Published online Oct 16, 2022. doi: 10.12998/wjcc.v10.i29.10735
Peer-review started: May 27, 2022
First decision: July 12, 2022
Revised: August 10, 2022
Accepted: September 8, 2022
Article in press: September 8, 2022
Published online: October 16, 2022
Processing time: 124 Days and 22.6 Hours
Dedicator of cytokinesis 8 (DOCK 8) deficiency, also known as autosomal recessive hyper immunoglobulin E (IgE) syndrome, is a combined immunodeficiency disease that was first recognized in 2009. It is caused by genetic alterations (mutations or deletions) in the DOCK 8 gene and is characterized by multiple allergies, elevated IgE levels, and susceptibility to viral and bacterial infections. Early diagnosis is critical to optimize the success of stem cell transplantation.
This study reports the case of a pediatric patient with DOCK 8 deficiency who had negative genetic testing using multiplex primary immunodeficiency (PID) panel and whole-exome sequencing (WES) with a next-generation sequencing method. He presented with chronic diarrhea and was managed as celiac disease based on previous negative workup for immunodeficiency and duodenal biopsy. He developed a generalized vesicular rash which was thought to be dermatitis herpetiformis associated with celiac disease. However, it turned out to be Eczema herpeticum based on positive herpes simplex virus from blood and lesions. The diagnosis was re-evaluated after the child was found to have multiple viral, bacterial, and parasitic co-infections (herpes simplex virus, cytomegalovirus, Epstein-Barr virus, Salmonella, and cryptosporidiosis). Re-evaluation with target gene testing with copy number variation (CNV) analysis and Multiplex Ligation Probe Amplification (MLPA) showed a large homozygous deletion in the DOCK 8 gene, confirming the diagnosis of DOCK 8 deficiency.
Targeted gene testing with CNV analysis might detect deletions that can be missed by WES for diagnosing patients with PID.
Core Tip: Diagnosis of primary immunodeficiency syndromes is challenging, especially those involving the innate immune system. Whole-exome sequencing (WES) is a promising method for diagnosis but has limitations. This article reports a case of delayed diagnosis of Dedicator of cytokinesis 8 deficiency based on negative WES results and highlights the importance of specific gene testing, when the clinical features are suggestive of specific disease, rather than depending on WES alone.
- Citation: Alshengeti A. Eczema herpeticum vs dermatitis herpetiformis as a clue of dedicator of cytokinesis 8 deficiency diagnosis: A case report. World J Clin Cases 2022; 10(29): 10735-10741
- URL: https://www.wjgnet.com/2307-8960/full/v10/i29/10735.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i29.10735
The dedicator of cytokinesis 8 (DOCK 8) gene, which is located on chromosome 9q24, was first discovered in 2009 by Zhang et al[1] after an extensive workup of 11 patients with recurrent sinopulmonary infections, viral skin infections, elevated immunoglobulin E (IgE), and multiple allergies. DOCK 8 deficiency, along with other genetic defects, causes rare autosomal recessive hyper-IgE syndromes, while signal transducer and activator of transcription 3 (STAT3) defect causes the most common classical autosomal dominant hyper IgE[1,2]. This report describes a case of DOCK 8 deficiency, the diagnosis of which was challenging.
An 8-year-old Yemeni boy was referred to our hospital for chronic diarrhea workup and management.
Upon admission to our hospital, he had diarrhea with 9-12 episodes of loose bowel motion per day with mucus but no blood. There was no history of vomiting, fever, cough or shortness of breath.
He had a history of recurrent skin abscesses, eczema, and multiple food allergies since the age of nine months. He was admitted at the ages of 18 mo and 3 years with community-acquired pneumonia. He had a history of recurrent chronic diarrhea that occurred 3-5 times per year and lasted 3-6 wk with increased baseline bowel habits (2-3 times/day). Two months before admission to our hospital, he developed severe diarrhea that led to hypovolemic shock and was admitted to the pediatric intensive care unit at the referring hospital. He stayed in the hospital for 45 days and lost approximately 7 kg of body weight (decreased from 21 kg to 14 kg).
The patient was born through a normal vaginal delivery with no major events or hospital admission in the first year of life. He had a history of poor weight gain since the age of three years. His developmental milestones were normal for his age. He had been vaccinated for up to 2 years of age.
The patient was evaluated at the age of 7 years for primary immunodeficiency, and the workup showed elevated IgE at a level of 2200 IU/mL (reference 25-550 IU/mL) with normal lymphocytes on flow cytometry. The primary immunodeficiency (PID) genetic testing panel, including the DOCK 8 gene, was negative for any pathogenic mutation. This panel uses a syndrome-based technique via simultaneous next-generation sequencing of the targeted genes (Saudi Mendeliome Group, Riyadh, Saudi Arabia, 2015)[3]. In addition, whole-exome sequencing (WES) was sent from the referring hospital before transfer and later turned negative for any pathogenic mutation (details regarding the WES methodology are outlined in Supplementary material 1.
The parents were first-degree relatives with another 10 and 3-year-old male and 5-year-old female healthy offspring.
On examination, he appeared ill and cachectic, with a loss of subcutaneous fat. His vital signs were normal without fever. His weight was 13.2 kg (below the 3rd percentile), height was 107 cm (below the 3rd percentile), and body mass index was 11.5 kg/m2. Systemic examination revealed dry eczematous skin with ulcerated lesions behind the left ear.
Initial and follow up laboratory investigation showed leukocytosis (15–32 × 109/L; normal range: 4–11 × 109/L), eosinophilia (0.9-3 × 109/L; normal range: 0.1-0.35 × 109/L), and mildly elevated liver enzymes. The workup for chronic diarrhea showed a positive stool culture for Salmonella spp., for which he received ciprofloxacin for 10 days. The patient was started on total parenteral nutrition and was scheduled for upper and lower GI endoscopy to rule out celiac disease and other pathologies.
Upper GI endoscopy with duodenal biopsy showed increased intraepithelial T-lymphocytes, consistent with a clinical history of celiac disease. In addition, there were increased kappa light chain-restricted plasma cells present in the lamina propria. The patient was diagnosed with celiac disease with celiac crisis based on his clinical presentation and duodenal biopsy findings. A gluten-free diet, along with other nutritional supplements, was initiated.
Abdominal ultrasound and chest radiography showed unremarkable findings.
The infectious disease team was consulted, as diarrhea persisted despite a gluten-free diet for three weeks, to rule out other infectious causes.
Stool testing for other pathogens revealed a positive Cryptosporidium antigen. The patient was administered paromomycin and azithromycin for 14 days. His diarrhea improved after 10 days of treatment; the stool frequency decreased to one to three times per day, with more formed stools. However, the repeat antigen test was positive. It was not clear whether the improvement in diarrhea was secondary to a gluten-free diet or antimicrobials for Cryptosporidium.
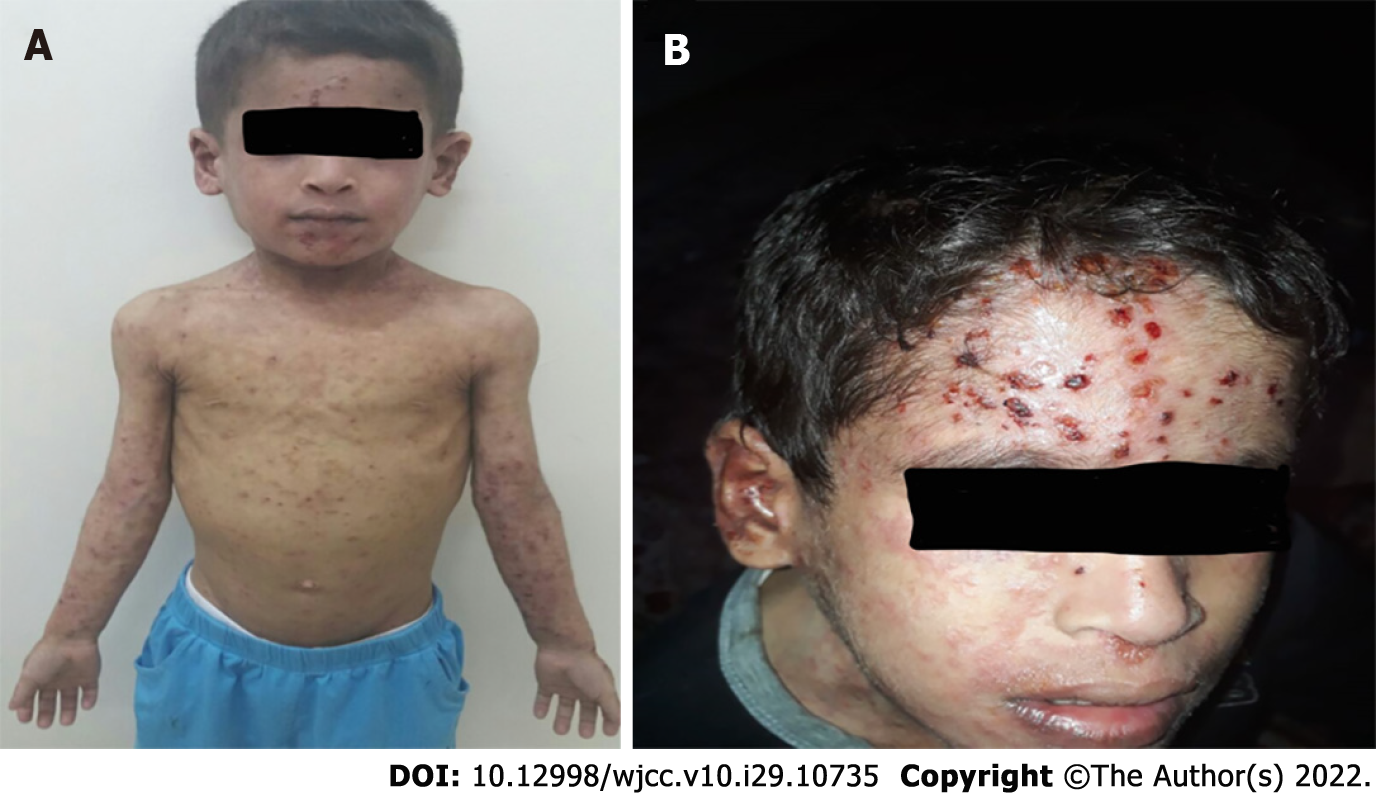
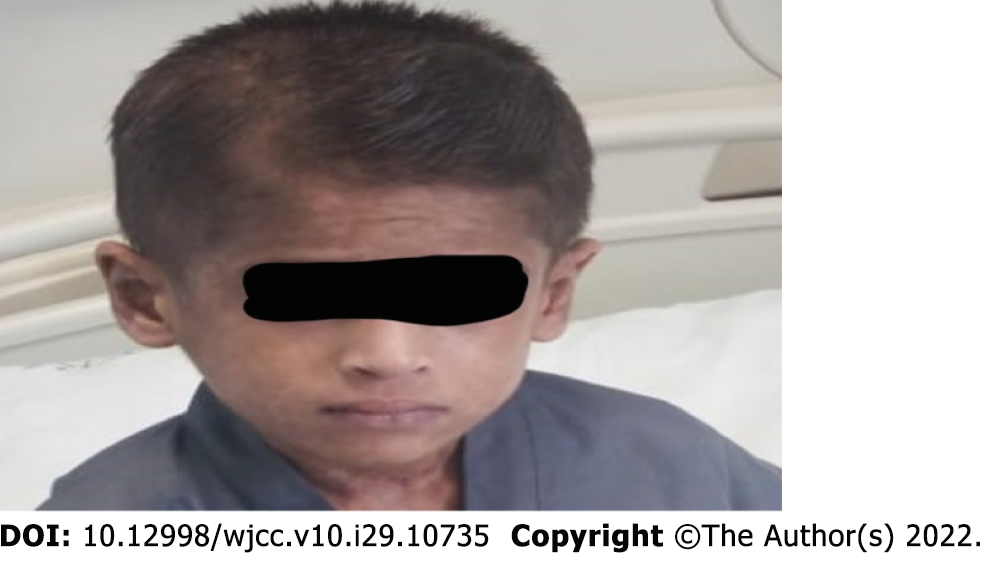
During the sixth week of admission, the patient developed a vesicular skin rash involving the face, upper limbs, and trunk (Figure 1A and B). The rash was itchy without fever. Based on the diagnosis of celiac disease, dermatitis herpetiformis was suspected. Therefore, dapsone treatment was initiated without improvement. At this time, a herpes simplex virus (HSV) swab from the lesion and serum for polymerase chain reaction (PCR) was sent, which turned positive for both. Viral work-up showed positive cytomegalovirus (CMV) and Epstein-Barr virus (EBV) viremia with low viral load. The skin rash resolved after treatment with intravenous acyclovir for 14 days (Figure 2). However, the patient continued to have recurrent HSV skin infections in subsequent months, which improved with valacyclovir prophylaxis. Repeated immune workup showed elevated IgE levels of 5500 IU/mL (reference 25-550 IU/mL), low T-lymphocytes (CD4+, CD8+, and NK), and high B-lymphocytes.
Based on these findings, the diagnosis of DOCK 8 deficiency was reconsidered despite negative previous genetic testing in the primary immunodeficiency panel and WES. Therefore, targeted gene sequencing for the DOCK 8 gene was performed using next-generation sequencing (NGS). NGS did not reveal any point mutations. However, due to the classic phenotype, a copy number variation (CNV) analysis of NGS data was performed which showed a large homozygous deletion in the DOCK 8 gene, including exons 23 to 48 (note that the DOCK 8 gene has 48 exons). This deletion was confirmed by Multiplex Ligation Probe Amplification (MLPA) (SALSA® MLPA® Probemix, MRC Holland, Amsterdam, The Netherlands). The CNV analysis data and the full result report are shown in Supplementary material 2 and 3, respectively.
Based on this finding, the patient was diagnosed with DOCK 8 deficiency.
The patient’s condition improved after the initiation of regular intravenous immunoglobulin (IVIG) therapy. The patient was referred to a bone marrow transplantation (BMT) center. He underwent hematopoietic stem cell transplantation (HSCT) from a partially mismatched sibling donor at the age of 10 years.
Unfortunately, the transplantation was complicated by engraftment failure, and the patient died from multiorgan failure after 6 wk.
This case report describes an unusual presentation of PID with viral, bacterial, and parasitic co-infections in a child. This led to a high index of suspicion and a final diagnosis of DOCK 8 deficiency secondary to a large deletion in the DOCK 8 gene. The diagnosis was delayed because of the negative results of the PID panel for point mutations, as well as negative WES. The diagnosis was made using target gene testing and CNV analysis.
The clinical and laboratory findings of the patient reported here are consistent with most cases reported in the literature[1,4]. Eczema, food allergies, recurrent and persistent viral infections, eosinophilia, elevated IgE, and low T lymphocyte levels are characteristics of DOCK 8 deficiency[1,4]. A unique feature of the patient reported here is multiple coinfections with different viruses (HSV, CMV, and EBV) and severe cryptosporidiosis at the same time. Although these infections have been reported among DOCK 8 patients separately, to our knowledge, coinfection has not been reported[4,5]. This coinfection was a clue to the diagnosis in this case.
The genetic deletion in our patient was detected by CNV analysis of the target gene NGS data and confirmed by MLPA after negative results for point mutation obtained from NGS. NGS is a new DNA sequencing method that can be used for whole-genome sequencing (WGS), WES, and targeted genes sequencing[6]. One of the known limitations of NGS in WES and targeted gene sequencing is the inability to detect deletions and duplications in the tested genes, as this requires a different analysis pipeline[6]. This limitation can be overcome by WGS, but it is expensive and time-consuming[6]. However, a new method for CNV analysis of NGS data using software is a promising technique for detecting this type of deletion[6]. Both point mutations and deletions have been reported in patients with DOCK 8 deficiency[7]. Therefore, testing for both deletion and point mutation should be performed when the phenotype is consistent with the genetic defect to overcome the false-negative result when only point mutation is tested, while the cause is a large deletion, as in our patient.
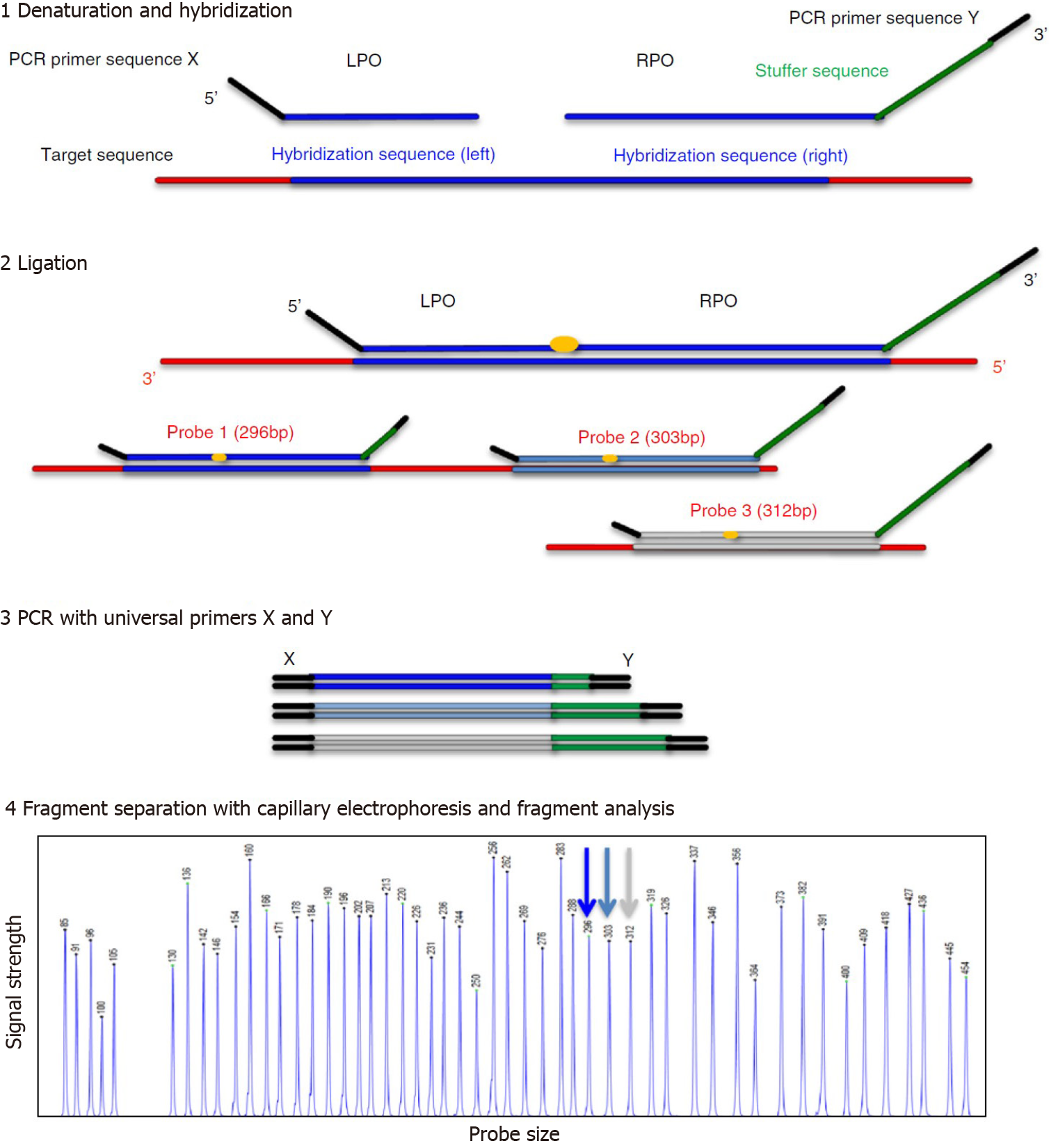
The CNV analysis of our patient’s NGS data was confirmed by MLPA. MLPA is a molecular technique that was developed by MRC Holland (Amsterdam, The Netherlands) in 2002, and can detect copy number variations (duplication or deletion), point mutations, and quantify the messenger RNA[8]. Figure 3 illustrates the MLPA method[9].
Early diagnosis and HSCT play critical roles in the treatment of patients with DOCK 8 deficiency. Evidence regarding BMT in patients with DOCK 8 deficiency is growing. A large retrospective multicenter study involving 81 patients with DOCK 8 deficiency showed that patients who underwent HSCT before age of 8 had better 2-year overall survival than those who were older than 8 (96% vs 78%)[10].
This case report shows the limitations of genetic testing and the essential rules of clinical findings to guide laboratory testing. Our study suggests that DOCK 8 deficiency should be considered in any child presenting with the typical symptoms, and both mutations and deletions should be tested to avoid delays in diagnosis and optimize outcomes with early HSCT.
The author would like to thank Dr. Abdulhadi Habeb, Dr. Yousef El-Hag, The Department of Pediatrics, and Mr. Abdurhaman Aboud, Laboratory Medicine Department, Prince Mohammad Bin Abdulaziz Hospital, The Ministry of National Guard Health Affairs, Al-Madinah, Saudi Arabia, for their support in genetic testing. Author would like to thank Suvi Savola, MRC, Holand and the Publisher Wolters Kluwer Health, Inc. for the permission to reuse figure 3.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Immunology
Country/Territory of origin: Saudi Arabia
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bredt LC, Brazil; Su YY, Taiwan S-Editor: Ma YJ L-Editor: A P-Editor: Ma YJ
| 1. | Zhang Q, Davis JC, Lamborn IT, Freeman AF, Jing H, Favreau AJ, Matthews HF, Davis J, Turner ML, Uzel G, Holland SM, Su HC. Combined immunodeficiency associated with DOCK8 mutations. N Engl J Med. 2009;361:2046-2055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 552] [Cited by in RCA: 536] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 2. | Alsum Z, Hawwari A, Alsmadi O, Al-Hissi S, Borrero E, Abu-Staiteh A, Khalak HG, Wakil S, Eldali AM, Arnaout R, Al-Ghonaium A, Al-Muhsen S, Al-Dhekri H, Al-Saud B, Al-Mousa H. Clinical, immunological and molecular characterization of DOCK8 and DOCK8-like deficient patients: single center experience of twenty-five patients. J Clin Immunol. 2013;33:55-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 3. | Saudi Mendeliome Group. Comprehensive gene panels provide advantages over clinical exome sequencing for Mendelian diseases. Genome Biol. 2015;16:134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 142] [Cited by in RCA: 150] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 4. | Aydin SE, Kilic SS, Aytekin C, Kumar A, Porras O, Kainulainen L, Kostyuchenko L, Genel F, Kütükcüler N, Karaca N, Gonzalez-Granado L, Abbott J, Al-Zahrani D, Rezaei N, Baz Z, Thiel J, Ehl S, Marodi L, Orange JS, Sawalle-Belohradsky J, Keles S, Holland SM, Sanal Ö, Ayvaz DC, Tezcan I, Al-Mousa H, Alsum Z, Hawwari A, Metin A, Matthes-Martin S, Hönig M, Schulz A, Picard C, Barlogis V, Gennery A, Ifversen M, van Montfrans J, Kuijpers T, Bredius R, Dückers G, Al-Herz W, Pai SY, Geha R, Notheis G, Schwarze CP, Tavil B, Azik F, Bienemann K, Grimbacher B, Heinz V, Gaspar HB, Aydin R, Hagl B, Gathmann B, Belohradsky BH, Ochs HD, Chatila T, Renner ED, Su H, Freeman AF, Engelhardt K, Albert MH; inborn errors working party of EBMT. DOCK8 deficiency: clinical and immunological phenotype and treatment options - a review of 136 patients. J Clin Immunol. 2015;35:189-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 206] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 5. | Su HC. Dedicator of cytokinesis 8 (DOCK8) deficiency. Curr Opin Allergy Clin Immunol. 2010;10:515-520. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 88] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 6. | Ellingford JM, Campbell C, Barton S, Bhaskar S, Gupta S, Taylor RL, Sergouniotis PI, Horn B, Lamb JA, Michaelides M, Webster AR, Newman WG, Panda B, Ramsden SC, Black GC. Validation of copy number variation analysis for next-generation sequencing diagnostics. Eur J Hum Genet. 2017;25:719-724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 75] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 7. | Engelhardt KR, McGhee S, Winkler S, Sassi A, Woellner C, Lopez-Herrera G, Chen A, Kim HS, Lloret MG, Schulze I, Ehl S, Thiel J, Pfeifer D, Veelken H, Niehues T, Siepermann K, Weinspach S, Reisli I, Keles S, Genel F, Kutukculer N, Camcioğlu Y, Somer A, Karakoc-Aydiner E, Barlan I, Gennery A, Metin A, Degerliyurt A, Pietrogrande MC, Yeganeh M, Baz Z, Al-Tamemi S, Klein C, Puck JM, Holland SM, McCabe ER, Grimbacher B, Chatila TA. Large deletions and point mutations involving the dedicator of cytokinesis 8 (DOCK8) in the autosomal-recessive form of hyper-IgE syndrome. J Allergy Clin Immunol. 2009;124:1289-302.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 402] [Cited by in RCA: 372] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 8. | Principle of MLPA. Accessed August 14 2022. Available from: https://www.mrcholland.com/technology/mlpa. |
| 9. | Hömig-Hölzel C, Savola S. Multiplex ligation-dependent probe amplification (MLPA) in tumor diagnostics and prognostics. Diagn Mol Pathol. 2012;21:189-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 103] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 10. | Aydin SE, Freeman AF, Al-Herz W, Al-Mousa HA, Arnaout RK, Aydin RC, Barlogis V, Belohradsky BH, Bonfim C, Bredius RG, Chu JI, Ciocarlie OC, Doğu F, Gaspar HB, Geha RS, Gennery AR, Hauck F, Hawwari A, Hickstein DD, Hoenig M, Ikinciogullari A, Klein C, Kumar A, Ifversen MRS, Matthes S, Metin A, Neven B, Pai SY, Parikh SH, Picard C, Renner ED, Sanal Ö, Schulz AS, Schuster F, Shah NN, Shereck EB, Slatter MA, Su HC, van Montfrans J, Woessmann W, Ziegler JB, Albert MH; Inborn Errors Working Party of the European Group for Blood and Marrow Transplantation and the European Society for Primary Immunodeficiencies. Hematopoietic Stem Cell Transplantation as Treatment for Patients with DOCK8 Deficiency. J Allergy Clin Immunol Pract. 2019;7:848-855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |