras-MAPK, PI3K-Akt, Hippo, and other signaling pathways via quercetin, kaempferol, ginsenoside Rh2, and other active compounds.
Regarding the detailed pharmacological mechanism of Wuzhuyu decoction in the treatment of HCC, we will conduct accurate target verification and clinical effectiveness verification in subsequent studies.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Emran TB, Bangladesh; Tanabe S, Japan S-Editor: Wang JJ L-Editor: Filipodia P-Editor: Yuan YY
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55701] [Article Influence: 7957.3] [Reference Citation Analysis (132)] |
| 2. | Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer. 2013;13:123-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 581] [Cited by in RCA: 632] [Article Influence: 52.7] [Reference Citation Analysis (0)] |
| 3. | Ganne-Carrié N, Nahon P. Hepatocellular carcinoma in the setting of alcohol-related liver disease. J Hepatol. 2019;70:284-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 250] [Article Influence: 41.7] [Reference Citation Analysis (0)] |
| 4. | Zhang W, He H, Zang M, Wu Q, Zhao H, Lu LL, Ma P, Zheng H, Wang N, Zhang Y, He S, Chen X, Wu Z, Wang X, Cai J, Liu Z, Sun Z, Zeng YX, Qu C, Jiao Y. Genetic Features of Aflatoxin-Associated Hepatocellular Carcinoma. Gastroenterology. 2017;153:249-262.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 99] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 5. | Couri T, Pillai A. Goals and targets for personalized therapy for HCC. Hepatol Int. 2019;13:125-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 375] [Article Influence: 62.5] [Reference Citation Analysis (0)] |
| 6. | Al-Salama ZT, Syed YY, Scott LJ. Lenvatinib: A Review in Hepatocellular Carcinoma. Drugs. 2019;79:665-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 198] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 7. | Xiang Y, Guo Z, Zhu P, Chen J, Huang Y. Traditional Chinese medicine as a cancer treatment: Modern perspectives of ancient but advanced science. Cancer Med. 2019;8:1958-1975. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 556] [Cited by in RCA: 493] [Article Influence: 82.2] [Reference Citation Analysis (1)] |
| 8. | Li M, Wang C. Traditional uses, phytochemistry, pharmacology, pharmacokinetics and toxicology of the fruit of Tetradium ruticarpum: A review. J Ethnopharmacol. 2020;263:113231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 9. | Liao JF, Chiou WF, Shen YC, Wang GJ, Chen CF. Anti-inflammatory and anti-infectious effects of Evodia rutaecarpa (Wuzhuyu) and its major bioactive components. Chin Med. 2011;6:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 72] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 10. | Yang X, Zhang Y, Huang Y, Wang Y, Qi X, Su T, Lu L. Evodiamine suppresses Notch3 signaling in lung tumorigenesis via direct binding to γ-secretases. Phytomedicine. 2020;68:153176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 11. | Cui G, Zhang W, Wang Q, Zhang A, Mu H, Bai H, Duan J. Extraction optimization, characterization and immunity activity of polysaccharides from Fructus Jujubae. Carbohydr Polym. 2014;111:245-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 12. | Ratan ZA, Youn SH, Kwak YS, Han CK, Haidere MF, Kim JK, Min H, Jung YJ, Hosseinzadeh H, Hyun SH, Cho JY. Adaptogenic effects of Panax ginseng on modulation of immune functions. J Ginseng Res. 2021;45:32-40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 65] [Article Influence: 16.3] [Reference Citation Analysis (1)] |
| 13. | Li X, Chu S, Lin M, Gao Y, Liu Y, Yang S, Zhou X, Zhang Y, Hu Y, Wang H, Chen N. Anticancer property of ginsenoside Rh2 from ginseng. Eur J Med Chem. 2020;203:112627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 122] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 14. | Mao QQ, Xu XY, Cao SY, Gan RY, Corke H, Beta T, Li HB. Bioactive Compounds and Bioactivities of Ginger (Zingiber officinale Roscoe). Foods. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 446] [Cited by in RCA: 489] [Article Influence: 81.5] [Reference Citation Analysis (0)] |
| 15. | Pereira MM, Haniadka R, Chacko PP, Palatty PL, Baliga MS. Zingiber officinale Roscoe (ginger) as an adjuvant in cancer treatment: a review. J BUON. 2011;16:414-424. [PubMed] |
| 16. | Li S, Zhang B. Traditional Chinese medicine network pharmacology: theory, methodology and application. Chin J Nat Med. 2013;11:110-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 625] [Article Influence: 56.8] [Reference Citation Analysis (0)] |
| 17. | Luo TT, Lu Y, Yan SK, Xiao X, Rong XL, Guo J. Network Pharmacology in Research of Chinese Medicine Formula: Methodology, Application and Prospective. Chin J Integr Med. 2020;26:72-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 438] [Article Influence: 73.0] [Reference Citation Analysis (0)] |
| 18. | Ru J, Li P, Wang J, Zhou W, Li B, Huang C, Guo Z, Tao W, Yang Y, Xu X, Li Y, Wang Y, Yang L. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1623] [Cited by in RCA: 3120] [Article Influence: 283.6] [Reference Citation Analysis (0)] |
| 19. | An L, Lin Y, Li L, Kong M, Lou Y, Wu J, Liu Z. Integrating Network Pharmacology and Experimental Validation to Investigate the Effects and Mechanism of Astragalus Flavonoids Against Hepatic Fibrosis. Front Pharmacol. 2020;11:618262. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 20. | UniProt Consortium T. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2018;46:2699. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1090] [Cited by in RCA: 1251] [Article Influence: 178.7] [Reference Citation Analysis (0)] |
| 21. | Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, Kaplan S, Dahary D, Warshawsky D, Guan-Golan Y, Kohn A, Rappaport N, Safran M, Lancet D. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics. 2016;54:1.30.1-1.30.33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1289] [Cited by in RCA: 2851] [Article Influence: 316.8] [Reference Citation Analysis (0)] |
| 22. | Amberger JS, Hamosh A. Searching Online Mendelian Inheritance in Man (OMIM): A Knowledgebase of Human Genes and Genetic Phenotypes. Curr Protoc Bioinformatics. 2017;58:1.2.1-1.2.12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 404] [Article Influence: 50.5] [Reference Citation Analysis (0)] |
| 23. | Rappaport N, Twik M, Plaschkes I, Nudel R, Iny Stein T, Levitt J, Gershoni M, Morrey CP, Safran M, Lancet D. MalaCards: an amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 2017;45:D877-D887. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 255] [Cited by in RCA: 410] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 24. | Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, Ronzano F, Centeno E, Sanz F, Furlong LI. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48:D845-D855. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 328] [Cited by in RCA: 1063] [Article Influence: 212.6] [Reference Citation Analysis (0)] |
| 25. | Wang Y, Zhang S, Li F, Zhou Y, Zhang Y, Wang Z, Zhang R, Zhu J, Ren Y, Tan Y, Qin C, Li Y, Li X, Chen Y, Zhu F. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020;48:D1031-D1041. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 257] [Cited by in RCA: 404] [Article Influence: 80.8] [Reference Citation Analysis (0)] |
| 26. | Davis AP, Grondin CJ, Johnson RJ, Sciaky D, Wiegers J, Wiegers TC, Mattingly CJ. Comparative Toxicogenomics Database (CTD): update 2021. Nucleic Acids Res. 2021;49:D1138-D1143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 619] [Cited by in RCA: 679] [Article Influence: 169.8] [Reference Citation Analysis (0)] |
| 27. | Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498-2504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24663] [Cited by in RCA: 33302] [Article Influence: 1585.8] [Reference Citation Analysis (0)] |
| 28. | Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607-D613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10161] [Cited by in RCA: 11669] [Article Influence: 1944.8] [Reference Citation Analysis (1)] |
| 29. | Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362-D368. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4345] [Cited by in RCA: 5062] [Article Influence: 562.4] [Reference Citation Analysis (0)] |
| 30. | Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8 Suppl 4:S11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1658] [Cited by in RCA: 3720] [Article Influence: 338.2] [Reference Citation Analysis (0)] |
| 31. | Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049-D1056. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2016] [Cited by in RCA: 2399] [Article Influence: 218.1] [Reference Citation Analysis (0)] |
| 32. | Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480-D484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3661] [Cited by in RCA: 4493] [Article Influence: 249.6] [Reference Citation Analysis (0)] |
| 33. | Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455-461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26714] [Cited by in RCA: 14362] [Article Influence: 957.5] [Reference Citation Analysis (0)] |
| 34. | Benson AB, D'Angelica MI, Abbott DE, Anaya DA, Anders R, Are C, Bachini M, Borad M, Brown D, Burgoyne A, Chahal P, Chang DT, Cloyd J, Covey AM, Glazer ES, Goyal L, Hawkins WG, Iyer R, Jacob R, Kelley RK, Kim R, Levine M, Palta M, Park JO, Raman S, Reddy S, Sahai V, Schefter T, Singh G, Stein S, Vauthey JN, Venook AP, Yopp A, McMillian NR, Hochstetler C, Darlow SD. Hepatobiliary Cancers, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2021;19:541-565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 354] [Cited by in RCA: 564] [Article Influence: 141.0] [Reference Citation Analysis (0)] |
| 35. | Liu X, Li M, Wang X, Dang Z, Yu L, Jiang Y, Yang Z. Effects of adjuvant traditional Chinese medicine therapy on long-term survival in patients with hepatocellular carcinoma. Phytomedicine. 2019;62:152930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 133] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 36. | Luo XY, Wu KM, He XX. Advances in drug development for hepatocellular carcinoma: clinical trials and potential therapeutic targets. J Exp Clin Cancer Res. 2021;40:172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 140] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 37. | Guo XX, Li XP, Zhou P, Li DY, Lyu XT, Chen Y, Lyu YW, Tian K, Yuan DZ, Ran JH, Chen DL, Jiang R, Li J. Evodiamine Induces Apoptosis in SMMC-7721 and HepG2 Cells by Suppressing NOD1 Signal Pathway. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 38. | Yang F, Shi L, Liang T, Ji L, Zhang G, Shen Y, Zhu F, Xu L. Anti-tumor effect of evodiamine by inducing Akt-mediated apoptosis in hepatocellular carcinoma. Biochem Biophys Res Commun. 2017;485:54-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 52] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 39. | Hu CY, Wu HT, Su YC, Lin CH, Chang CJ, Wu CL. Evodiamine Exerts an Anti-Hepatocellular Carcinoma Activity through a WWOX-Dependent Pathway. Molecules. 2017;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 40. | Fernández-Palanca P, Fondevila F, Méndez-Blanco C, Tuñón MJ, González-Gallego J, Mauriz JL. Antitumor Effects of Quercetin in Hepatocarcinoma In Vitro and In Vivo Models: A Systematic Review. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 41. | Wu L, Li J, Liu T, Li S, Feng J, Yu Q, Zhang J, Chen J, Zhou Y, Ji J, Chen K, Mao Y, Wang F, Dai W, Fan X, Wu J, Guo C. Quercetin shows anti-tumor effect in hepatocellular carcinoma LM3 cells by abrogating JAK2/STAT3 signaling pathway. Cancer Med. 2019;8:4806-4820. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 112] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 42. | Wu H, Pan L, Gao C, Xu H, Li Y, Zhang L, Ma L, Meng L, Sun X, Qin H. Quercetin Inhibits the Proliferation of Glycolysis-Addicted HCC Cells by Reducing Hexokinase 2 and Akt-mTOR Pathway. Molecules. 2019;24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 78] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 43. | Xu T, Huang S, Huang Q, Ming Z, Wang M, Li R, Zhao Y. Kaempferol attenuates liver fibrosis by inhibiting activin receptor-like kinase 5. J Cell Mol Med. 2019;23:6403-6410. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 44. | Mylonis I, Lakka A, Tsakalof A, Simos G. The dietary flavonoid kaempferol effectively inhibits HIF-1 activity and hepatoma cancer cell viability under hypoxic conditions. Biochem Biophys Res Commun. 2010;398:74-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 82] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 45. | Yao W, Ba Q, Li X, Li H, Zhang S, Yuan Y, Wang F, Duan X, Li J, Zhang W, Wang H. A Natural CCR2 Antagonist Relieves Tumor-associated Macrophage-mediated Immunosuppression to Produce a Therapeutic Effect for Liver Cancer. EBioMedicine. 2017;22:58-67. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 130] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 46. | Nair B, Anto RJ, M S, Nath LR. Kaempferol-Mediated Sensitization Enhances Chemotherapeutic Efficacy of Sorafenib Against Hepatocellular Carcinoma: An In Silico and In Vitro Approach. Adv Pharm Bull. 2020;10:472-476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 47. | Li Q, Li B, Dong C, Wang Y, Li Q. 20(S)-Ginsenoside Rh2 suppresses proliferation and migration of hepatocellular carcinoma cells by targeting EZH2 to regulate CDKN2A-2B gene cluster transcription. Eur J Pharmacol. 2017;815:173-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 48. | Yang Z, Zhao T, Liu H, Zhang L. Ginsenoside Rh2 inhibits hepatocellular carcinoma through β-catenin and autophagy. Sci Rep. 2016;6:19383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 55] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 49. | Nisha R, Kumar P, Gautam AK, Bera H, Bhattacharya B, Parashar P, Saraf SA, Saha S. Assessments of in vitro and in vivo antineoplastic potentials of β-sitosterol-loaded PEGylated niosomes against hepatocellular carcinoma. J Liposome Res. 2021;31:304-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 50. | Li Z, Yang M, Peng Y, Gao M, Yang B. Rutaecarpine ameliorated sepsis-induced peritoneal resident macrophages apoptosis and inflammation responses. Life Sci. 2019;228:11-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 51. | Cui B, Liu S, Wang Q, Lin X. Effect of β-carotene on immunity function and tumour growth in hepatocellular carcinoma rats. Molecules. 2012;17:8595-8603. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 52. | Soussi T, Wiman KG. TP53: an oncogene in disguise. Cell Death Differ. 2015;22:1239-1249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 170] [Cited by in RCA: 226] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 53. | Aubrey BJ, Strasser A, Kelly GL. Tumor-Suppressor Functions of the TP53 Pathway. Cold Spring Harb Perspect Med. 2016;6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 200] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 54. | Wu Y, Zhang Y, Qin X, Geng H, Zuo D, Zhao Q. PI3K/AKT/mTOR pathway-related long non-coding RNAs: roles and mechanisms in hepatocellular carcinoma. Pharmacol Res. 2020;160:105195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 99] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 55. | Delire B, Stärkel P. The Ras/MAPK pathway and hepatocarcinoma: pathogenesis and therapeutic implications. Eur J Clin Invest. 2015;45:609-623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 194] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 56. | Boopathy GTK, Hong W. Role of Hippo Pathway-YAP/TAZ Signaling in Angiogenesis. Front Cell Dev Biol. 2019;7:49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 253] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 57. | Alwhaibi A, Verma A, Adil MS, Somanath PR. The unconventional role of Akt1 in the advanced cancers and in diabetes-promoted carcinogenesis. Pharmacol Res. 2019;145:104270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 60] [Article Influence: 10.0] [Reference Citation Analysis (0)] |