Published online Jul 16, 2023. doi: 10.12998/wjcc.v11.i20.4800
Peer-review started: March 22, 2023
First decision: April 11, 2023
Revised: April 23, 2023
Accepted: May 19, 2023
Article in press: May 19, 2023
Published online: July 16, 2023
Processing time: 101 Days and 18.3 Hours
The prognosis of gastric cancer is extremely poor. Metabolic reprogramming involving lipids has been associated with cancer occurrence and progression.
To illustrate fatty acid metabolic mechanisms in gastric cancer, detect core genes, develop a prognostic model, and provide treatment options.
Raw data from The Cancer Genome Atlas and Gene Expression Omnibus databases were collected and analyzed. Differentially expressed fatty acid metabolism genes were identified and incorporated into a risk model based on least absolute shrinkage and selection operator regression analysis. Then, patients from The Cancer Genome Atlas were assigned to high- and low-risk cohorts according to the mean value of the risk score as the threshold, which was verified in the Gene Expression Omnibus database. Relationships between chemotherapeutic sensitivity and tumor microenvironment features were assessed.
An integrated evaluation was performed in this study. Fatty acid metabolism-related genes were used to construct the risk model. Patients classified into the high-risk cohort were considered to be resistant to chemotherapy based on results of the “pRRophetic” R package. Patients in the high-risk cohort were associated with type I/II interferon activation, increased inflammation level, immune cell infiltration, and tumor immune dysfunction based on the exclusion algorithm, indicating the potential benefit of immunotherapy in these patients.
We constructed a fatty acid-related risk score model to assess the comprehensive fatty acid features in gastric cancer and validated its vital role in prognosis, chemotherapy sensitivity, and immunotherapy.
Core Tip: We established a prognostic risk model using data collected from The Cancer Genome Atlas database, explored the function of the risk model, and identified the relationship between the risk model and clinical features. The findings of our study provide innovative therapeutic options in clinical practice.
- Citation: Fu Y, Wang B, Fu P, Zhang L, Bao Y, Gao ZZ. Delineation of fatty acid metabolism in gastric cancer: Therapeutic implications. World J Clin Cases 2023; 11(20): 4800-4813
- URL: https://www.wjgnet.com/2307-8960/full/v11/i20/4800.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i20.4800
Gastric cancer (GC) is associated with poor overall survival and is one of the most common types of fatal cancer worldwide[1,2]. Regional differences in the incidence of GC have been reported. Globally, the 5-year overall survival rate is reported to be less than 25%, given the late-stage presentation, metastasis, tumor heterogeneity, and the intrinsic therapeutic resistance of GC[3]. Joshi et al[3] investigated GC biology and found that, compared with cases in Western countries, GC cases in eastern Asia were characterized by lower proportions of signet ring cell histology. However, the 5-year overall survival rates remain low. Identifying the novel mechanisms underlying GC is of critical importance to develop treatment strategies that can be used in clinical practice.In general, cancer is characterized by the interactions between solitary metabolic features and the surrounding complex tumor environment[4]. Carbohydrates, lipids, and amino acids are the main nutrients in cancer cells, among which lipids are crucial mediators in energy metabolism and signal transduction. Importantly, fatty acids are the basic components of diverse types of lipids[5,6]. Furthermore, metabolic reprogramming has been identified as a critical hallmark of tumor cell proliferation and differentiation. Ma et al[7] described various transcription factors controlling gene expression programs including fatty acid metabolism (FAM)-related genes. Khan et al[8] confirmed that SIRT6 could regulate fatty acid transport by suppressing PPAR signaling, thereby promoting tumor progression. Luo et al[9] demonstrated that the critical role of fatty acid oxidation in CD8+ T cell memory formation, which is crucial in the tumor microenvironment. Garcia et al[10] found that elevated levels of fatty acid may accelerate cancer progression through endoplasmic reticulum stress by modulating the forkhead box (FOX)O3-FOXM1 axis. In addition, Zhang et al[11] demonstrated that fatty acid-induced CD36 expression promoted the progression of GC by activating the nuclear factor-κB pathway and direct binding of CD36 at S468 and T470. Adenosine triphosphate citrate lyase, acetyl-CoA synthases, acetyl-CoA carboxylase, and fatty acid synthase are crucial enzymes in fatty acid synthesis[12]. Although various molecules have been identified in GC metastasis, uncovering novel links between fatty acids and the tumor environment are necessary for a comprehensive understanding of GC and will contribute to accurate diagnosis, prognosis prediction, and recurrence risk and metastasis assessments in patients with GC. In this study, we constructed a novel prognostic risk model for GC based on FAM-related genes. We examined the mechanisms underlying FAM in GC, the relationship between the risk of GC and tumor microenvironment characteristics, and treatment strategies for GC.
Raw data for GC were downloaded from The Cancer Genome Atlas (TCGA) project (https://portal.gdc.cancer.gov/) and the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). Transcription profiling data from TCGA-stomach adenocarcinoma project were collected in the stomach carcinoma-counts workflow type. Simple nucleotide variation data were also retrieved as masked somatic mutation data from TCGA-GDC portal. Clinical information of patients with GC including age, sex, tumor stage, tumor grade, T/N/M (tumor, node, metastasis) stages, and survival information was also obtained. The microarray data from the GSE84437 dataset in the GEO GPL6947 platform were also acquired. The annotation platform was implemented to convert the Entrez Gene identifiers (IDs) of each case to gene symbols.
Generally, FAM-related genes were acquired, as previously described. Differentially expressed genes (DEGs) in normal and tumor cases were identified using the “limma” statistical package in R (version 4.1.3; R Foundation for Statistical Computing, Vienna, Austria). A false discovery rate (FDR) < 0.05 was considered the cutoff value. Conversion from Entrez Gene IDs to gene symbols was processed using the “org.Hs.eg.db” R package. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were conducted to detect the biological characteristics using the “clusterProfiler” R package. Finally, enrichment analysis results were visualized using the “enrich plot” and “ggplot2” packages. A P value less than 0.05 (P < 0.05) was considered statistically significant.
Clinical samples from TCGA database were defined as the training cohort, and the GSE84437 dataset from the GEO database was defined as the validation cohort. We combined the expression level of FAM-related genes with clinical information using the patient IDs. Genes related to survival were based on DEGs and analyzed using univariable cox regression, in line with the validation cohort. Then, genes with statistically significant values (< 0.05) were selected for further analysis. Prognostic genes were generated through least absolute shrinkage and selection operator (LASSO) cox regression analysis using the “glmnet” package, and a prognostic model was constructed. The risk score was calculated as described previously. All samples were segmented into high-risk and low-risk cohorts based on the median value of the risk scores. Kaplan–Meier analysis and the log-rank test were applied to determine the survival difference between low- and high-risk score groups. Finally, the “survivalROC” package was used to assess the predictive accuracy of the prognostic risk model. All data were verified in the validation group.
The Ggplot2 and limma R packages were utilized to conduct principal component analysis (PCA) of gene expression profiles to construct a risk model from TCGA database. Accordingly, PCA was performed on FAM-related gene expression. Two-dimensional diagrams were then visualized to present the results.In addition, gene set variation analysis (GSVA), a non-parametric method implementing gene sets from the molecular signature (https://www.gsea-msigdb.org/gsea/msigdb) database was applied to identify the biological function and pathways between high- and low-risk groups. An FDR < 0.05 was considered statistically significant.
The expression profiles of DEGs between high- and low-risk models were analyzed. The gene symbols were merged into an online STRING database (https://cn.string-db.org/) to establish the protein-protein interaction (PPI) network with interaction scores > 0.7. The PPI network was then presented using Cytoscape software (version: 3.9.2, The Cytoscape Consortium, San Diego, CA, United States). Next, CytoHubba (version 0.1) was applied to generate hub genes from all the DEGs. Based on the previous genes, GO and KEGG enrichment analyses were used to illustrate the biological functions of the identified hub genes. Finally, the differences in the degree of immune cell infiltration based on the expression of hub genes were determined.
R packages including “pRRophetic” and “rms” were applied to demonstrate the predictive value of the clinical features. The Wilcoxon rank sum and Kruskall–Wallis tests were used to perform group comparisons. Kaplan–Meier analysis was used to assess survival. Univariable and multivariable regression models were applied to identify independent factors related to overall survival in GC. Receiver operating characteristic (ROC) curves were drawn to verify the value of the risk model. All analyses were performed using R (version 4.1.3). Statistical significance was defined as P < 0.05.
We conducted differential expression analysis of FAM-related genes using the limma R package. In total, 113 DEGs with |logFC|= 1.5 and FDR ≤ 0.05 were identified in TCGA cohort, of which, 60 genes were upregulated, and 53 genes were downregulated, as shown in Supplementary Figure 1A. Thereafter, functional analysis was conducted for the DEGs. FAM-related genes were enriched in fatty acid metabolic, acyl-CoA metabolic, and fatty acid biosynthetic processes in GO as well as KEGG analyses (Supplementary Figure 1B-C).
Clinical cases from TCGA database cohort were included in the training set. Univariable cox regression analysis was conducted among the 113 DEGs related to FAM. Overall, statistically significant links to GC prognosis were detected in 17 genes (P < 0.05; Figure 1A). Next, the relationship among the 17 FAM-related genes was calculated using a correlation test (Figure 1B). In total, FAM-related gene mutations occurred in 65 patients, equating to a frequency of 16.5% (Figure 1C). The cyclooxygenase 1 (PTGS1) gene had the highest mutation frequency among patients with GC. Furthermore, a co-occurrence mutation was demonstrated between PTGS1 and other FAM-related genes. LASSO regression analysis was applied to generate the risk score model. Collectively, six genes [monoamine oxidase A, CD36, fatty acid elongase 2, alcohol dehydrogenase 4, gamma-glutamyltransferase 5, ELOVL4] were included in the risk model (Figure 1D-E). The risk score for each patient was calculated based on the model and the patients were categorized into high- and low-risk groups (Figure 1F-G).
We defined TCGA-GC cases as the training cohort and GEO cases as the validation cohort. In total, 339 patients with GC were recruited from TCGA database. Of these, 169 were considered to have high risk and 170 were considered to have low risk using the median risk score value. The relationship between clinical characteristics and risk model score was investigated, including age, sex, tumor grade, and T/M/N stages. No significant differences between groups were detected according to sex, M stage, and tumor grade (Figure 2A-C). Nonetheless, higher risk scores were associated with higher T (tumor invasion) and N (lymphoid metastasis) stages and with older age (Figure 2D-F). Furthermore, patients with higher risk scores had lower survival rates (Figure 2G), which was validated in the validation group based on patient data derived from the GEO database (GSE84437) (Figure 2H), including 231 patients classified as high-risk cohort and 202 patients classified as low-risk cohort. Moreover, time- and variable-dependent ROC analysis was performed to verify the results (Figure 2I-J). Univariable and multivariable regression models were constructed according to age, sex, tumor grade, tumor stage, and risk score. The risk score was an independent prognostic predictor of overall survival (Figure 2K-L).
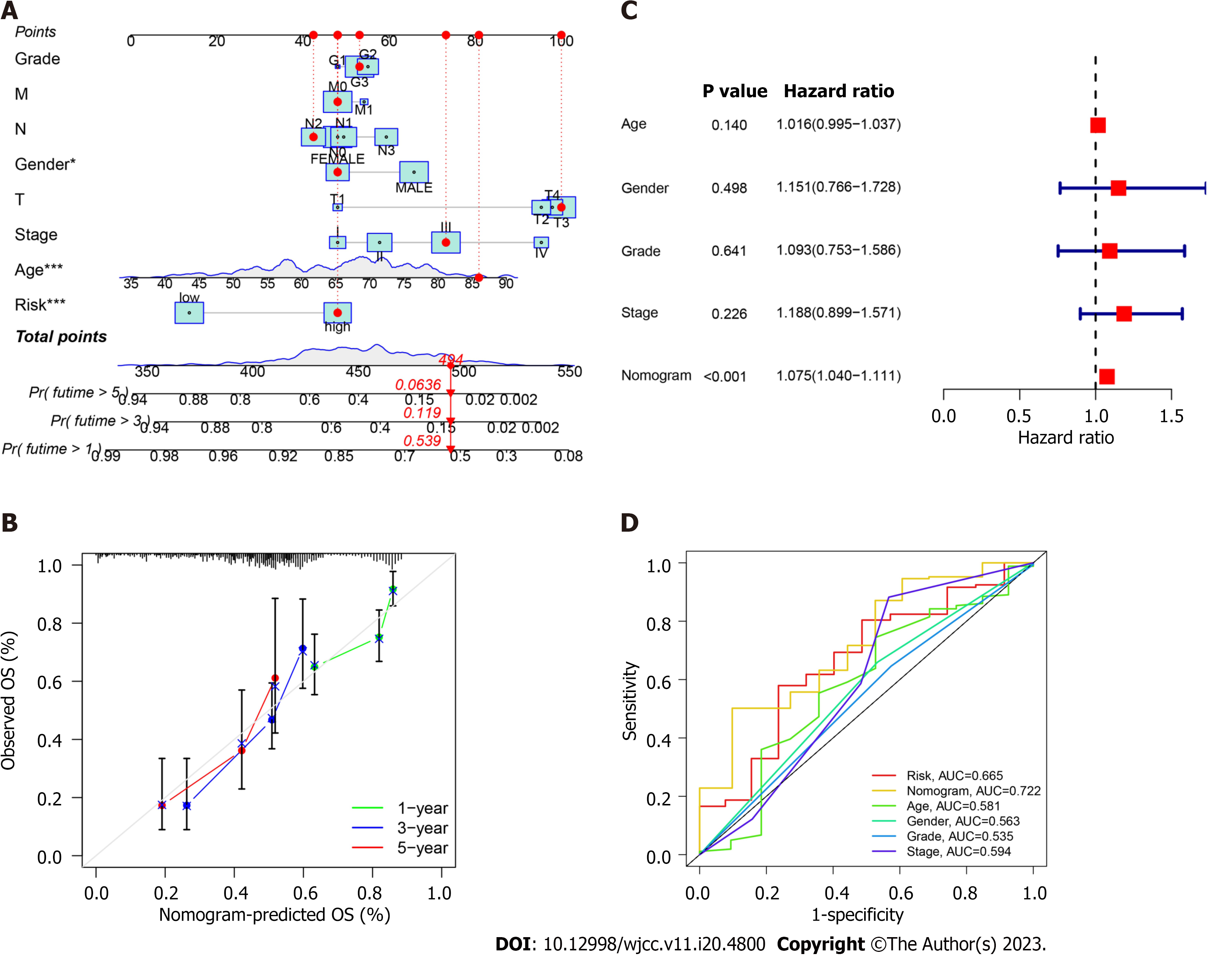
We constructed a nomogram involving tumor grade, T/N/M stages, sex, tumor stage, and risk score to predict overall survival in patients with GC (Figure 3A). The calibration curves at 1-, 3-, and 5 years demonstrated that the nomogram was relevant to overall survival in patients with GC (Figure 3B). Multivariable regression analysis revealed nomogram score as an independent index (Figure 3C), which may be a superior indicator than other clinical features used in clinical practice (Figure 3D).
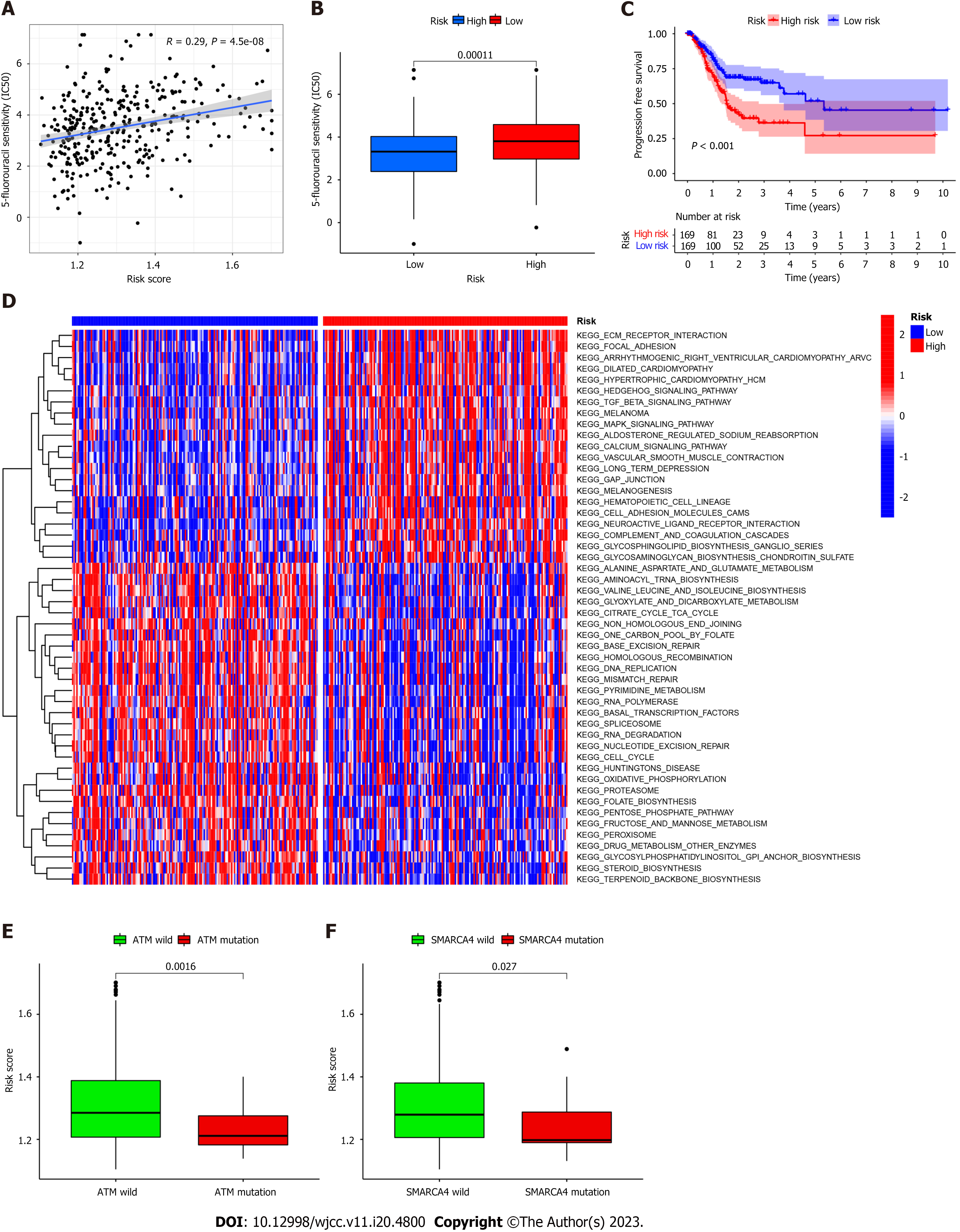
The FAM-related risk score is associated with overall survival in patients with GC; therefore, demonstrating the association between risk score and chemotherapy response is important. The “pRRophetic” R package was used to calculate the half maximal inhibitory concentration, which was applied to evaluate the sensitivity of the risk score and response to 5-Fluorouracil (5-FU). As shown in Figure 4A-B, the high-risk group was more sensitive to 5-FU in TCGA cohort. Furthermore, a negative relationship was noted between risk score and progression-free survival, indicating that the high-risk group may be more sensitive to chemotherapy (Figure 4C). However, its exclusive molecular type and tumor mutation burden facilitated the progression of tumor by activation of focal adhesion pathways, transformation of growth factor-beta signaling, and contraction of vascular smooth muscle (Figure 4D), ultimately shortening survival. These findings are consistent with previously reported results[12,13]. In line with previous studies, GSVA was performed to investigate the biological features between high- and low-risk groups by implementing “c2.cp.kegg.v7.2” in R Studio acquired from the Molecular Signature Database. As shown in Figure 4D, DNA replication, mismatch repair, and steroid biosynthesis were highly enriched in the low-risk group. Some molecules incorporating the transcription activator BRG1 (SMARCA4) and ataxia telangiectasia mutated (ATM) proteins are reportedly correlated with immune activation (Figure 4E-F). Our results indicate that ATM mutation is associated with a positive response to immunotherapy.
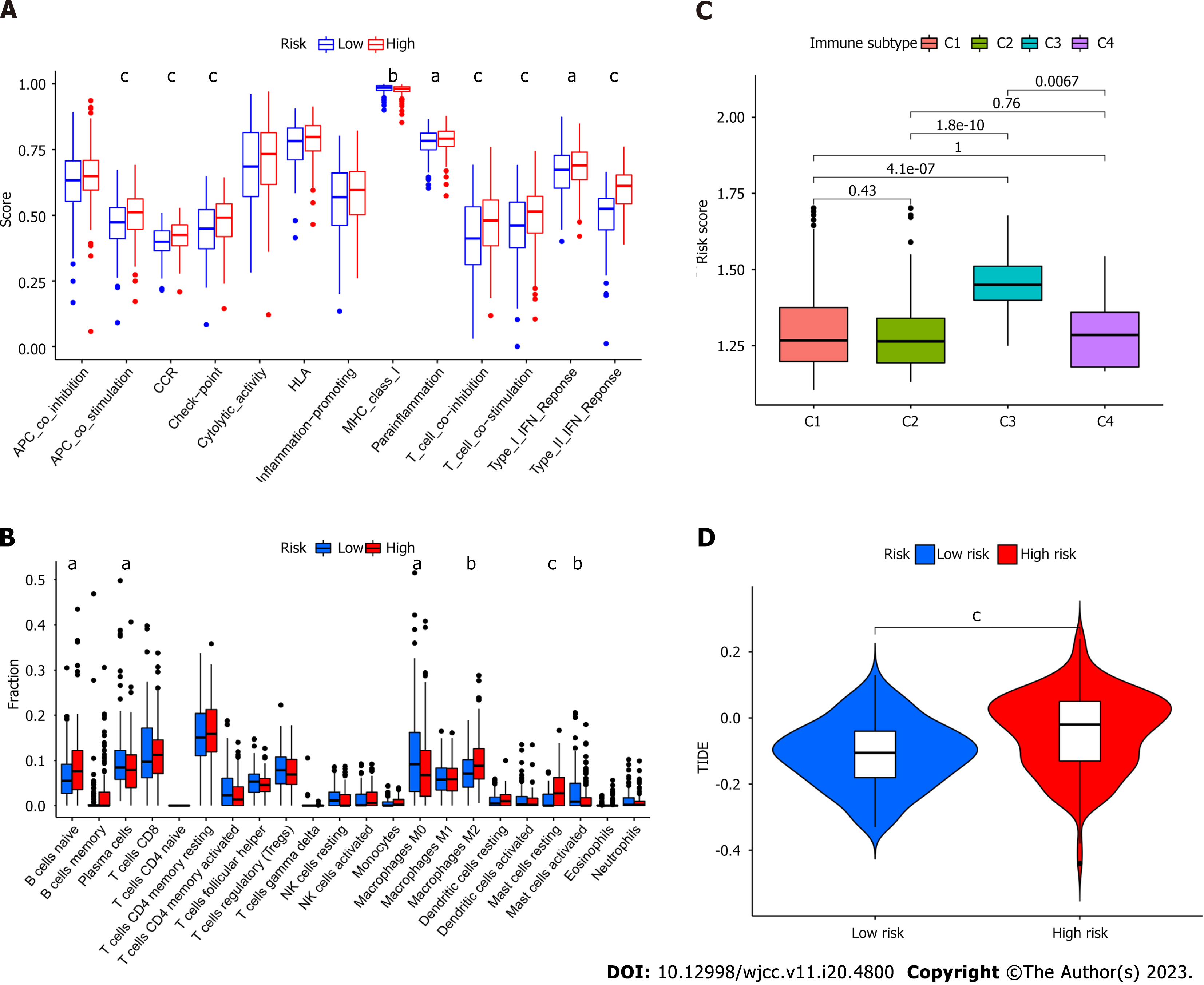
Regarding immune cell infiltration, distinctive enrichment of macrophage M2 was detected in the high-risk group, in line with the previous survival analysis (Figure 5A). Additionally, para-inflammation, T cell co-inhibition, T cell co-stimulation, type I interferon (IFN) response, and type II IFN response were activated in the high-risk group (Figure 5B). Different immune subtypes were identified (Figure 5C) and validated using the TIDE (Tumor Immune Dysfunction and Exclusion) algorithm.
The expression profiles of DEGs were collected and analyzed to establish a PPI network using a STRING online database, which was then visualized using Cytoscape. As shown in Figure 6A, upregulated genes in the high-risk group are labeled in red, and those in the low-risk group are labeled in blue. Thereafter, the CytoHubba plug-in package was applied to generate the hub genes of FAM-related DEGs. In general, 10 genes (integrin beta (ITGB)-3, integrin alpha (ITGA)-1, fibronectin (FN)-1, fibroblast growth factor (FGF)-2, von Willebrand factor (VWF), ITGA5, ITGA4, ITGA8, thrombospondin (THBS)-1, ITGA9) were selected (Figure 6B). Functional enrichment analysis was performed using the “GOplot” R package, and these genes were involved in the integrin-mediated signaling pathway, cell-substrate adhesion, cell-matrix adhesion (Figure 6C), extracellular matrix-receptor, phosphoinositide 3-kinase-Akt signaling pathway, focal adhesion, and regulation of actin cytoskeleton (Figure 6D). Finally, survival and correlation analyses were conducted to determine the key regulators including FGF2 and THBS1, which were verified to be negatively associated with survival, consistent with previous studies (Figure 6E-F).
Cancer proliferation and progression are associated with metabolic reprogramming[14-17]. The metabolism alteration theory was first proposed by Otto Warburg in the 1920s and is known as the “Warburg phenomenon,” characterized by increased glycolysis in the context of an environment lacking oxygen[18,19]. Lactic acids generated from glycolysis favored tumor evasion and metastasis and restrained the antitumor immune effect[7,8,20]. In addition, glucose was converted into nicotinamide adenine dinucleotide phosphate by cancer cells to cope with the complex tumor environment thereby promoting fatty acid synthesis[21-23]. Meanwhile, the medium pyruvate generated from glycolysis was manipulated by the cancer cells to enter a truncated tricarboxylic acid cycle, thereby creating acetyl-CoA that was utilized for long-chain fatty acid synthesis. Collectively, this process highlights the involvement of FAM in cancer development and metastasis.Few studies have identified the role of FAM in GC. Cui et al concluded that abnormal FAM had discernible effects on GC growth[24,25]. Aberrant expression of FAM-related genes was associated with chemotherapeutic drug resistance and recurrence. Sterol O-acyltransferase (SOAT)1 was highly expressed in GC tissues and negatively related to GC prognosis via regulation of sterol regulatory element-binding protein (SREBP)-1 and SREBP2 expression, which propelled lymph-angiogenesis through increased expression of vascular endothelial growth factor C[26]. However, other innovative mechanisms of FAM in GC warrant further investigation. In this study, we explored and elaborated on the relationship between FAM-related genes and GC using TCGA and GEO databases. FAM-related genes incorporated into the present study were obtained as previously described. We conducted a differential analysis to identify DEGs. A total of 113 FAM-related genes were screened to detect genes involved in GC progression, tumor microenvironment involvement, and prognosis. In total, 17 genes were included to establish a predictive/prognostic risk model using cox regression and then verified using LASSO regression analysis. TCGA cohort was considered as the training set and the GEO cohort as the validation set. The overall survival prediction of the risk score (each sample from TCGA cohort had a risk score, which was used to classify patients into high-risk and low-risk cohorts) was constructed in TCGA database and verified in the GEO database. The risk score generated from this model was confirmed as an independent and accurate prognostic index.
Chemotherapeutic sensitivity between high-risk and low-risk groups was analyzed to evaluate the predictive value of the risk model in drugs screen in GC treatment. The increased risk score was positively associated with 5-FU resistance, consistent with previous studies[27]. In parallel, patients in the high-risk score group had reduced survival rates, suggesting that the risk model based on FAM-related genes may be applied to determine the optimal treatment (chemotherapy vs radiation therapy) for patients with GC. Moreover, patients from the high-risk score group were enriched in stroma immune activation, indicating cytol-toxic drug resistance, consistent with previously reported studies[28-30]. Furthermore, high-risk score patients exhibited activation of the type I IFN response, T cell co-inhibition, type II IFN response, and para-inflammation function, demonstrating that patients with high-risk scores may benefit from immunotherapy, on account of the TIDE analysis. Altogether, the predictive/prognostic risk model established in this study may be valuable in clinical practice.
Significant differences in gene expression were detected between the high-risk and low-risk core cohorts. Therefore, we further evaluated FAM-related genes from the risk model. Expression of fatty acid binding protein 4 (FABP4), THBS1, and other marker levels were significantly different between the two cohorts. Various studies have suggested that pro- or antitumor manifestation of FABP4 was associated with various cancer types[31-33]. Further, FABP4 reportedly promotes breast cancer cell invasion by regulating the interleukin-6/signal transducer and activator of the transcription-3 axes. Moreover, its inhibition by BMS309403 (inhibitory molecule of FABP4) can prevent the proliferation and metastasis of ovarian cancer. Additionally, increased expression of FABP4 has been associated with a worse prognosis in GC, although Chen et al[34] came to a contrary conclusion. On the contrary, increased FABP4 expression prohibited tumor progression and metastasis in hepatocellular carcinoma. Accordingly, THBS1 was identified as a hub gene that can serve as an indicator for GC diagnosis. However, its predictive and prognostic role in patients with GC remains elusive. Consequently, demonstrating the role of FABP4 and THBS1 in GC in future studies is important and requires urgent attention.
We constructed a fatty acid risk score model to assess the intact fatty acid features in GC. This risk model combined with clinicopathological characteristics, prognosis, chemotherapy sensitivity, and immune cell function, can be used to develop personalized treatment strategies for patients with GC. This study may sharpen and enrich the rationale used in prediction models to optimize the care of patients with GC.
the interactions between solitary metabolic features and the surrounding complex tumor environment characterize cancer. fatty acids are the essential components of diverse types of lipids, which are crucial mediators in energy metabolism and signal transduction. Although various molecules have been identified in gastric cancer (GC) metastasis, uncovering novel links between fatty acids and the tumor environment is necessary to comprehensively understand gastric cancer.
we constructed a novel prognostic risk model for GC based on fatty acid metastasis (FAM) -related genes. We examined the mechanisms underlying FAM in GC, the relationship between the risk of GC and tumor microenvironment characteristics, and treatment strategies for GC.
we want to construct novel links between fatty acids and the tumor environment to contribute to accurate diagnosis, prognosis prediction, and recurrence risk and metastasis assessments in patients with GC.
Data download and analysis; Detection of differentially expressed genes and functional enrichment analysis between normal and cancer samples; Establishment and validation of prognostic risk model; Principal component analysis and gene set variation analysis; Construction of protein-protein interaction network
Functional analysis of FAM-related genes between normal and cancer samples from TCGA database; Construction and validation of the prognostic risk model in GC; Correlation between risk model score and clinical features; Establishment of predictive nomogram in patients with GC; A FAM-related model predicting response to chemotherapy and GSVA in high- and low-risk groups; Immune differences between high- and low-risk groups; Functional enrichment analysis of FAM-related DEGs in the low- and high-risk score groups based on protein-protein interaction.
A fatty acid risk score model to assess the intact fatty acid features in GC was constructed. This risk model combined with clinicopathological characteristics, prognosis, chemotherapy sensitivity, and immune cell function.
We established a prognostic risk model using data collected from The Cancer Genome Atlas database, explored the function of the risk model, and identified the relationship between the risk model and clinical features.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Liang W, China; Shalaby MN, Egypt S-Editor: Liu JH L-Editor: A P-Editor: Chen YX
| 1. | Sexton RE, Al Hallak MN, Diab M, Azmi AS. Gastric cancer: a comprehensive review of current and future treatment strategies. Cancer Metastasis Rev. 2020;39:1179-1203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 449] [Article Influence: 89.8] [Reference Citation Analysis (0)] |
| 2. | Thrift AP, El-Serag HB. Burden of Gastric Cancer. Clin Gastroenterol Hepatol. 2020;18:534-542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 432] [Cited by in RCA: 952] [Article Influence: 190.4] [Reference Citation Analysis (1)] |
| 3. | Joshi SS, Badgwell BD. Current treatment and recent progress in gastric cancer. CA Cancer J Clin. 2021;71:264-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 382] [Cited by in RCA: 1058] [Article Influence: 264.5] [Reference Citation Analysis (0)] |
| 4. | Ding C, Shan Z, Li M, Chen H, Li X, Jin Z. Characterization of the fatty acid metabolism in colorectal cancer to guide clinical therapy. Mol Ther Oncolytics. 2021;20:532-544. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 67] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 5. | Fhu CW, Ali A. Fatty Acid Synthase: An Emerging Target in Cancer. Molecules. 2020;25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 234] [Article Influence: 46.8] [Reference Citation Analysis (1)] |
| 6. | Lee JY, Nam M, Son HY, Hyun K, Jang SY, Kim JW, Kim MW, Jung Y, Jang E, Yoon SJ, Kim J, Seo J, Min JK, Oh KJ, Han BS, Kim WK, Bae KH, Song J, Huh YM, Hwang GS, Lee EW, Lee SC. Polyunsaturated fatty acid biosynthesis pathway determines ferroptosis sensitivity in gastric cancer. Proc Natl Acad Sci U S A. 2020;117:32433-32442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 316] [Article Influence: 63.2] [Reference Citation Analysis (0)] |
| 7. | Ma S, Zhou B, Yang Q, Pan Y, Yang W, Freedland SJ, Ding LW, Freeman MR, Breunig JJ, Bhowmick NA, Pan J, Koeffler HP, Lin DC. A Transcriptional Regulatory Loop of Master Regulator Transcription Factors, PPARG, and Fatty Acid Synthesis Promotes Esophageal Adenocarcinoma. Cancer Res. 2021;81:1216-1229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 57] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 8. | Khan D, Ara T, Ravi V, Rajagopal R, Tandon H, Parvathy J, Gonzalez EA, Asirvatham-Jeyaraj N, Krishna S, Mishra S, Raghu S, Bhati AS, Tamta AK, Dasgupta S, Kolthur-Seetharam U, Etchegaray JP, Mostoslavsky R, Rao PSM, Srinivasan N, Sundaresan NR. SIRT6 transcriptionally regulates fatty acid transport by suppressing PPARγ. Cell Rep. 2021;35:109190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 39] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 9. | Luo Y, Wang H, Liu B, Wei J. Fatty Acid Metabolism and Cancer Immunotherapy. Curr Oncol Rep. 2022;24:659-670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 34] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 10. | Garcia KA, Costa ML, Lacunza E, Martinez ME, Corsico B, Scaglia N. Fatty acid binding protein 5 regulates lipogenesis and tumor growth in lung adenocarcinoma. Life Sci. 2022;301:120621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 11. | Zhang C, Liao Y, Liu P, Du Q, Liang Y, Ooi S, Qin S, He S, Yao S, Wang W. FABP5 promotes lymph node metastasis in cervical cancer by reprogramming fatty acid metabolism. Theranostics. 2020;10:6561-6580. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 119] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 12. | Zhao Q, Cao L, Guan L, Bie L, Wang S, Xie B, Chen X, Shen X, Cao F. Immunotherapy for gastric cancer: dilemmas and prospect. Brief Funct Genomics. 2019;18:107-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 103] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 13. | Yang H, Deng Q, Ni T, Liu Y, Lu L, Dai H, Wang H, Yang W. Targeted Inhibition of LPL/FABP4/CPT1 fatty acid metabolic axis can effectively prevent the progression of nonalcoholic steatohepatitis to liver cancer. Int J Biol Sci. 2021;17:4207-4222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 51] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 14. | Zeng D, Wu J, Luo H, Li Y, Xiao J, Peng J, Ye Z, Zhou R, Yu Y, Wang G, Huang N, Rong X, Sun L, Sun H, Qiu W, Xue Y, Bin J, Liao Y, Li N, Shi M, Kim KM, Liao W. Tumor microenvironment evaluation promotes precise checkpoint immunotherapy of advanced gastric cancer. J Immunother Cancer. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 142] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 15. | Yoon H, Lee S. Fatty Acid Metabolism in Ovarian Cancer: Therapeutic Implications. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 52] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 16. | Yang F, Gan L, Pan J, Chen Y, Zhang H, Huang L. Integrated Single-Cell RNA-Sequencing Analysis of Gastric Cancer Identifies FABP1 as a Novel Prognostic Biomarker. J Oncol. 2022;2022:4761403. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 17. | Takei S, Kawazoe A, Shitara K. The New Era of Immunotherapy in Gastric Cancer. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 107] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 18. | Ohshima K, Morii E. Metabolic Reprogramming of Cancer Cells during Tumor Progression and Metastasis. Metabolites. 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 125] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 19. | Mei D, Qi Y, Xia Y, Ma J, Hu H, Ai J, Chen L, Wu N, Liao D. Microarray profile analysis identifies ETS1 as potential biomarker regulated by miR-23b and modulates TCF4 in gastric cancer. World J Surg Oncol. 2021;19:311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 20. | Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell. 2008;13:472-482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1553] [Cited by in RCA: 1696] [Article Influence: 99.8] [Reference Citation Analysis (0)] |
| 21. | Jiang M, Wu N, Xu B, Chu Y, Li X, Su S, Chen D, Li W, Shi Y, Gao X, Zhang H, Zhang Z, Du W, Nie Y, Liang J, Fan D. Fatty acid-induced CD36 expression via O-GlcNAcylation drives gastric cancer metastasis. Theranostics. 2019;9:5359-5373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 111] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 22. | Iwamoto H, Abe M, Yang Y, Cui D, Seki T, Nakamura M, Hosaka K, Lim S, Wu J, He X, Sun X, Lu Y, Zhou Q, Shi W, Torimura T, Nie G, Li Q, Cao Y. Cancer Lipid Metabolism Confers Antiangiogenic Drug Resistance. Cell Metab. 2018;28:104-117.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 215] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 23. | Heravi G, Jang H, Wang X, Long Z, Peng Z, Kim S, Liu W. Fatty acid desaturase 1 (FADS1) is a cancer marker for patient survival and a potential novel target for precision cancer treatment. Front Oncol. 2022;12:942798. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 26] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 24. | Cui MY, Yi X, Zhu DX, Wu J. The Role of Lipid Metabolism in Gastric Cancer. Front Oncol. 2022;12:916661. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 33] [Reference Citation Analysis (0)] |
| 25. | Cui MY, Yi X, Zhu DX, Wu J. Aberrant lipid metabolism reprogramming and immune microenvironment for gastric cancer: a literature review. Transl Cancer Res. 2021;10:3829-3842. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 26. | Zhu T, Wang Z, Zou T, Xu L, Zhang S, Chen Y, Chen C, Zhang W, Wang S, Ding Q, Xu G. SOAT1 Promotes Gastric Cancer Lymph Node Metastasis Through Lipid Synthesis. Front Pharmacol. 2021;12:769647. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 27. | Qi Y, Chen D, Lu Q, Yao Y, Ji C. Bioinformatic Profiling Identifies a Fatty Acid Metabolism-Related Gene Risk Signature for Malignancy, Prognosis, and Immune Phenotype of Glioma. Dis Markers. 2019;2019:3917040. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 28. | Xie J, Fu L, Jin L. Immunotherapy of gastric cancer: Past, future perspective and challenges. Pathol Res Pract. 2021;218:153322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 29. | Valdez-Salazar HA, Ares MA, Fernández FJ, Ibarra JA, Torres J, Bustamante VH, De la Cruz MA. Long-chain fatty acids alter transcription of Helicobacter pylori virulence and regulatory genes. PeerJ. 2021;9:e12270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 30. | Chen QY, Huang XB, Zhao YJ, Wang HG, Wang JB, Liu LC, Wang LQ, Zhong Q, Xie JW, Lin JX, Lu J, Cao LL, Lin M, Tu RH, Zheng CH, Li P, Huang CM. The peroxisome proliferator-activated receptor agonist rosiglitazone specifically represses tumour metastatic potential in chromatin inaccessibility-mediated FABP4-deficient gastric cancer. Theranostics. 2022;12:1904-1920. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 31. | Jung Y, Cho SM, Kim S, Cheong JH, Kwon HJ. Functional inhibition of fatty acid binding protein 4 ameliorates impaired ciliogenesis in GCs. Biochem Biophys Res Commun. 2021;539:28-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 32. | He Y, Song H, Jiang Y, Ren W. Identification of Immune-Related Prognostic Markers in Gastric Cancer. J Healthc Eng. 2022;2022:7897274. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 33. | Guo Y, Wang ZW, Su WH, Chen J, Wang YL. Prognostic Value and Immune Infiltrates of ABCA8 and FABP4 in Stomach Adenocarcinoma. Biomed Res Int. 2020;2020:4145164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 34. | Chen Y, Yuan H, Yu Q, Pang J, Sheng M, Tang W. Bioinformatics Analysis and Structure of Gastric Cancer Prognosis Model Based on Lipid Metabolism and Immune Microenvironment. Genes (Basel). 2022;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |