Published online Jul 6, 2023. doi: 10.12998/wjcc.v11.i19.4553
Peer-review started: April 24, 2023
First decision: May 8, 2023
Revised: May 27, 2023
Accepted: June 13, 2023
Article in press: June 13, 2023
Published online: July 6, 2023
Processing time: 67 Days and 4.8 Hours
To analyze the potential action mechanism of Huangqin decoction (HQD) in colorectal cancer (CRC) treatment on the basis of network pharmacology and molecular docking.
To investigate the molecular mechanisms of HQD for CRC treatment by using network pharmacology and molecular docking.
All HQD active ingredients were searched using the Systematic Pharmacology and Traditional Chinese Medicine Systems Pharmacology databases and the Bioinformatics Analysis Tool for Molecular Mechanisms in traditional Chinese medicine. Then, the targets of the active ingredients were screened. The abbreviations of protein targets were obtained from the UniProt database. A “drug–com
In total, 280 potential drug-active ingredients were present in HQD, including 1474 targets of the drug-active ingredients. The main active ingredients identified were betulin, tetrahydropalmatine, and quercetin. In total, 10249 CRC-related targets and 1014 drug-disease intersecting targets were identified, including 28 core targets of action such as Jun proto-oncogene, AP-1 transcription factor subunit, signal transducer and activator of transcription 3, tumor protein p53, vascular endothelial growth factor, and AKT serine/threonine kinase 1. The gene ontology enrichment functional analysis yielded 503 enrichment results, including 406 biological processes that were mainly related to the positive regulation of both gene expression and transcription and cellular response to hypoxia, etc. In total, 38 cellular components were primarily related to polymer complexes, transcription factor complexes, and platelet alpha granule lumen. Then, 59 molecular functions were closely related to the binding of enzymes, homologous proteins, and transcription factors. The Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis yielded 139 enrichment results, involving epidermal growth factor receptor tyrosine kinase inhibitor resistance and HIF-1 and mitogen-activated protein kinase signaling pathways.
HQD can play a role in CRC treatment through the “multi-component-target–pathway”. The active ingredients betulin, tetrahydropalmatine, and quercetin may act on targets such as Jun proto-oncogene, AP-1 transcription factor subunit, signal transducer and activator of transcription 3, tumor protein p53, vascular endothelial growth factor, and AKT serine/threonine kinase 1, which in turn regulate HIF-1 and mitogen-activated protein kinase signaling pathways in CRC treatment. The molecular docking junction clarified that all four key target proteins could bind strongly to the main HQD active ingredients. This indicates that HQD could slow down CRC progression by modulating multiple targets and signaling pathways.
Core Tip: Huangqin decoction may treat colorectal cancer through multi-component targeting and modulation of multiple signaling pathways.
- Citation: Li YJ, Tang DX, Yan HT, Yang B, Yang Z, Long FX. Network pharmacology and molecular docking-based analyses to predict the potential mechanism of Huangqin decoction in treating colorectal cancer. World J Clin Cases 2023; 11(19): 4553-4566
- URL: https://www.wjgnet.com/2307-8960/full/v11/i19/4553.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i19.4553
The incidence of colorectal cancer (CRC) has continued to increase, accounting for nearly 10% of cancer patients. CRC is the second most common cause of cancer death[1,2]. Many pathogenic factors could cause CRC such as obesity, poor diet, alcohol addiction, and genetic mutations or familial inheritance[1-4]. According to domestic statistics, CRC incidence in China has increased each year from 1990 to 2019. Using the ARIMA model, the standardized CRC incidence in China is predicted to reach 33.93/100000 in 2025, bringing a huge burden to the Chinese healthcare system[5]. Radical surgical resection, chemotherapy, radiotherapy, and targeted therapy are the current main treatment modalities for CRC[6]. However, these treatments are generally deficient as they are prone to recurrence and side effects. Therefore, finding effective drugs with few side effects in CRC treatment is valuable.
CRC is classified as “viscera toxin” and “intestinal accumulation” in traditional Chinese medicine (TCM). In the early CRC stage, clearing heat, regulating qi, dispelling dampness, eliminating phlegm, removing blood stasis, detoxifying, soft, and firm dispersing, and other treatment methods are used in TCM for dispelling evil spirits[7]. Huangqin Decoction (HQD) is derived from “Treatise on Febrile Diseases: Taiyang and Shaoyang combined disease, diarrhea, treat with HQD.” The HQD recipe consists of Scutellariae Radix (3 taels), Paeonia lactiflora Pall. (2 taels), Glycyrrhiza uralensis Fisch. (2 taels), and Ziziphus jujuba Mill. (12 pieces). Scutellariae Radix for the king, bitter, and cold can clear heat. Paeonia lactiflora Pall. can relieve pain. HQD can supply the qi. The combination of various drugs can clear heat and relieve pain[8]. HQD has been proven to treat CRC clinically with a remarkable effect, but the exact mechanism has not been elucidated.
TCM is based on the formulation principles of “king, minister, adjuvant, and ambassador” and incompatibility taboo. With the characteristics of the multi-component-target-pathway and synergistic treatment, TCM has great potential to improve cancer symptoms. Network pharmacology is an emerging discipline based on systems biology theories. It analyses and calculates the relationship between relevant drugs and diseases through drug-compound-target interaction networks[9]. By inferring the treatment effects and regulatory mechanisms of drugs in disease through multiple targets, it also provides new methods for TCM application, thereby facilitating the promotion of the development and heritage of Chinese herbs and compounds. Using informatics, molecular docking predicts the potential binding conformation of ligands and receptors. It also verifies the binding affinity of core components and targets. We here constructed a drug-compound-target network according to the HQD active ingredients by using network pharmacology and molecular docking and thus analyzed and calculated the molecular mechanism and regulation signal pathways of HQD for CRC treatment. The study findings also provided a certain theoretical basis for future studies.
In this study, Traditional Chinese Medicine Systems Pharmacology and Bioinformatics Analysis Tool for Molecular Mechanisms in traditional Chinese medicine databases were used to search the constituent herbs of HQD and their chemical and pharmacological data. Duplicate compounds were combined and removed to gather information about the HQD active ingredients. The targets of all these active ingredients were further searched and screened by referring to Traditional Chinese Medicine Systems Pharmacology and UniProt to realize the herb targets of HQD. All database URLs in this study are shown in Table 1.
| Database | Address |
| TCMSP | http://www.tcmspw.com/tcmsp.php |
| BATMAN-TCM | http://bionet.ncpsb.org.cn/batman-tcm/ |
| Genecards | https://www.genecards.org |
| TTD | http://bidd.nus.edu.sg/group/ttd/ttd.asp |
| OMIM | https://omim.org/ |
| DisGeNET | https://www.disgenet.org/home/ |
| Wayne tool | http://jvenn.toulouse.inra.fr/app/example.html |
| String | https://string-db.org/ |
| DAVID 6.8 | http://david.ncifcrf.gov |
| Omicstudio | https://www.omicstudio.cn/tool |
| PDB | https://www.rcsb.org |
| PubChem | https://pubchem.ncbi.nlmnih.gov |
We analyzed the obtained components and targets and constructed a network of “drug-compound-target” by using Cytoscape 3.7.2. In this network, nodes represent drugs, compounds, and targets, and edges represent the relationship between the three nodes. We analyzed the number of connections of each node by calculating the “degree” value.
The term “colorectal cancer” was searched, and potential targets were screened for in GeneCards, TTD, OMIM, and DisGeNET databases. All CRC-related targets were acquired after removing duplicate targets. HQD action targets and CRC-related targets were imported into the online Wayne tool to map the intersection of HQD and CRC targets and visualize the results.
To elucidate the functional interactions between the screened proteins, the intersecting targets were put into the STRING database to construct the protein-protein interaction (PPI) network. This PPI network was then put into Cytoscape 3.7.2, and the CytoNCA plugin was applied to filter core proteins based on betweenness (BC), closeness (CNC), degree (DC), eigenvector (EC), local average connectivity (LAC)-based method, and network (NC) 2 times the median value of the core proteins.
We obtained information about the primary biological processes and signaling pathways by importing the screened core targets into the DAVID 6.8 database for gene ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. Then, the enrichment results were visualized using the LianChuan BioCloud platform.
The three-dimensional (3D) structure of the core target proteins was first obtained from the PDB database. The MOL2 file of the core active compounds was then obtained from the PubChem platform. Chem 3D was used to optimize the total amount of core component energy. PyMOL software was used to optimize the number of core target proteins removed for water molecule hydrogenation and charge calculation. Molecular docking and binding activity analyses were performed using the AutoDock Vina program. Finally, docking sites were visualized on the PLIP platform. When the binding energy of the molecular docking conformation was lower, the binding conformation was more stable. This reflects the greater likelihood of binding between the receptor molecule and ligand. Therefore, only the highest absolute value of binding energy for each molecular docking pair was retained for the docking results.
We identified the HQD active ingredients and the potential therapeutic targets and mechanisms of HQD in CRC treatment by conducting network pharmacology and functional gene pathway analyses.
Among the active ingredients obtained, 46 were Scutellariae Radix, 28 were Paeonia lactiflora Pall., 160 were Glycyrrhiza uralensis Fisch., and 58 were Ziziphus jujuba Mill. Then, 280 compounds and 1474 potential targets were obtained for the action of the ingredients after removing the duplicate information.
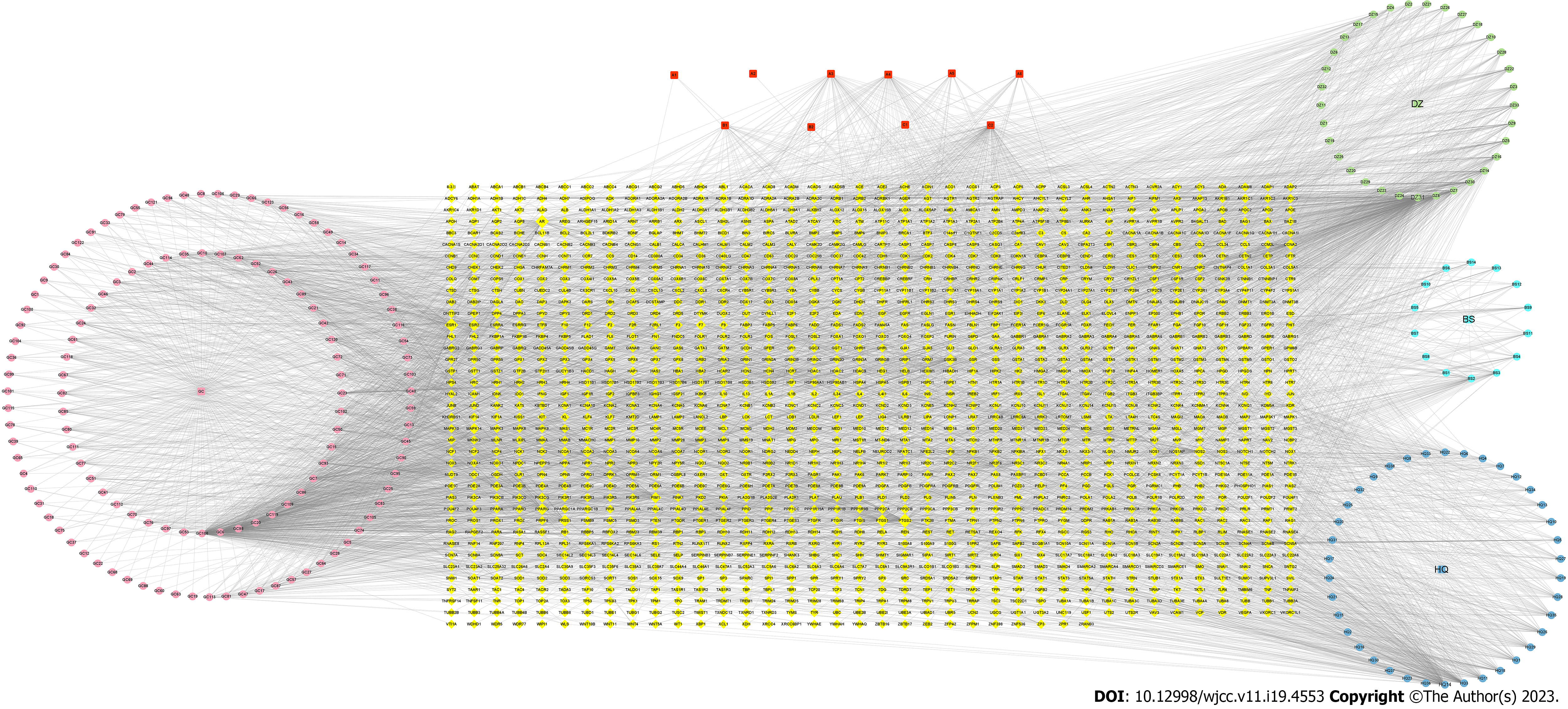
The network “drug-compound-target” was built using Cytoscape 3.7.1 software. It included 1695 nodes and 5567 edges (Figure 1), with red squares representing drug co-components. The specific information is presented in Table 2. The HQD compounds with high activity are betulin, tetrahydropalmatine, and quercetin, which may be crucial for CRC treatment.
| Numbers | Mol ID | Molecule name | OB (%) | DL |
| A1 | MOL000359 | Sitosterol | 36.91 | 0.75 |
| A2 | MOL000211 | Mairin | 55.38 | 0.78 |
| A3 | MOL000422 | Kaempferol | 41.88 | 0.24 |
| A4 | MOL008583 | Beta-sitosterol | 15.00 | 0.81 |
| A5 | MOL000492 | (+)-Catechin | 54.83 | 0.24 |
| A6 | MOL000096 | (-)-Catechin | 49.68 | 0.24 |
| B1 | MOL002320 | γ-Sitosterol | 36.91 | 0.75 |
| B2 | MOL002307 | 20-Hexadecanoylingenol | 28.20 | 0.68 |
| C1 | MOL004349 | Ruvoside | 18.13 | 0.63 |
| C2 | MOL000098 | Quercetin | 46.43 | 0.28 |
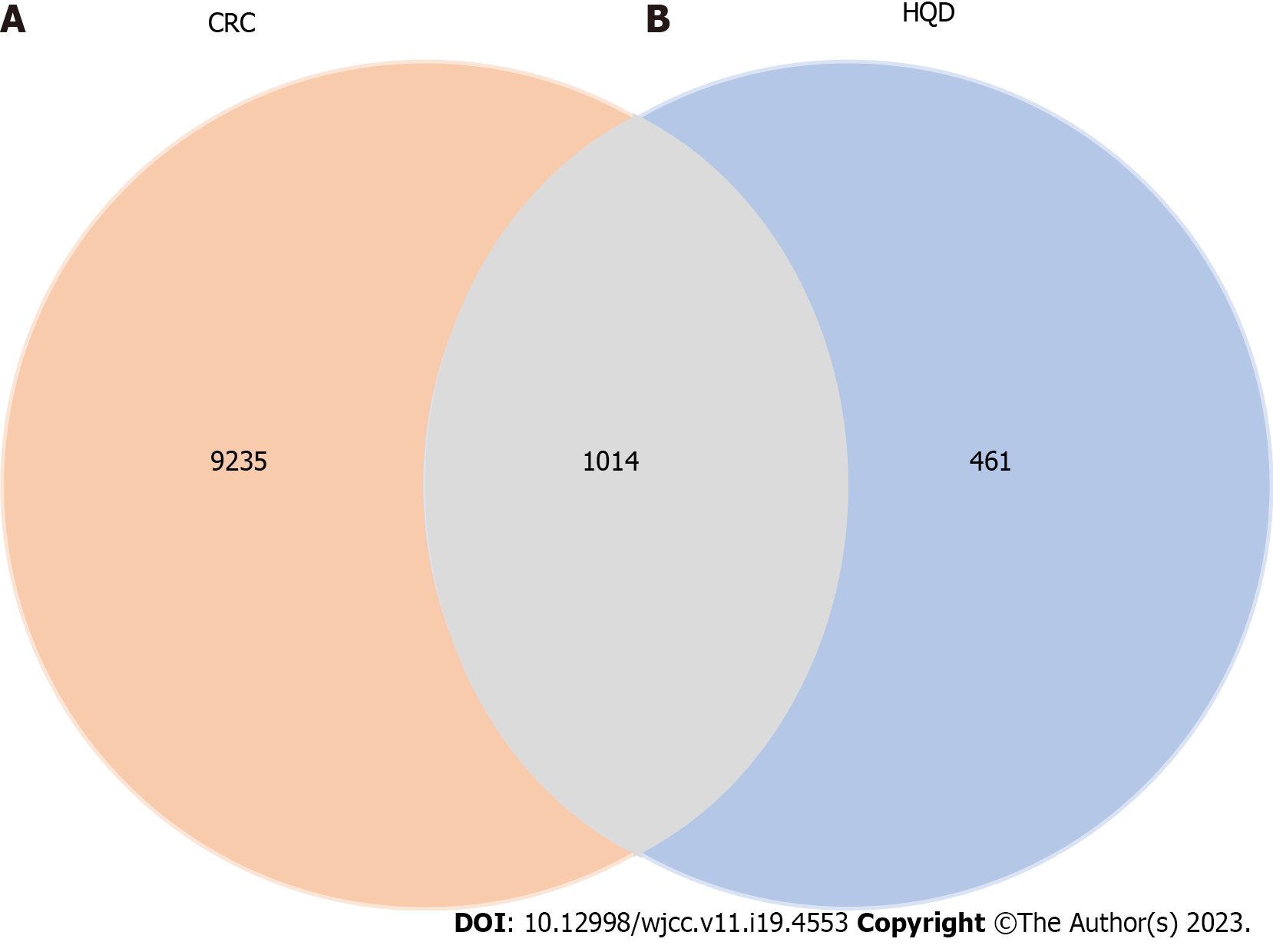
We obtained 10249 CRC-related targets for its treatment from the disease databases of Genecards, OMIM, DisGeNET, and TTD. The targets of both CRC and the HQD active ingredients were imported into the Venn diagram editing website, and then 1014 potential targets of HQD in CRC treatment were identified (Figure 2).
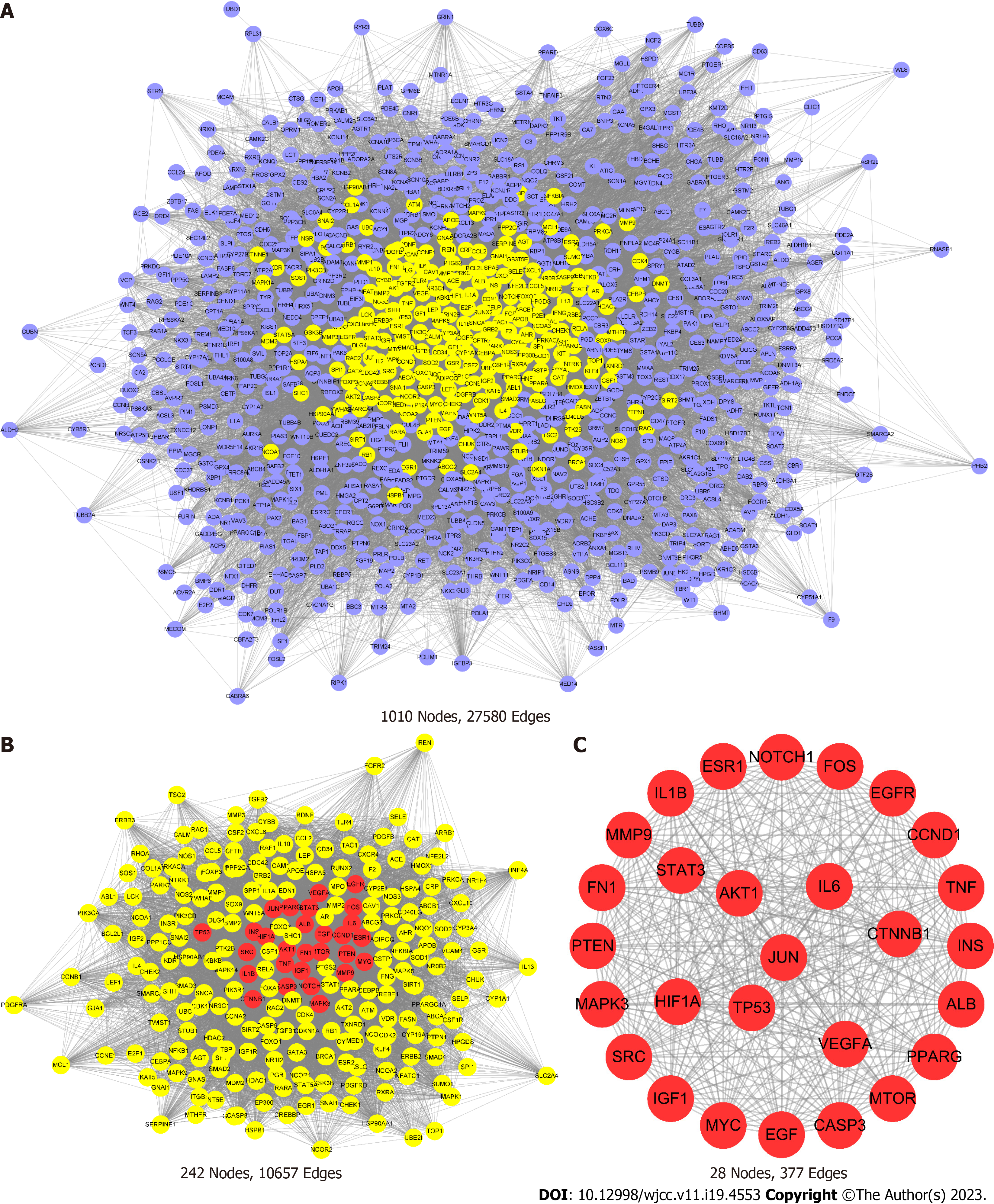
The STRING was used to obtain key proteins for CRC treatment with HQD. The analysis data were imported into Cytoscape 3.7.2. Based on the BC, CNC, DC, EC, LAC, and NC values of topological parameters calculated using CytoNCA, 28 targets that play key roles in CRC, such as Jun proto-oncogene, AP-1 transcription factor subunit (JUN), signal transducer and activator of transcription 3 (STAT3), tumor protein p53 (TP53), vascular endothelial growth factor (VEGFA), and AKT serine/threonine kinase 1 (AKT1) were screened (Figure 3). These targets could be considered the HQD core targets in CRC treatment.
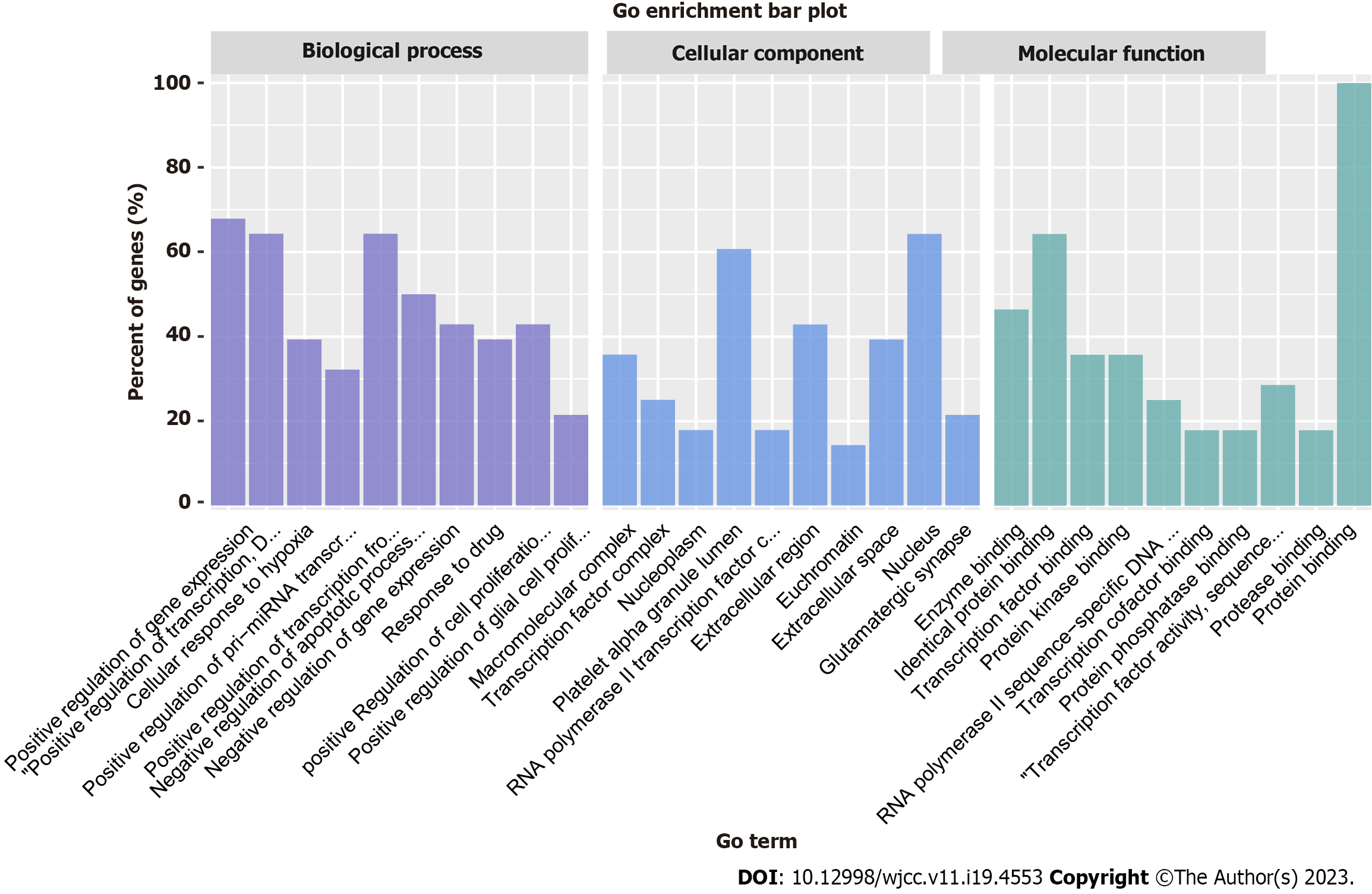
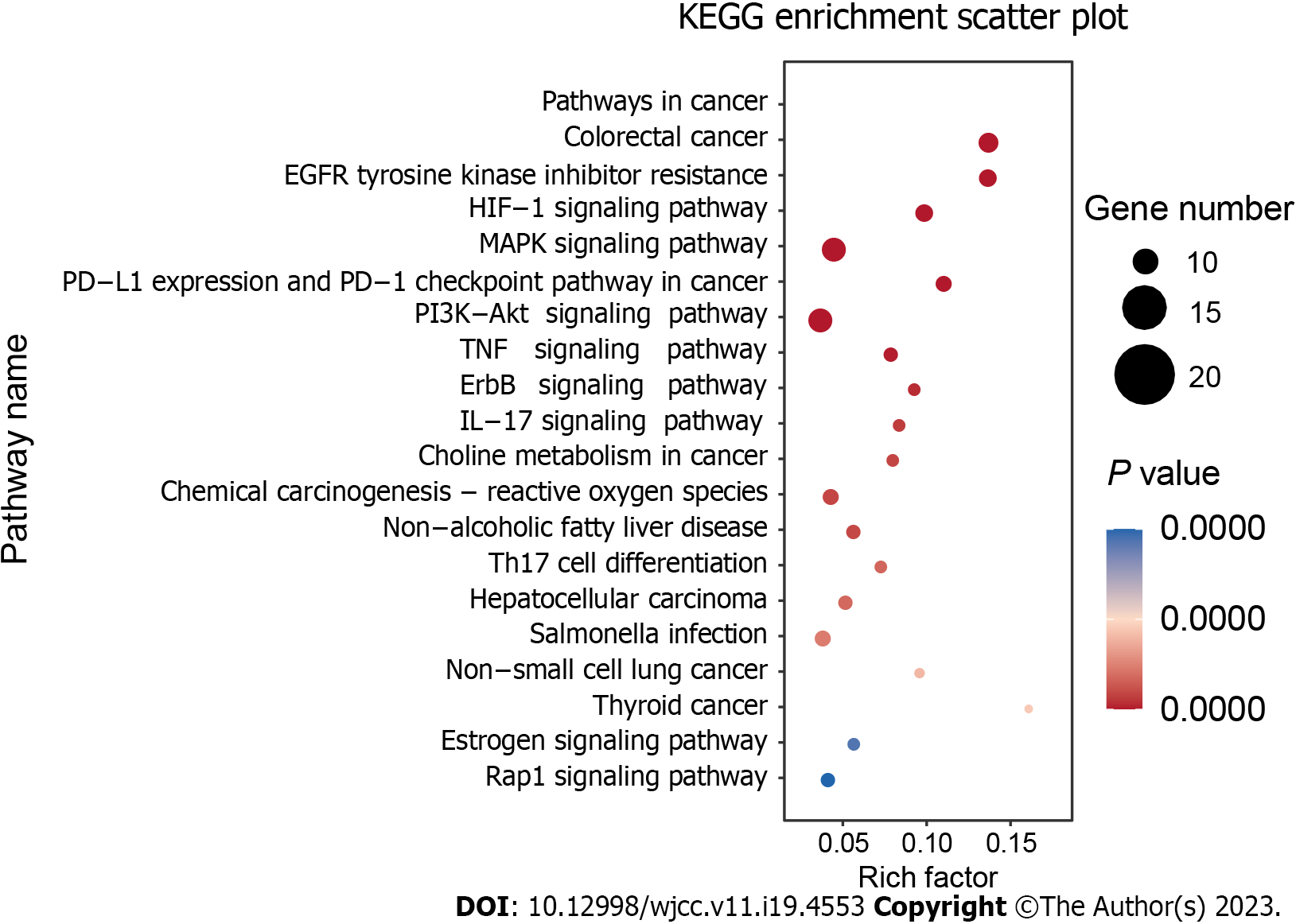
We imported 28 core targets into the DAVID 6.8 database to identify the relevant HQD biological functions in CRC treatment by analyzing GO functional enrichment. This analysis revealed 503 enrichment results, including 406 biological processes, 38 cellular components, and 59 molecular functions. The biological processes were mainly related to the positive regulation of gene expression and transcription and cellular response to hypoxia. The cellular components were principally related to the macromolecular complex, transcription factor complex, platelet alpha granule lumen, etc. The molecular functions primarily involved the binding of enzymes, identical proteins, and transcription factors. The GO enrichment results were sorted from the smallest to the largest P value. The top 10 entries were selected to create a histogram (Figure 4). To determine the potential mechanism underlying CRC treatment with HQD, we performed the KEGG pathway enrichment analysis and obtained 139 HQD-associated signaling pathways for CRC treatment. The top 20 pathways were visualized in descending order of the P value (Figure 5). These mainly included signaling pathways such as epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor resistance, HIF-1, and mitogen-activated protein kinase (MAPK), suggesting that HQD can treat CRC through these pathways.
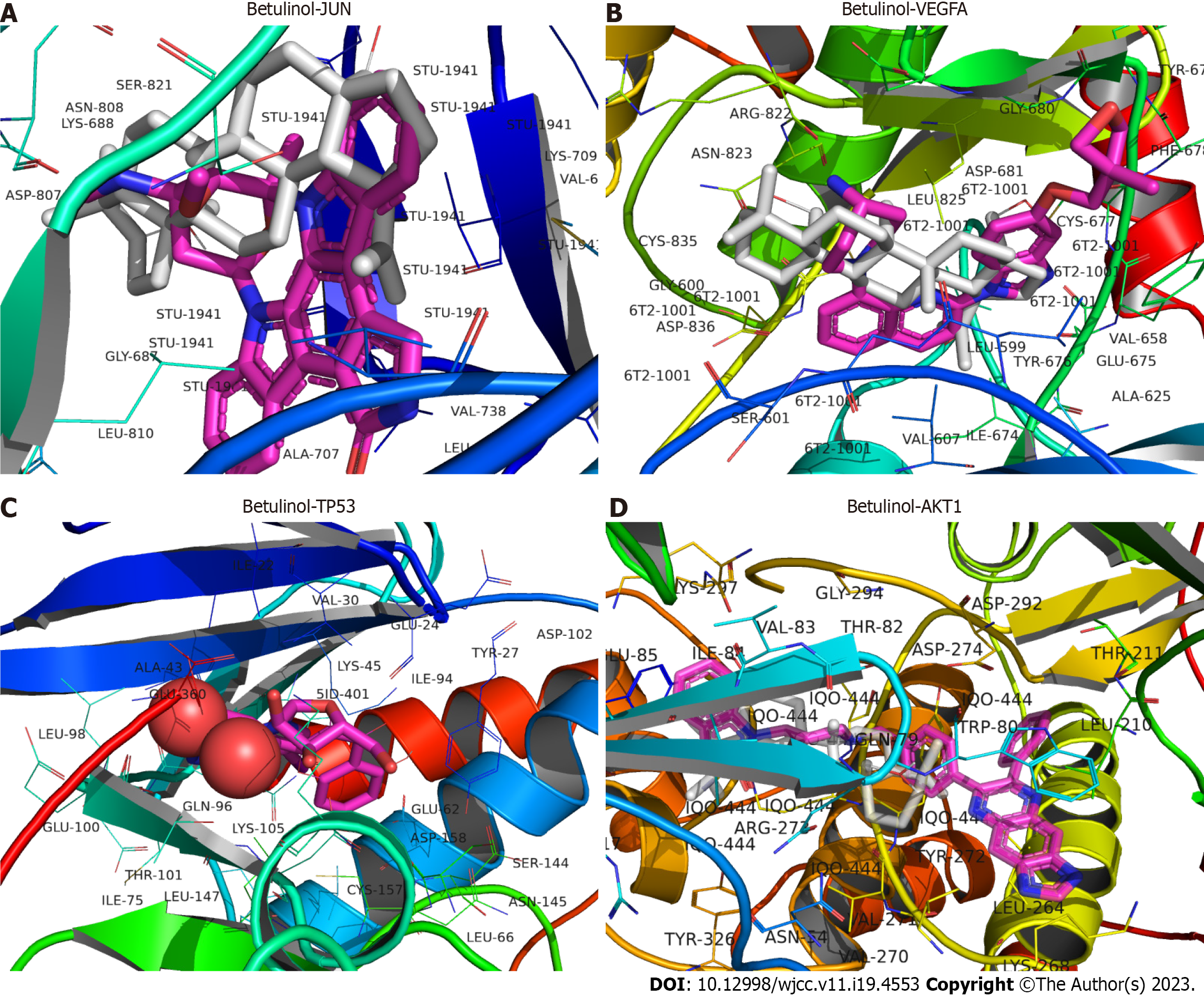
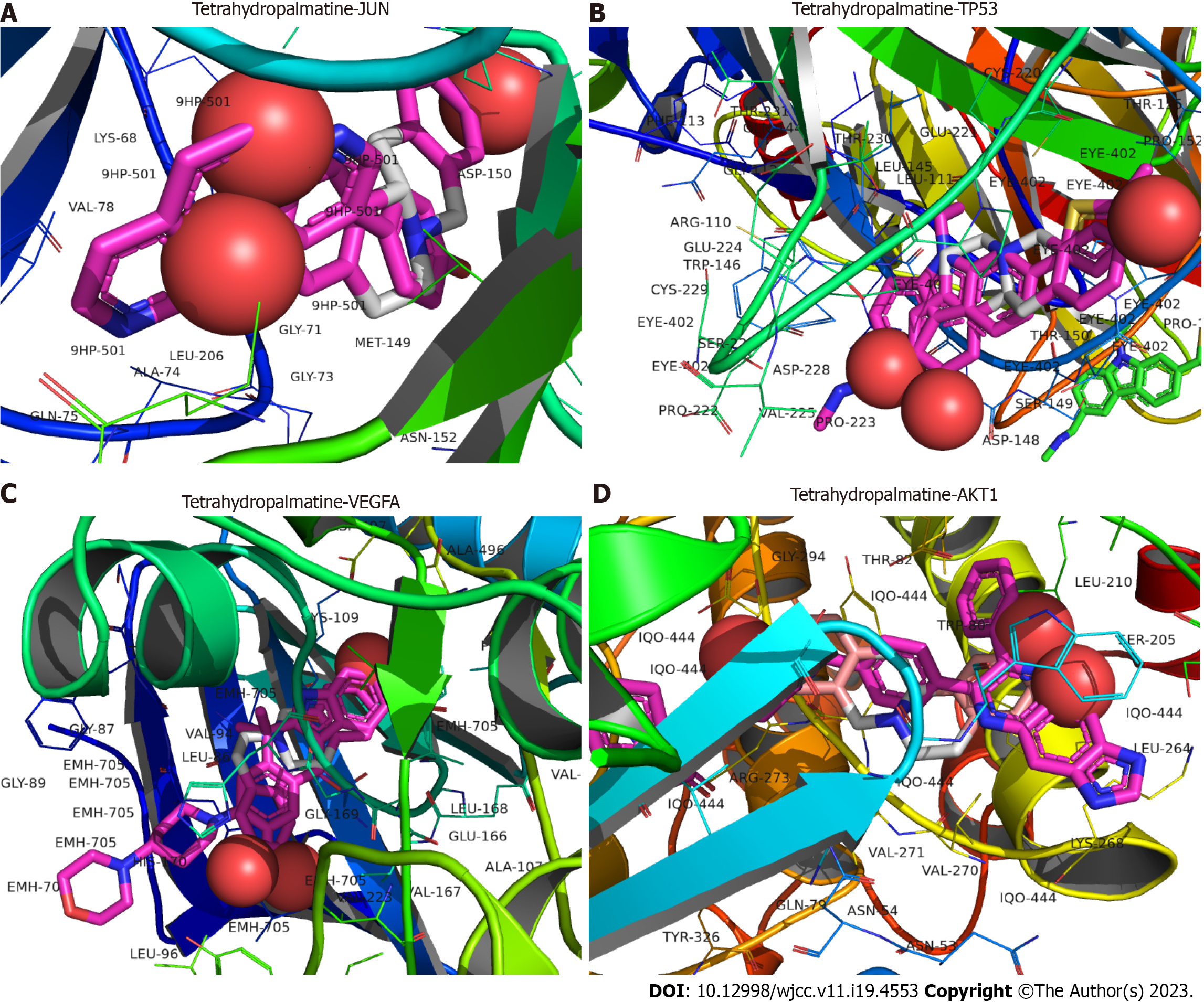
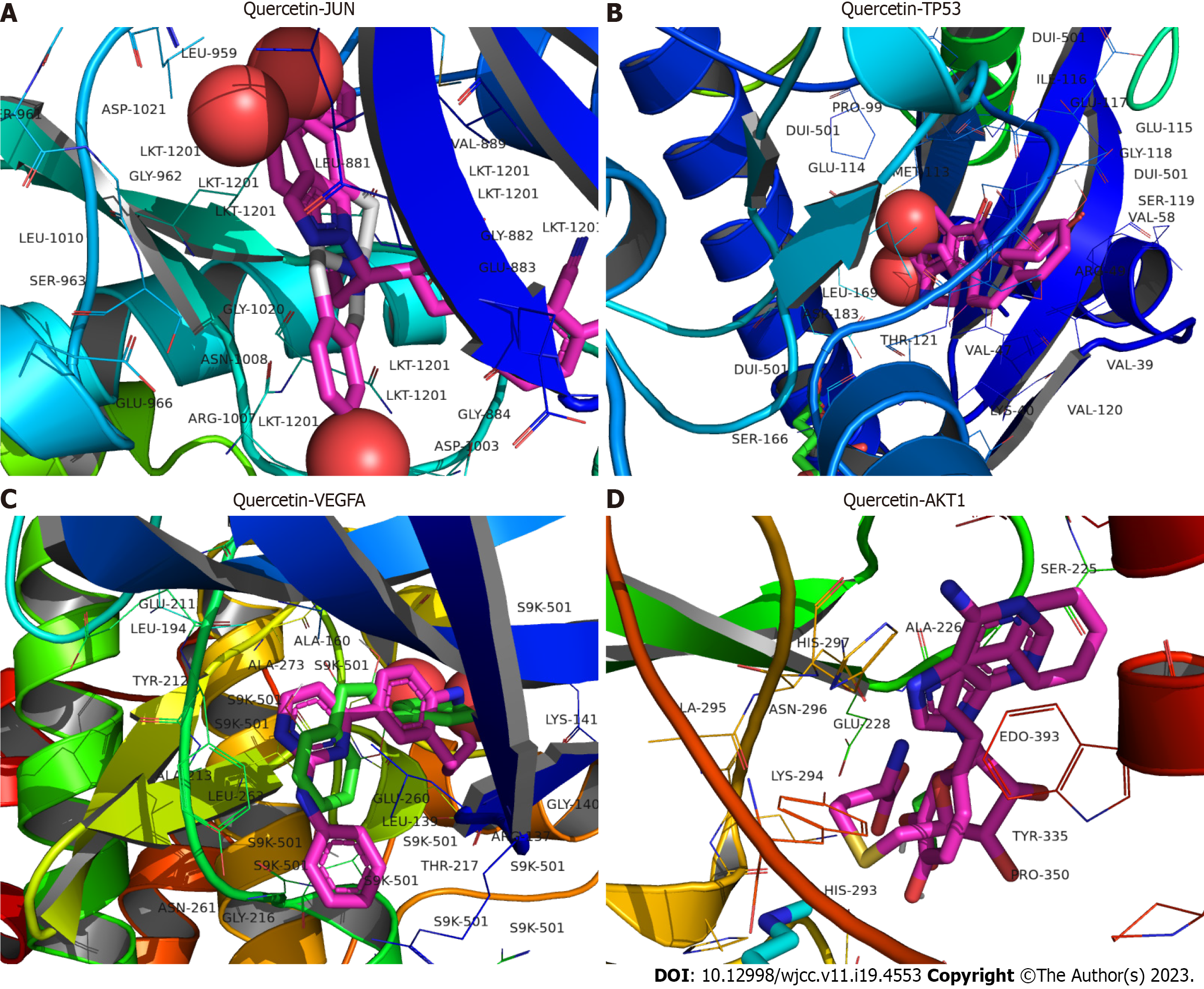
Five key target proteins (JUN, STAT3, TP53, VEGFA, and AKT1) were molecularly docked to three core components (betulin, tetrahydropalmatine, and quercetin) to assess the binding capacity of the target and ligand proteins. The binding capacity was predicted using AutoDock software, with binding energies typically < -5.0 kcal·mol−1 indicating significant binding activity between the molecules. All three active components exhibited a strong binding capacity to JUN, TP53, VEGFA, and AKT1 (Table 3), with birudinol failing to bind well to STAT3. The 3D model plots of the three active ingredients docked with JUN, TP53, VEGFA, and AKT1 were visualized using PyMOL and PLIP software (Figures 6-8).
| Active ingredients | Binding energy (kcal/mol) | |||
| JUN | TP53 | VEGFA | AKT1 | |
| Betulinol | -8.4 | -6 | -8.7 | -10.3 |
| Tetraydropalmatine | -7.2 | -6.1 | -9.4 | -9.6 |
| Quercetin | -6.4 | -8.3 | -8.1 | -8.3 |
According to TCM, the diseased region in CRC is the large intestine. Moreover, CRC is closely related to the spleen and stomach. “Miraculous Pivot” mentioned that “The injury of the stomach, the blood overflow in the parenteral, the parenteral cold, the juice foam and blood phase kneading, the combination of condensation cannot be dispersed, and the product into.” Several clinical studies have retrospectively investigated the TCM symptoms of CRC patients and their associated factors. These studies have found that the most common TCM classification of CRC is the syndrome of damp heat accumulation[10-12]. HQD was first mentioned by Zhang Zhongjing in “Treatise on Febrile Diseases.” According to this monograph, HQD could clear away heat and relieve pain and is a classic therapeutic formula for damp heat in Chinese medicine.
We conducted a network pharmacological analysis to determine active compounds, potential targets, and action mechanisms of HQD in CRC treatment. In total, 280 core ingredients of HQD were screened. The topological analysis of the “drug-compound-target” network revealed that the top-ranked compounds were betulin, tetrahydropalmatine, and quercetin. These may be the potential compounds of HQD in CRC treatment. Among them, betulinol induced apoptosis of CRC cells through the MAPK signaling pathway and significantly inhibited malignant metastasis of CT26 cells in the lungs[13]. The therapeutic efficacy of tetrahydropalmatine has been demonstrated in various cancers[14,15], suggesting that tetrahydropalmatine can be used as a potential novel therapeutic agent for CRC patients. Quercetin, a flavanol compound, has various biological activities, such as antioxidant activity. Quercetin could interfere with CRC development in various ways, such as inhibiting growth and proliferation and affecting the cycle of CRC cells. Thus, it could induce apoptosis and inhibit metastasis and invasion of CRC[16-19]. The aforementioned results proved that the main active ingredients of HQD play a crucial role in CRC treatment, suggesting that HQD is an effective therapeutic formula for CRC treatment.
On the basis of BC, CNC, DC, EC, LAC, and NC, the top five key targets, namely JUN, STAT3, TP53, VEGFA, and AKT1, were finally screened through cascading analysis of key targets using the PPI network. JUN is a transcription factor that recognizes and binds to the enhanced heptameric motif 5’-TGA[CG]TCA-3’. JUN can upregulate HuR expression by inhibiting miR-22 transcriptional regulation, which in turn leads to proliferation, migration, and tumor growth in CRC cells[20].
STAT3, a member of the STAT cytoplasmic transcription factor family, is the family member most associated with CRC development[21]. Large amounts of STAT3 are activated in CRC to promote tumor growth, invasion, migration[22], and proliferation of cancer cells. These features suggest that STAT3 can be used as a therapeutic target in CRC.
TP53 encodes the tumor suppressor p53, which is the major driver gene for suppressing the development of various cancers. The TP53 mutation has been observed in up to 80% of patients with advanced metastatic CRC[23], and this mutation can lead to poor prognosis in various cancer patients, including CRC patients[24].
VEGFA, the subtype of the vascular endothelial growth factor (VEGF), has a critical role in angiogenesis. According to some studies, tumor growth, spread, and metastasis depend on angio
AKT is a member of the serine/threonine kinase proto-oncogene family that regulates different inflammatory and metabolism-related signaling pathways and is involved in cancer cell proliferation, survival, and metabolic processes[27]. Approximately 70% of CRC exhibits highly activated AKT, which is closely associated with cancer development.
During molecular docking, the three core ingredients of HQD exhibited strong affinity to the four key target proteins, confirming the targeting effect of HQD in CRC treatment. The predicted results thus theoretically elucidate the ameliorative effect of HQD on CRC and help in the further investigation of the action mechanism of HQD or its bioactive compounds as an alternative treatment for CRC. The GO functional enrichment suggested that CRC treatment with HQD was mainly related to the positive regulation of gene expression and transcription as well as a cellular response to hypoxia. The KEGG pathway revealed that the pathways most closely related to CRC were EGFR tyrosine kinase inhibitor resistance, and HIF-1 and MAPK signaling pathways.
EGFR is closely associated with CRC recurrence and deterioration. Studies have demonstrated that miR-323a-3p can promote the apoptosis of CRC cells by directly targeting EGFR signaling[28]. Acquired resistance to 5-fluorouracil is a clinical challenge for CRC treatment. The HIF-1α and 5-fluorouracil combination significantly enhances the antitumor effects of 5-fluorouracil[29]. Therefore, targeting HIF-1α could act as an effective therapeutic strategy for 5-fluorouracil-resistant CRC. The MAPK signaling pathway is activated by peptide growth factors, cytokines, oxidative stress, etc. This pathway then regulates tumor cell proliferation, differentiation, survival, and death[30]. Several MAPK, ERK, c-Jun, and JNK subfamilies are involved in CRC pathogenesis[31], with ERK/MAPK playing a crucial role in cell proliferation[30]. Therefore, EGFR tyrosine kinase inhibitor resistance and HIF-1 and MAPK signaling pathways are closely linked to CRC development.
In conclusion, by analyzing “multi-components-targets-biological pathways,” this study found that HQD could exert therapeutic effects. Our findings also provide a clue regarding the theoretical basis for the active ingredients of HQD that are most effective in CRC treatment. However, because of the deficiencies of network pharmacology and molecular docking, follow-up in vivo and in vitro experiments are warranted to verify the action mechanism of HQD in CRC treatment.
Colorectal cancer (CRC) is the second most common cause of cancer death in recent years. CRC is a danger to human health.
The side effects caused by radiotherapy severely hinder the treatment progress of CRC patients. Huangqin decoction (HQD) can improve the immunity of CRC patients and enhance their quality of life.
This study exploited network pharmacology and molecular docking to uncover the potential targets and mechanisms of HQD for CRC treatment. It also provided a molecular biological basis for CRC treatment with HQD in a clinical setting.
This study involved preliminary exploration of the potential targets and mechanisms of HQD for CRC treatment by using network pharmacology and molecular docking.
The active ingredients betulin, tetrahydropalmatine, and quercetin in HQD may act on targets such as Jun proto-oncogene, AP-1 transcription factor subunit, signal transducer and activator of transcription 3, tumor protein p53, vascular endothelial growth factor, and AKT serine/threonine kinase 1, which in turn regulate HIF-1 and mitogen-activated protein kinase signaling pathways in CRC treatment.
HQD could treat CRC by regulating HIF-1 and mitogen-activated protein kinase signaling pathways.
At present, many shortcomings still exist in studying the action mechanisms of herbal medicine for various diseases on the basis of network pharmacology. Future research should be focused on in vivo and in vitro experiments to verify the key targets and type pathways and then combined proteomics and metabolomics to more systematically elucidate the action mechanism of HQD in CRC treatment.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Batra SK, United States; Chiang SF, Taiwan S-Editor: Ma YJ L-Editor: Filipodia P-Editor: Zhao S
| 1. | Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2268] [Cited by in RCA: 3246] [Article Influence: 649.2] [Reference Citation Analysis (2)] |
| 2. | Biller LH, Schrag D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA. 2021;325:669-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 1398] [Article Influence: 349.5] [Reference Citation Analysis (0)] |
| 3. | Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019;16:713-732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 777] [Cited by in RCA: 1555] [Article Influence: 259.2] [Reference Citation Analysis (2)] |
| 4. | de la Chapelle A, Peltomäki P. Genetics of hereditary colon cancer. Annu Rev Genet. 1995;29:329-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 5. | Liu Y, Zhang C, Wang Q, Wu K, Sun Z, Tang Z, Zhang B. Temporal Trends in the Disease Burden of Colorectal Cancer with Its Risk Factors at the Global and National Level from 1990 to 2019, and Projections Until 2044. Clin Epidemiol. 2023;15:55-71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 37] [Reference Citation Analysis (0)] |
| 6. | Zheng Y, Fu Y, Wang PP, Ding ZY. Neoantigen: A Promising Target for the Immunotherapy of Colorectal Cancer. Dis Markers. 2022;2022:8270305. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 7. | Sun L, Mao JJ, Yan Y, Xu Y, Yang Y. Patient Reported Traditional Chinese Medicine Spleen Deficiency Syndrome (TCM-SDS) Scale for Colorectal Cancer: Development and Validation in China. Integr Cancer Ther. 2021;20:15347354211020105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 8. | Xu DD, Hou XY, Wang O, Wang D, Li DT, Qin SY, Lv B, Dai XM, Zhang ZJ, Wan JB, Xu FG. A four-component combination derived from Huang-Qin Decoction significantly enhances anticancer activity of irinotecan. Chin J Nat Med. 2021;19:364-375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 9. | Pei T, Zheng C, Huang C, Chen X, Guo Z, Fu Y, Liu J, Wang Y. Systematic understanding the mechanisms of vitiligo pathogenesis and its treatment by Qubaibabuqi formula. J Ethnopharmacol. 2016;190:272-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 67] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 10. | Wang YN, Zou M, Wang D, Zhang ZK, Qu LP, Xu J, Shi CD, Gao F. An exploratory study on TCM syndrome differentiation in preoperative patients with colorectal cancer assisted by laboratory indicators. Heliyon. 2022;8:e10207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 11. | Lu Y, Zhou C, Zhu M, Fu Z, Shi Y, Li M, Wang W, Zhu S, Jiang B, Luo Y, Su S. Traditional chinese medicine syndromes classification associates with tumor cell and microenvironment heterogeneity in colorectal cancer: a single cell RNA sequencing analysis. Chin Med. 2021;16:133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 12. | Wang P, Ding S, Sun L, Feng Y, Guo K, Zhu Y, Huang D, Ruan S. Characteristics and differences of gut microbiota in patients with different Traditional Chinese Medicine Syndromes of Colorectal Cancer and normal population. J Cancer. 2020;11:7357-7367. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 13. | Han YH, Mun JG, Jeon HD, Kee JY, Hong SH. Betulin Inhibits Lung Metastasis by Inducing Cell Cycle Arrest, Autophagy, and Apoptosis of Metastatic Colorectal Cancer Cells. Nutrients. 2019;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 14. | Xia X, He J, Liu B, Shao Z, Xu Q, Hu T, Yu C, Liu X, Liao Y, Liu N, Huang H. Targeting ERα degradation by L-Tetrahydropalmatine provides a novel strategy for breast cancer treatment. Int J Biol Sci. 2020;16:2192-2204. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 15. | Gong J, Xing C, Wang LY, Xie SS, Xiong WD. L-Tetrahydropalmatine enhances the sensitivity of human ovarian cancer cells to cisplatin via microRNA-93/PTEN/Akt cascade. J BUON. 2019;24:701-708. [PubMed] |
| 16. | Yang L, Liu Y, Wang M, Qian Y, Dong X, Gu H, Wang H, Guo S, Hisamitsu T. Quercetin-induced apoptosis of HT-29 colon cancer cells via inhibition of the Akt-CSN6-Myc signaling axis. Mol Med Rep. 2016;14:4559-4566. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 17. | Al-Ghamdi MA, Al-Enazy A, Huwait EA, Albukhari A, Harakeh S, Moselhy SS. Enhancement of Annexin V in response to combination of epigallocatechin gallate and quercetin as a potent arrest the cell cycle of colorectal cancer. Braz J Biol. 2021;83:e248746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 18. | Raja SB, Rajendiran V, Kasinathan NK, P A, Venkatabalasubramanian S, Murali MR, Devaraj H, Devaraj SN. Differential cytotoxic activity of Quercetin on colonic cancer cells depends on ROS generation through COX-2 expression. Food Chem Toxicol. 2017;106:92-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 62] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 19. | Han M, Song Y, Zhang X. Quercetin Suppresses the Migration and Invasion in Human Colon Cancer Caco-2 Cells Through Regulating Toll-like Receptor 4/Nuclear Factor-kappa B Pathway. Pharmacogn Mag. 2016;12:S237-S244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 20. | Liu Y, Chen X, Cheng R, Yang F, Yu M, Wang C, Cui S, Hong Y, Liang H, Liu M, Zhao C, Ding M, Sun W, Liu Z, Sun F, Zhang C, Zhou Z, Jiang X. The Jun/miR-22/HuR regulatory axis contributes to tumourigenesis in colorectal cancer. Mol Cancer. 2018;17:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 100] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 21. | Corvinus FM, Orth C, Moriggl R, Tsareva SA, Wagner S, Pfitzner EB, Baus D, Kaufmann R, Huber LA, Zatloukal K, Beug H, Ohlschläger P, Schütz A, Halbhuber KJ, Friedrich K. Persistent STAT3 activation in colon cancer is associated with enhanced cell proliferation and tumor growth. Neoplasia. 2005;7:545-555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 279] [Cited by in RCA: 331] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 22. | Xiong H, Zhang ZG, Tian XQ, Sun DF, Liang QC, Zhang YJ, Lu R, Chen YX, Fang JY. Inhibition of JAK1, 2/STAT3 signaling induces apoptosis, cell cycle arrest, and reduces tumor cell invasion in colorectal cancer cells. Neoplasia. 2008;10:287-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 349] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 23. | Brannon AR, Vakiani E, Sylvester BE, Scott SN, McDermott G, Shah RH, Kania K, Viale A, Oschwald DM, Vacic V, Emde AK, Cercek A, Yaeger R, Kemeny NE, Saltz LB, Shia J, D'Angelica MI, Weiser MR, Solit DB, Berger MF. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol. 2014;15:454. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 244] [Cited by in RCA: 286] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 24. | Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2:a001008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1046] [Cited by in RCA: 1472] [Article Influence: 98.1] [Reference Citation Analysis (0)] |
| 25. | Peeters M, Price T. Biologic therapies in the metastatic colorectal cancer treatment continuum--applying current evidence to clinical practice. Cancer Treat Rev. 2012;38:397-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 26. | Ellis LM, Takahashi Y, Liu W, Shaheen RM. Vascular endothelial growth factor in human colon cancer: biology and therapeutic implications. Oncologist. 2000;5 Suppl 1:11-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 149] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 27. | Li D, Wang G, Jin G, Yao K, Zhao Z, Bie L, Guo Y, Li N, Deng W, Chen X, Chen B, Liu Y, Luo S, Guo Z. Resveratrol suppresses colon cancer growth by targeting the AKT/STAT3 signaling pathway. Int J Mol Med. 2019;43:630-640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 28. | Zhang Y, Liang S, Xiao B, Hu J, Pang Y, Liu Y, Yang J, Ao J, Wei L, Luo X. MiR-323a regulates ErbB3/EGFR and blocks gefitinib resistance acquisition in colorectal cancer. Cell Death Dis. 2022;13:256. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 29. | Dong S, Liang S, Cheng Z, Zhang X, Luo L, Li L, Zhang W, Li S, Xu Q, Zhong M, Zhu J, Zhang G, Hu S. ROS/PI3K/Akt and Wnt/β-catenin signalings activate HIF-1α-induced metabolic reprogramming to impart 5-fluorouracil resistance in colorectal cancer. J Exp Clin Cancer Res. 2022;41:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 257] [Article Influence: 85.7] [Reference Citation Analysis (0)] |
| 30. | Kim EK, Choi EJ. Pathological roles of MAPK signaling pathways in human diseases. Biochim Biophys Acta. 2010;1802:396-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1432] [Cited by in RCA: 1807] [Article Influence: 120.5] [Reference Citation Analysis (0)] |
| 31. | Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005;6:322-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 577] [Cited by in RCA: 854] [Article Influence: 42.7] [Reference Citation Analysis (0)] |