Published online Sep 16, 2022. doi: 10.12998/wjcc.v10.i26.9254
Peer-review started: March 16, 2022
First decision: April 10, 2022
Revised: April 21, 2022
Accepted: August 12, 2022
Article in press: August 12, 2022
Published online: September 16, 2022
Processing time: 169 Days and 7.9 Hours
Gene expression of inflammatory cytokines may take part in the pathophysiology of different forms of gastroesophageal reflux disease (GERD).
To explore gene expression of inflammatory cytokines in esophageal mucosa in patients with erosive esophagitis (EE) and non-erosive forms of GERD (NERD) and its association with data of esophageal multichannel intraluminal impedance-pH (MII-pH) measurements.
This was a single-center prospective study. Esophageal mucosa samples were taken from the lower part of the esophagus during endoscopy. Expression of interleukin (IL)-1β, IL-10, IL-18, tumor necrosis factor α (TNFA), toll-like receptor 4 (TLR4), GATA binding protein 3 (GATA3), differentiation cluster 68 (CD68) and β-2 microglobulin genes in esophageal mucosa was assessed with ImmunoQuantex assays. MII-pH measurements were performed on all the participants. Diagnosis of GERD was confirmed by the results of the MII-pH data. Based on the endoscopy, patients were allocated to the groups of EE and NERD. The control group consisted of non-symptomatic subjects with normal endoscopy and MII-pH results. We used nonparametric statistics to compare the differences between the groups. Association of expression of the mentioned genes with the results of the MII-pH data was assessed with Spearman’s rank method.
Data from 60 patients with GERD and 10 subjects of the control group were available for the analysis. Higher expression of IL-18 (5.89 ± 0.4 vs 5.28 ± 1.1, P = 0.04) and GATA3 (2.92 ± 0.86 vs 2.23 ± 0.96, P = 0.03) was found in the EE group compared to NERD. Expression of IL-1β, IL-18, TNFA, and TLR4 was lower (P < 0.05) in the control group compared to EE and NERD. Esophageal acid exposure correlated with the expression of IL-1β (Spearman’s rank r = 0.29), IL-18 (r = 0.31), TNFA (r = 0.35), GATA3 (r = 0.34), TLR4 (r = 0.29), and CD68 (r = 0.37). Mean esophageal рН correlated inversely with the expression of IL-18, TNFA, GATA3, TLR4, and CD68. No association of gene expression with the number of gastroesophageal refluxes was found.
In patients with EE, local expression of IL-18 and GATA3 was higher compared to subjects with NERD. Esophageal acid exposure correlated directly with expression of IL-1β, IL-18, TNFA, TLR4, CD68, and β-2 microglobulin genes. Inverse correlation was revealed between expression of IL-18, TNFA, GATA3, TLR4, and CD68 and mean esophageal pH.
Core Tip: Local expression of cytokines may be involved in pathophysiology of different forms of gastroesophageal reflux disease. We found a different profile of local expression of cytokines in subgroups of patients with erosive esophagitis and non-erosive forms of gastroesophageal reflux disease. For the first time we have revealed a correlation between gene expression of interleukin-18, tumor necrosis factor alpha, GATA binding protein 3, toll-like receptor 4, differentiation cluster 68 and mean esophageal pH and an association of acid exposure with expression of interleukin-1β, interleukin-18, tumor necrosis factor alpha, toll-like receptor 4, differentiation cluster 68 and β-2 microglobulin.
- Citation: Morozov S, Sentsova T. Local inflammatory response to gastroesophageal reflux: Association of gene expression of inflammatory cytokines with esophageal multichannel intraluminal impedance-pH data. World J Clin Cases 2022; 10(26): 9254-9263
- URL: https://www.wjgnet.com/2307-8960/full/v10/i26/9254.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i26.9254
Gastroesophageal reflux disease (GERD) is a common disorder that significantly impairs patient quality of life due to persistent symptoms caused by the reflux of gastric content into the esophagus. Besides typical symptoms like heartburn and regurgitation, it has a vast spectrum of manifestations[1]. Pathogenesis of GERD is complex, with a paramount role of impaired esophageal-gastric junction motility, delayed gastric emptying, and damage of the esophageal mucosa with aggressive content of the refluxate[2]. Traditionally, the grade of esophageal mucosa damage and intensity of symptoms were thought to be directly related to the acidity of the reflux content[3]. However, recent data suggest that this relationship is not linear, and the spectrum of manifestations may be genetically determined or be a result of a balance in factors responsible for a local inflammatory response in the esophageal mucosa, perception of refluxate, and processing of signals by the peripheral and central nervous systems[4,5]. For example, it was shown that demographic profiles of patients with GERD differ. Severe grades of esophagitis are commonly found in elderly men of white race and are associated with abdominal-type obesity, while persistent symptoms of the disease in the absence of esophageal mucosa lesions are more typical for young women[6]. When not treated, esophageal mucosa breaks remain stable with time in most of the cases[7]. This allows GERD to be distinguished into different forms: erosive esophagitis (EE), Barrett’s esophagus, and non-erosive form of GERD (NERD), which despite the common etiology, may differ by pathophysiology.
Experimental studies suggest that an exposure of refluxate does not lead to a chemical burn but induces inflammatory cytokine synthesis [like interleukin (IL)-1β and IL-8], followed by migration of lymphocytes and neutrophils into the mucosa and its further damage[8]. These data formed the basis of the theory of “cytokine sizzle” as the basis of the pathogenesis of GERD[9]. The endoscopic phenotype seen in a patient is a result of the balance between proinflammatory and anti-inflammatory factors that counteract. This balance is genetically determined, as cytokine production depends on the expression of the related genes. Regulation of the gene expression is complex, and its dependence on the acidity of the refluxate and types of gastroesophageal refluxes (GER) is insufficiently studied. Moreover, cytokine profiles studied in humans are few, and reports on the involvement of IL-18, transcription factor GATA binding protein 3 (GATA3), toll-like receptor 4 (TLR4), and differentiation cluster 68 (CD68) in local inflammatory response in subjects with different forms of GERD are still lacking.
The aims of the present work were to study: (1) Gene expression of cytokines in esophageal mucosa of patients with EE and NERD; and (2) Association of the expression with the data of esophageal multichannel intraluminal impedance-pH (MII-pH) measurements.
The study was approved by the Ethics Committee of the Federal Research Center of Nutrition and Biotechnology. The data of complex examinations of patients served as the source for the study.
Inclusion criteria in the present study including: (1) Males and females, ≥ 18 years and ≤ 80 years; and (2) Written informed consent to participate in this study.
(1) The use of medications that could influence manifestations of GERD, damage esophageal mucosa, affect esophageal motility or influence cytokine response. This included, but was not limited to the use of the following drugs: Glucocorticosteroids, nonsteroid anti-inflammatory drugs (except topical agents more than 2 wk before the enrollment), calcium channel antagonist, nitrates, and agents that affect adrenergic or acetylcholine receptors, or have a local irritating effect on the gastrointestinal mucosa. The use of antisecretory agents (proton pump inhibitors or H2-histamine receptor blockers) was not allowed for at least 4 wk before the enrollment; (2) History of chest and/or abdominal surgery in the anamnesis (excluding appendectomy and cholecystectomy performed at least 6 mo prior to the enrollment); (3) History of esophageal varicose bleeding or presence of esophageal varices on endoscopy; (4) Impossibility to perform at least one type of the examinations required by the study protocol; and (5) Active systemic connective tissue or autoimmune disorders, decompensated condition of any organ or system, and general condition of a patient that made him inapplicable for the study at the discretion of the investigator.
The data of the subjects were not included in the final analysis in the case when the data of examinations required by the study protocol were missing or incomplete.
(1) Clinical data (presence of heartburn and/or acid regurgitation at least once per week; presence of symptoms for more than 6 mo that have stayed relevant for at least the last 3 mo; history of treatment with proton pump inhibitors with positive effect in the past); (2) GERD-Q score ≥ 8 (validated language-specific version of the international questionnaire was used)[10,11]; and (3) Results of 24 h esophageal MII-pH according to the Lyon consensus[12].
Twenty-four hour MII-pH recordings: Twenty-four hour MII-pH recordings were performed with the use of a pH-impedance recorder Ohmega (MMS, Enschede, The Netherlands) and the manufacturer’s software (Solar Gastro; MMS) and standard pH-impedance catheters with 2 pH and 6 impedance channels (Unisensor, AG, Wiesendangen, Switzerland). The distal pH-sensor was placed 5 cm above the esophagogastric junction, based on the results of high-resolution esophageal manometry. Values of acid exposure time in the lower esophagus more than 6% during the 24 h and/or number of GER > 80/d were considered abnormal. When acid exposure time and number of refluxes were lower than the above mentioned limits, association of symptoms with GER was taken into account (sensitivity index > 80%, symptom index > 50%)[12].
Endoscopic evaluation: Endoscopic evaluation of esophageal, stomach, and duodenal mucosa was necessary to exclude concomitant pathology and to establish the presence of esophagitis. Olympus Exera II CV-180 panendoscope (Olympus, Tokyo, Japan) was used to perform endoscopy. EE was described according to the Los Angeles classification, 1999[13].
Based on the results, patients with GERD were allocated either to the group of EE or to the group of NERD. The control group consisted of the subjects eligible for the study according to the inclusion/ exclusion criteria who did not experience symptoms of GERD and had normal esophageal endoscopy and data of MII-pH measurements.
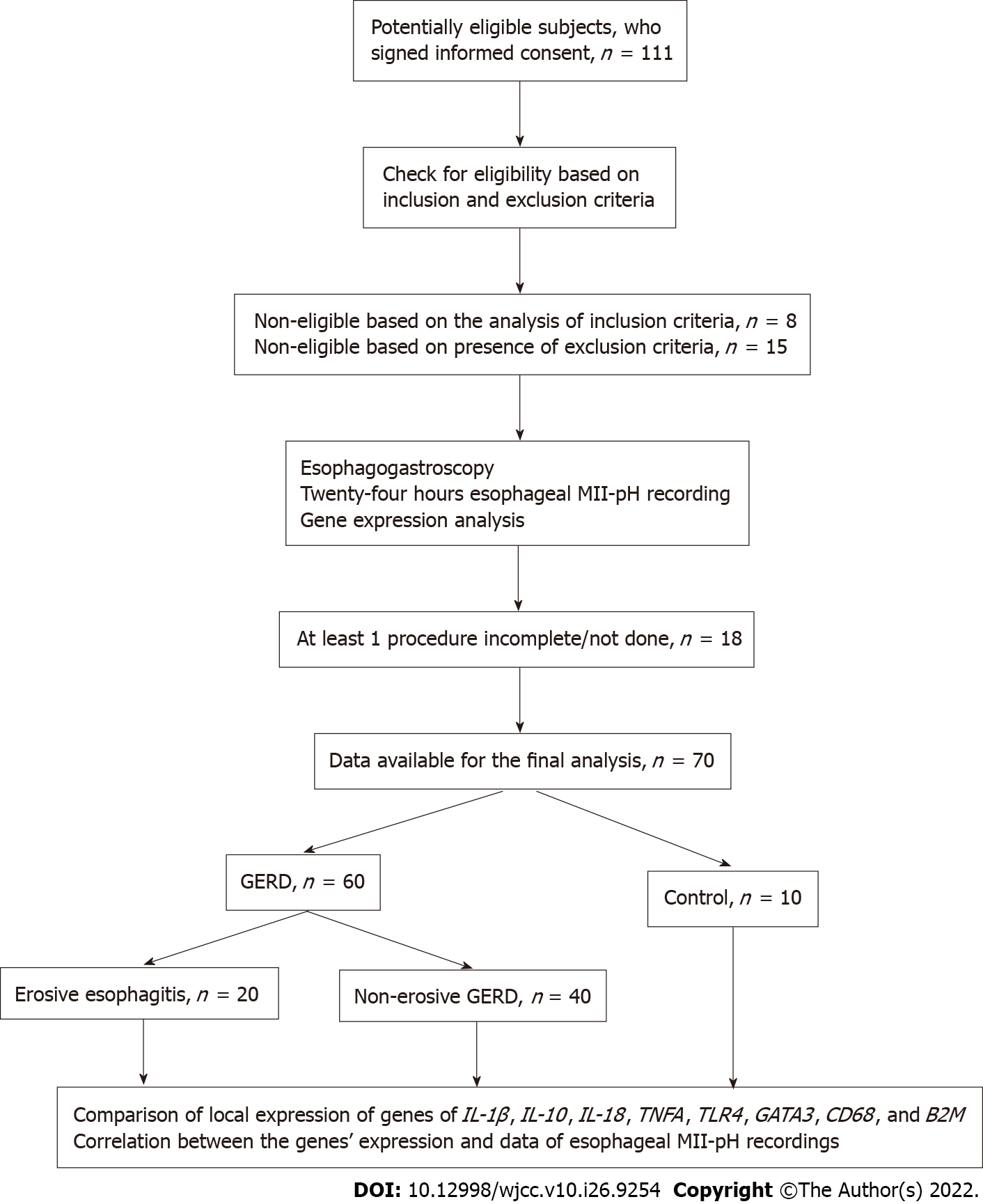
Gene expression: Samples of esophageal mucosa were taken from the distal part of the esophagus (5 cm above the esophagogastric junction) with sterile forceps during endoscopy. In the case of EE, the samples were obtained from the margin of erosions. The tissues were immediately placed in 1.5 mL plastic tubes containing 500 μL RNA stabilizer (IntactRNA; Evrogen, Moscow, Russia) and stored up to 2 wk at -20 °C. RNA purification was performed as previously described with the use of PREP-NA extraction kit (DNA-Technology, Moscow, Russia) followed by ImmunoQuantex assay (DNA-Technology)[14,15]. This test system provides a reverse transcription reaction to obtain an mRNA template complementary to DNA from the previously isolated pool of DNA and RNA. Further, this mRNA template is amplified by real-time PCR. The reverse transcription reaction (synthesis of cDNA on the obtained RNA) was carried out in a volume of 40 μL. Specific oligonucleotides and M-MuLV reverse transcriptase (Evrogen) were used as primers for reverse transcription. We traced reproducibility and linearity of real time quantitative PCR assays to ensure quality control. The assessment of gene expression was based on the comparison of threshold cycles method [2 delta Cq method with normalization to reference gene β-2 microglobulin (B2M)]. The relative value of gene X expression was calculated by the formula: Expression (X) = 2-[Cp (x) - NF]. After the amplification stage, the mRNA expression level was calculated (based on the indicator of the cycle) for the following genes: IL-1β, IL-10, IL-18, tumor necrosis factor α (TNFA), TLR4, GATA3, CD68, and B2M. Study design, subject flow, and allocation chart are shown in Figure 1.
Statistical processing: Statistical processing of the results was carried out with commercially available software: MS Excel 2016 (Microsoft, Redmond, WA, United States) and Statistica 10 (StatSoft, Tulsa, OK, United States). The data are presented as means and standard deviation. Nonparametric statistics (Mann-Whitney’s U criteria, Spearman’s rank R) were used to compare the results between the groups. Values of P < 0.05 were considered significant.
Overall, 111 patients were enrolled; 23 subjects were not eligible according to inclusion/exclusion criteria. Incomplete results were obtained in 18 patients. The data of 60 patients with GERD [26 (43.3%) men and 34 (56.7%) women], mean age of 54.6 ± 15.6 years (18.3% Asians, 81.7% Caucasian) and of 10 patients in the control group (60% men; 46.2 ± 13.0-years-old; 20% Asians) were available for the final analysis (Figure 1). Those with EE were younger: 47.5 ± 13 years vs 58.2 ± 15.8 years, P = 0.007. No significant difference in distribution by sex was found between the groups. The relative number of women in EE group was 65% and was 40% in the NERD group, P = 0.06.
The results of the MII-pH and gene expression analysis are shown in Tables 1 and 2. The total number of GER, number of acid GERs, and acid exposure time were higher in the EE group compared to the NERD group. However, the number of weak-acid, non-acid refluxes, and mean pH values in the distal part of the esophagus did not differ in the GERD groups.
| Control, n = 10 | EE, n = 20 | NERD, n = 40 | P value1 | |
| Total number of GERs, n, mean ± SD | 25.1 ± 13.7 | 75.7 ± 27.5 | 59.1 ± 30.6 | 0.01 |
| Acid GERs, n, mean ± SD | 19.8 ± 11.4 | 40.3 ± 26.4 | 19.8 ± 9.7 | 0.004 |
| Weak-acid GERs, n, mean ± SD | 3.4 ± 2.5 | 25.4 ± 22.9 | 27.0 ± 25.1 | 0.97 |
| Non-acid GERs, n, mean ± SD | 1.9 ± 1.7 | 0.9 ± 1.0 | 2.4 ± 6.3 | 0.73 |
| High GERs, %, mean ± SD | 8.9 ± 8.0 | 46.5 ± 21.7 | 38.0 ± 21.4 | 0.28 |
| AET, %, mean ± SD | 2.3 ± 1.6 | 12.1 ± 3.7 | 9.1 ± 4.8 | 0.001 |
| Mean pH, mean ± SD | 6.3 ± 0.5 | 5.4 ± 0.4 | 5.8 ± 1.0 | 0.075 |
| Control, n = 10 | EE, n = 20 | NERD, n = 40 | |
| IL-1β, log, mean ± SD | 1.1 ± 0.2b,c | 3.7 ± 0.4 | 3.8 ± 0.6 |
| IL-10, log, mean ± SD | 0.9 ± 0.1 | 1.4 ± 1.1 | 1.5 ± 1.8 |
| IL-18, log, mean ± SD | 2.1 ± 0.3b,c | 5.9 ± 0.4 | 5.3 ± 1.1a |
| TNFA, log, mean ± SD | 0.2 ± 0.1b,c | 3.2 ± 0.4 | 3.1 ± 0.9 |
| TLR4, log, mean ± SD | 0.9 ± 0.2b,c | 2.6 ± 0.7 | 2.5 ± 1.2 |
| GATA3, log, mean ± SD | 1.9 ± 0.2b | 2.9 ± 0.9 | 2.2 ± 1.0a |
| CD68, log, mean ± SD | 3.2 ± 1.3c | 4.9 ± 0.6 | 5.0 ± 1.1 |
| B2M, log, mean ± SD | 4.0 ± 1.2 | 5.6 ± 0.6 | 5.5 ± 1.1 |
We found direct correlation between gene expression of IL-1β, IL-18, TNFA, TLR4, CD68, and B2M and acid exposure time in the distal esophagus (Table 3). There was an inverse correlation between the expression of IL-18, TNFA, GATA3, TLR4, and CD68 and mean pH values.
| No. of GERs | Acid GERs | Weak-acid GERs | Non-acid GERs | High GERs | Mean pH | AET | |
| IL-1β | -0.02 | 0.14 | -0.11 | 0.04 | -0.03 | -0.24 | 0.29a |
| IL-10 | -0.15 | -0.02 | -0.16 | 0.02 | 0.01 | -0.21 | 0.18 |
| IL-18 | 0.01 | 0.06 | -0.02 | 0.01 | 0.01 | -0.28a | 0.31a |
| TNFA | -0.03 | 0.08 | -0.12 | 0.01 | -0.12 | -0.33a | 0.35a |
| GATA3 | 0.04 | 0.15 | -0.13 | -0.1 | 0.02 | -0.28a | 0.34a |
| TLR4 | -0.05 | 0.04 | -0.15 | 0.06 | -0.04 | -0.28a | 0.29a |
| CD68 | 0.01 | 0.05 | -0.03 | -0.01 | -0.08 | -0.39a | 0.37a |
| B2M | 0.04 | 0.10 | -0.04 | 0.17 | 0.09 | -0.26 | 0.34a |
In this study, we showed an association of a local inflammatory response of the esophageal mucosa with data of MII-pH recordings in patients with different forms of GERD that was confirmed according to the current standards. Expression of inflammatory cytokines was assessed in a number of previously published works[16-18]. In most of them, symptoms and endoscopic data were used to confirm GERD. Nowadays, this approach is not fully reliable because esophageal erosions may be caused by different factors, and subjects with other diseases (eosinophilic esophagitis, depression) often experience similar symptoms[12]. In our study, diagnosis of GERD was confirmed by the data of MII-pH measurements. The use of this technique allowed us to confirm the presence of pathological reflux and to analyze the association of gene expression of inflammatory cytokines with different types of GER.
According to the results, acid exposure time (proportion of time with pH < 4.0 per day at 5 cm above upper border of gastroesophageal sphincter) and, to a lesser extent, mean esophageal pH were the factors associated with gene expression of inflammatory cytokines. Zavala-Solares et al[19] succeeded in showing that the expression of IL-1β and TNFA were higher when an abnormal acid exposure in the esophagus (pH < 4 more than 4.2% of time per day) was present. Unfortunately, in the mentioned study diagnosis of GERD was based predominantly on symptoms and endoscopy and did not include esophageal impedance monitoring. Similar to our results, they failed to reveal association of IL-10 expression with acid exposure in the esophagus[19]. This cytokine plays an anti-inflammatory and modulatory effect on inflammation and reduces production of TNFA, IL-1β, IL-12, and secretion of interferon gamma[20]. The lack of an inhibitory effect may cause an imbalance in the inflammatory response and predominance of proinflammatory factors, which leads to tissue damage[21].
To our knowledge, a negative correlation of mean pH values at the lower esophagus with the expression of IL-18, TNFA, GATA3, TLR4, and CD68 has not been reported yet. The obtained results may be important from both scientific and practical viewpoints. Indeed, it is difficult to explain the presence of endoscopic or histological changes within the mucosa only by aggressive nature of the reflux content. Significant overlap in the acid exposure in the esophagus was revealed in subjects with different forms of GERD[22,23]. Similarly, in the present study the mean pH values in the esophagus did not differ in subjects with EE and NERD. This suggests that a different response may be caused by genetically determined local inflammatory response[24]. Increased gene expression of inflammatory cytokines causes higher local concentrations of the cytokines and triggers a cascade of corresponding reactions that impair integrity of the mucosa[17,24]. The correlation analysis does not provide information on the direction of influence. However, previous studies with the use of proton pump inhibitors (lansoprazole 30 mg/d for 8 wk) showed a significant decrease in gene expression of another inflammatory cytokine (IL-8) after the treatment[16,25].
It was reported that GATA3, TNFA, and IL-10 may increase secretion of immunoglobulin E and stimulate migration of eosinophils[26,27]. It may have diagnostic value and help explain why some of the patients with eosinophilic esophagitis respond to therapy with proton pump inhibitors[28,29]. Eosinophilic inflammation may develop in response to the overexpression of these cytokines caused by GER.
The obtained results may be of use for the search of new treatment options in cases where refractory symptoms of GERD are present[30]. In such cases, the use of immune modulators affecting local immune response may help to achieve better results[26].
The limitation of the present study is a relatively small sample size. However, strict eligibility criteria could minimize the risk of inclusion of subjects with non-confirmed diagnosis in the GERD group. We did not divide subgroups depending on the severity of esophagitis. These subgroups could help us to obtain more details, but at the same time, this approach increases the risk of type II error.
The presence of bile acids in the reflux content may lead to greater impairment of esophageal mucosal integrity and less effective chemical clearance[31,32]. We did not detect bile in the refluxate in the present study; however, it seems very promising to analyze the association of cytokine gene expression with reflux of bile acids.
Our study provides additional information on the pathogenesis of GERD. However, larger studies are necessary to confirm the results.
In patients with EE, local expression of IL-18 and GATA3 was higher compared to subjects with NERD. Direct correlation was found between the local expression of IL-1β, IL-18, TNFA, TLR4, CD68, and B2M and acid exposure time in the distal esophagus. Expression of IL-18, TNFA, GATA3, TLR4, and CD68 correlated inversely with mean pH values in the distal part of the esophagus. No correlation was found between the gene expression of cytokines in the esophageal mucosa and the number of GER.
Gastroesophageal reflux disease (GERD) has a number of manifestations, including erosive esophagitis (EE) and non-erosive form of GERD (NERD). Similar levels of intraesophageal acidity are often found in subjects with EE and NERD and little is known about the reasons.
Several reports suggest involvement of inflammatory cytokines in the pathogenesis of different forms of GERD. Local inflammatory response of the esophageal mucosa to gastroesophageal reflux (GER) may depend on gene expression of cytokines. However, data on the dependence of the expression on esophageal acidity are still lacking.
The objective of the study was to explore gene expression of inflammatory cytokines in esophageal mucosa in patients with EE and NERD and its association with data of esophageal multichannel intraluminal impedance-pH measurements.
The data were obtained in a single-center prospective study. We analyzed the expression of interleukin (IL)-1β, IL-10, IL-18, tumor necrosis factor α (TNFA), toll-like receptor 4 (TLR4), GATA binding protein 3 (GATA3), differentiation cluster 68 (CD68), and β-2 microglobulin genes in esophageal mucosa samples obtained during endoscopy. All the subjects underwent multichannel intraluminal impedance-pH measurements. Based on the presence of abnormal results, they were allocated either to the GERD group or to the control group. The GERD group was further divided into the EE group and the NERD group. Spearman’s rank correlation analysis was used to analyze association of cytokine expression with esophageal acidity and number and types of GER.
We found higher expression of IL-18 and GATA3 genes in the EE group compared to the NERD group. Expression of IL-1β, IL-18, TNFA, and TLR4 was lower (P < 0.05) in the control group compared to EE and NERD. Esophageal acid exposure correlated with the expression of IL-1β (Spearman’s rank r = 0.29), IL-18 (r = 0.31), TNFA (r = 0.35), GATA3 (r = 0.34), and TLR4 (r = 0.29), CD68 (r = 0.37). Mean esophageal pH correlated inversely with the expression of IL-18, TNFA, GATA3, TLR4, and CD68. No association of the gene expression with number of GER was found.
In patients with EE, local expression of IL-18 and GATA3 was higher compared to subjects with NERD. Esophageal acid exposure correlated directly with expression of IL-1β, IL-18, TNFA, TLR4, CD68, and β-2 microglobulin. An inverse correlation was revealed between the expression of IL-18, TNFA, GATA3, TLR4, and CD68 and mean esophageal acidity. Expression levels of IL-10 did not differ significantly in the studied groups and did not correlate with esophageal acid exposure and number of GER.
Larger studies are necessary to confirm the obtained results. Assessment of the association of the local inflammatory response with concentration of bile acids within the refluxate seems promising.
The authors acknowledge the participants of the study. We would like to thank Dr. Donnikov A MD, PhD (DNK-Technologiya) and Dr. Vorozhko I, MD, PhD (Laboratory of Clinical Biochemistry and Immunology) for their help with genetic testing. We also express gratitude to Professor Isakov V (Federal Research Center of Nutrition and Biotechnology) for his support of the research.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author’s Membership in Professional Societies: European Society of Neurogastroenterology and Motility, No. 22458; International Society for Diseases of the Esophagus, No. 45108704.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Russia
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bortolotti M, Italy; Xu G, China S-Editor: Yan JP L-Editor: Filipodia P-Editor: Yan JP
| 1. | Vakil N, van Zanten SV, Kahrilas P, Dent J, Jones R; Global Consensus Group. The Montreal definition and classification of gastroesophageal reflux disease: a global evidence-based consensus. Am J Gastroenterol. 2006;101:1900-20; quiz 1943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2368] [Cited by in RCA: 2445] [Article Influence: 128.7] [Reference Citation Analysis (2)] |
| 2. | Iwakiri K, Fujiwara Y, Manabe N, Ihara E, Kuribayashi S, Akiyama J, Kondo T, Yamashita H, Ishimura N, Kitasako Y, Iijima K, Koike T, Omura N, Nomura T, Kawamura O, Ohara S, Ozawa S, Kinoshita Y, Mochida S, Enomoto N, Shimosegawa T, Koike K. Evidence-based clinical practice guidelines for gastroesophageal reflux disease 2021. J Gastroenterol. 2022;57:267-285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 127] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 3. | Mastracci L, Grillo F, Parente P, Unti E, Battista S, Spaggiari P, Campora M, Scaglione G, Fassan M, Fiocca R. Gastro-esophageal reflux disease and Barrett’s esophagus: an overview with an histologic diagnostic approach. Pathologica. 2020;112:117-127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 4. | Kasyap AK, Sah SK, Chaudhary S. Clinical spectrum and risk factors associated with asymptomatic erosive esophagitis as determined by Los Angeles classification: A cross-sectional study. PLoS One. 2018;13:e0192739. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 5. | Katzka DA, Pandolfino JE, Kahrilas PJ. Phenotypes of Gastroesophageal Reflux Disease: Where Rome, Lyon, and Montreal Meet. Clin Gastroenterol Hepatol. 2020;18:767-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 100] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 6. | Savarino E, Tutuian R, Zentilin P, Dulbecco P, Pohl D, Marabotto E, Parodi A, Sammito G, Gemignani L, Bodini G, Savarino V. Characteristics of reflux episodes and symptom association in patients with erosive esophagitis and nonerosive reflux disease: study using combined impedance-pH off therapy. Am J Gastroenterol. 2010;105:1053-1061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 178] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 7. | Bajbouj M, Reichenberger J, Neu B, Prinz C, Schmid RM, Rösch T, Meining A. A prospective multicenter clinical and endoscopic follow-up study of patients with gastroesophageal reflux disease. Z Gastroenterol. 2005;43:1303-1307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 8. | Yamaguchi T, Yoshida N, Tomatsuri N, Takayama R, Katada K, Takagi T, Ichikawa H, Naito Y, Okanoue T, Yoshikawa T. Cytokine-induced neutrophil accumulation in the pathogenesis of acute reflux esophagitis in rats. Int J Mol Med. 2005;16:71-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 9. | Souza RF, Huo X, Mittal V, Schuler CM, Carmack SW, Zhang HY, Zhang X, Yu C, Hormi-Carver K, Genta RM, Spechler SJ. Gastroesophageal reflux might cause esophagitis through a cytokine-mediated mechanism rather than caustic acid injury. Gastroenterology. 2009;137:1776-1784. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 279] [Cited by in RCA: 284] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 10. | Jonasson C, Wernersson B, Hoff DA, Hatlebakk JG. Validation of the GerdQ questionnaire for the diagnosis of gastro-oesophageal reflux disease. Aliment Pharmacol Ther. 2013;37:564-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 187] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 11. | Morozov SV. GERD-Q questionnaire – a novel tool for gastroesophageal reflux disease diagnosis. Acad J West Sib. 2014;3:26-28. |
| 12. | Gyawali CP, Kahrilas PJ, Savarino E, Zerbib F, Mion F, Smout AJPM, Vaezi M, Sifrim D, Fox MR, Vela MF, Tutuian R, Tack J, Bredenoord AJ, Pandolfino J, Roman S. Modern diagnosis of GERD: the Lyon Consensus. Gut. 2018;67:1351-1362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 672] [Cited by in RCA: 936] [Article Influence: 133.7] [Reference Citation Analysis (0)] |
| 13. | Lundell LR, Dent J, Bennett JR, Blum AL, Armstrong D, Galmiche JP, Johnson F, Hongo M, Richter JE, Spechler SJ, Tytgat GN, Wallin L. Endoscopic assessment of oesophagitis: clinical and functional correlates and further validation of the Los Angeles classification. Gut. 1999;45:172-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1518] [Cited by in RCA: 1649] [Article Influence: 63.4] [Reference Citation Analysis (1)] |
| 14. | Fonseca-Camarillo G, Furuzawa-Carballeda J, Iturriaga-Goyon E, Yamamoto-Furusho JK. Differential Expression of IL-36 Family Members and IL-38 by Immune and Nonimmune Cells in Patients with Active Inflammatory Bowel Disease. Biomed Res Int. 2018;2018:5140691. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 15. | Kozlov IB, Vatazin AV, Kildushevsky AV, Zulkarnaev A, Fedulkina VA, Faenko AP, Yazdovskiy VV, Gudima GO, Kofiadi IA. Analysis of expression of immune system genes that are responsible for activation and inhibition of T-cell immune response in renal transplant recipients after extracorporeal photochemotherapy. Immuno. 41:20-30. [DOI] [Full Text] |
| 16. | Yoshida N, Uchiyama K, Kuroda M, Sakuma K, Kokura S, Ichikawa H, Naito Y, Takemura T, Yoshikawa T, Okanoue T. Interleukin-8 expression in the esophageal mucosa of patients with gastroesophageal reflux disease. Scand J Gastroenterol. 2004;39:816-822. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 72] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 17. | Mönkemüller K, Wex T, Kuester D, Fry LC, Peitz U, Beyer M, Roessner A, Malfertheiner P. Interleukin-1beta and interleukin-8 expression correlate with the histomorphological changes in esophageal mucosa of patients with erosive and non-erosive reflux disease. Digestion. 2009;79:186-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 18. | Rieder F, Cheng L, Harnett KM, Chak A, Cooper GS, Isenberg G, Ray M, Katz JA, Catanzaro A, O’Shea R, Post AB, Wong R, Sivak MV, McCormick T, Phillips M, West GA, Willis JE, Biancani P, Fiocchi C. Gastroesophageal reflux disease-associated esophagitis induces endogenous cytokine production leading to motor abnormalities. Gastroenterology. 2007;132:154-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 108] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 19. | Zavala-Solares MR, Fonseca-Camarillo G, Valdovinos M, Granados J, Grajales-Figueroa G, Zamora-Nava L, Aguilar-Olivos N, Valdovinos-García LR, Yamamoto-Furusho JK. Gene expression profiling of inflammatory cytokines in esophageal biopsies of different phenotypes of gastroesophageal reflux disease: a cross-sectional study. BMC Gastroenterol. 2021;21:201. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 20. | Akdis CA, Joss A, Akdis M, Faith A, Blaser K. A molecular basis for T cell suppression by IL-10: CD28-associated IL-10 receptor inhibits CD28 tyrosine phosphorylation and phosphatidylinositol 3-kinase binding. FASEB J. 2000;14:1666-1668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 129] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 21. | Wang Y, Yu P, Li Y, Zhao Z, Wu X, Zhang L, Feng J, Hong JS. Early-Released Interleukin-10 Significantly Inhibits Lipopolysaccharide-Elicited Neuroinflammation In Vitro. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 22. | Shapiro M, Green C, Faybush EM, Esquivel RF, Fass R. The extent of oesophageal acid exposure overlap among the different gastro-oesophageal reflux disease groups. Aliment Pharmacol Ther. 2006;23:321-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 23. | Böhmer AC, Schumacher J. Insights into the genetics of gastroesophageal reflux disease (GERD) and GERD-related disorders. Neurogastroenterol Motil. 2017;29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 24. | Fitzgerald RC, Onwuegbusi BA, Bajaj-Elliott M, Saeed IT, Burnham WR, Farthing MJ. Diversity in the oesophageal phenotypic response to gastro-oesophageal reflux: immunological determinants. Gut. 2002;50:451-459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 210] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 25. | Isomoto H, Wang A, Mizuta Y, Akazawa Y, Ohba K, Omagari K, Miyazaki M, Murase K, Hayashi T, Inoue K, Murata I, Kohno S. Elevated levels of chemokines in esophageal mucosa of patients with reflux esophagitis. Am J Gastroenterol. 2003;98:551-556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 96] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 26. | Barnes PJ. Targeting cytokines to treat asthma and chronic obstructive pulmonary disease. Nat Rev Immunol. 2018;18:454-466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 285] [Article Influence: 47.5] [Reference Citation Analysis (0)] |
| 27. | Hamaguchi M, Fujiwara Y, Takashima T, Hayakawa T, Sasaki E, Shiba M, Watanabe T, Tominaga K, Oshitani N, Matsumoto T, Higuchi K, Arakawa T. Increased expression of cytokines and adhesion molecules in rat chronic esophagitis. Digestion. 2003;68:189-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 44] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 28. | Vinit C, Dieme A, Courbage S, Dehaine C, Dufeu CM, Jacquemot S, Lajus M, Montigny L, Payen E, Yang DD, Dupont C. Eosinophilic esophagitis: Pathophysiology, diagnosis, and management. Arch Pediatr. 2019;26:182-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 29. | Molina-Infante J, Gonzalez-Cordero PL, Lucendo AJ. Proton pump inhibitor-responsive esophageal eosinophilia: still a valid diagnosis? Curr Opin Gastroenterol. 2017;33:285-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 30. | Rettura F, Bronzini F, Campigotto M, Lambiase C, Pancetti A, Berti G, Marchi S, de Bortoli N, Zerbib F, Savarino E, Bellini M. Refractory Gastroesophageal Reflux Disease: A Management Update. Front Med (Lausanne). 2021;8:765061. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 43] [Article Influence: 10.8] [Reference Citation Analysis (1)] |
| 31. | Pardon NA, Vicario M, Vanheel H, Vanuytsel T, Ceulemans LJ, Vieth M, Jimenez M, Tack J, Farré R. A weakly acidic solution containing deoxycholic acid induces esophageal epithelial apoptosis and impairs integrity in an in vivo perfusion rabbit model. Am J Physiol Gastrointest Liver Physiol. 2016;310:G487-G496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 32. | de Bortoli N, Gyawali CP, Frazzoni M, Tolone S, Frazzoni L, Vichi E, Visaggi P, Bellini M, Marabotto E, Penagini R, Savarino V, Marchi S, Savarino EV. Bile reflux in patients with nerd is associated with more severe heartburn and lower values of mean nocturnal baseline impedance and chemical clearance. Neurogastroenterol Motil. 2020;32:e13919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |