Published online Aug 26, 2022. doi: 10.12998/wjcc.v10.i24.8641
Peer-review started: December 14, 2021
First decision: May 30, 2022
Revised: June 1, 2022
Accepted: July 18, 2022
Article in press: July 18, 2022
Published online: August 26, 2022
Processing time: 244 Days and 21.7 Hours
Confined placental mosaicism (CPM) is one of the major reasons for discrepancies between the results of non-invasive prenatal testing (NIPT) and fetal karyotype analysis.
We encountered a primiparous singleton pregnant woman with a rare CPM consisting of 47,XY,+21; 47,XXY; and 46,XY, who obtained a false-positive result on NIPT with a high risk for trisomy 21. Copy-number variation sequencing on amniotic fluid cells, fetal tissue, and placental biopsies showed that the fetal karyotype was 47,XXY, while the placenta was a rare mosaic of 47,XY,+21; 47,XXY; and 46,XY.
The patient had a rare CPM consisting of 47,XY,+21; 47,XXY; and 46,XY, which caused a discrepancy between the result of NIPT and the actual fetal karyotype. It is important to remember that NIPT is a screening test, not a diagnostic test. Any positive result should be confirmed with invasive testing, and routine ultrasound examination is still necessary after a negative result.
Core Tip: We identified that the patient had a rare confined placental mosaicism consisting of 47,XY,+21; 47,XXY; and 46,XY, which caused a discrepancy between non-invasive prenatal testing (NIPT) and fetal karyotype. Although NIPT has high sensitivity and specificity, false negatives and false positives are still possible. It is important to remember that NIPT is just a screening test, and any positive results need to be confirmed with invasive testing. Patients with negative NIPT results still require follow-up ultrasound examination.
- Citation: Li Z, Lai GR. Discrepancy between non-invasive prenatal testing result and fetal karyotype caused by rare confined placental mosaicism: A case report. World J Clin Cases 2022; 10(24): 8641-8647
- URL: https://www.wjgnet.com/2307-8960/full/v10/i24/8641.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i24.8641
Currently, non-invasive prenatal testing (NIPT) using next-generation sequencing on a sample of cell-free fetal DNA (cffDNA) from maternal plasma is widely used as a screening test for common fetal aneuploidies (e.g., trisomy 21, 18, and 13; sex chromosome aneuploidies)[1]. This method of aneuploidy screening is not only non-invasive, but also highly accurate, with the sensitivity and specificity for pooled common aneuploidies as high as 99%[1,2]. NIPT offers higher accuracy when compared with serologic screening tests[3], thereby reducing the use of invasive diagnostic procedures that may result in miscarriage or intrauterine infection. However, NIPT is still a screening test and not a diagnostic test. As the cffDNA in maternal plasma originates from apoptotic placental trophoblast cells, it mainly consists of placental DNA[4,5], and the results may not represent the actual fetal karyotype. One of the most common reasons for false results on NIPT is a confined placental mosaicism (CPM)[6]. We report our experience with a patient whose NIPT result indicated a high risk for trisomy 21, but in whom the actual fetal karyotype was 47,XXY. The reason for this discrepancy was the presence of a CPM; the placenta was a rare mosaic of 47,XY,+21; 47,XXY; and 46,XY.
The patient was a 26-year-old primiparous woman with a singleton pregnancy. At 15 + 1 wk, the second-trimester serologic screening showed an elevated risk for Down’s syndrome, at 1 in 146 [alpha-fetoprotein: 0.67 multiples of the median (MoM); free β human chorionic gonadotropin: 3.18 MoM; unconjugated estradiol: 0.76 MoM]. The patient requested further testing.
The patient has no present illness.
The patient has no past illness.
The patient denied any personal or family history.
The patient’s basic vital signs were within normal limits. She requested NIPT before amniocentesis.
Maternal plasma was collected for NIPT at 15 + 3 wk. We followed the standard method for performing NIPT, which has been described previously[7]. The NIPT results showed a high risk for trisomy 21, with a Z-score of 16.21 for chromosome 21; however, there was a low risk for sex chromosome aneuploidy (the Z-score of chromosome X and Y was -12.88 and 79.64, respectively).
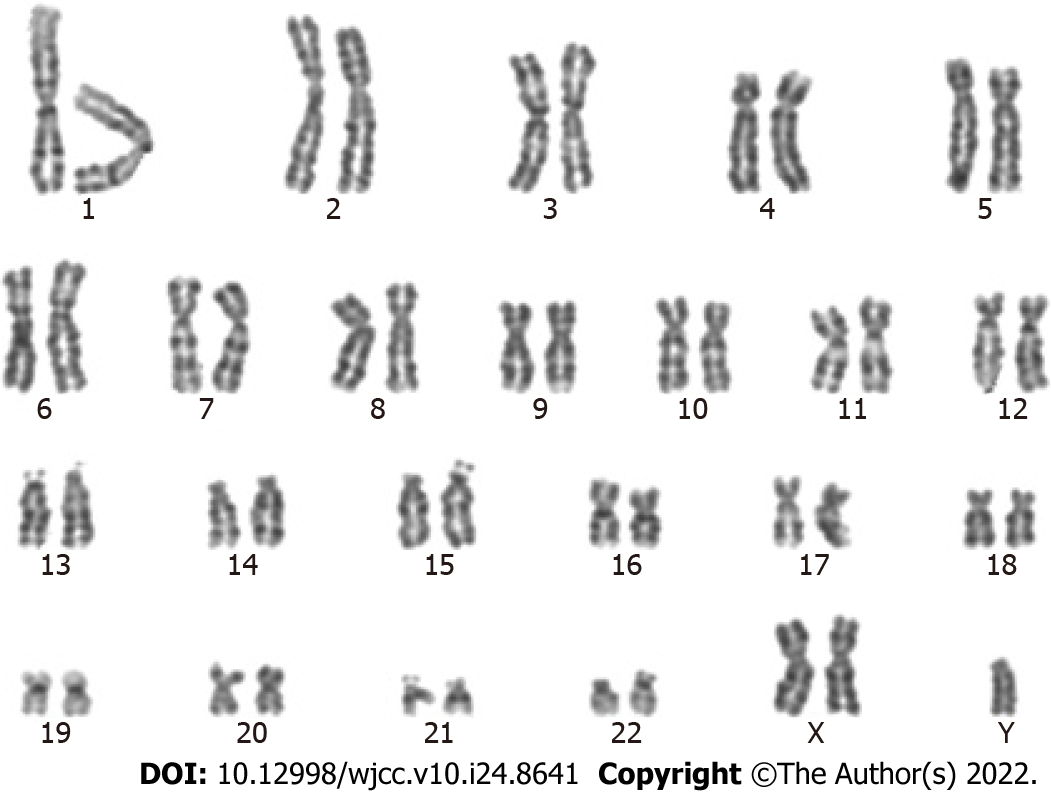
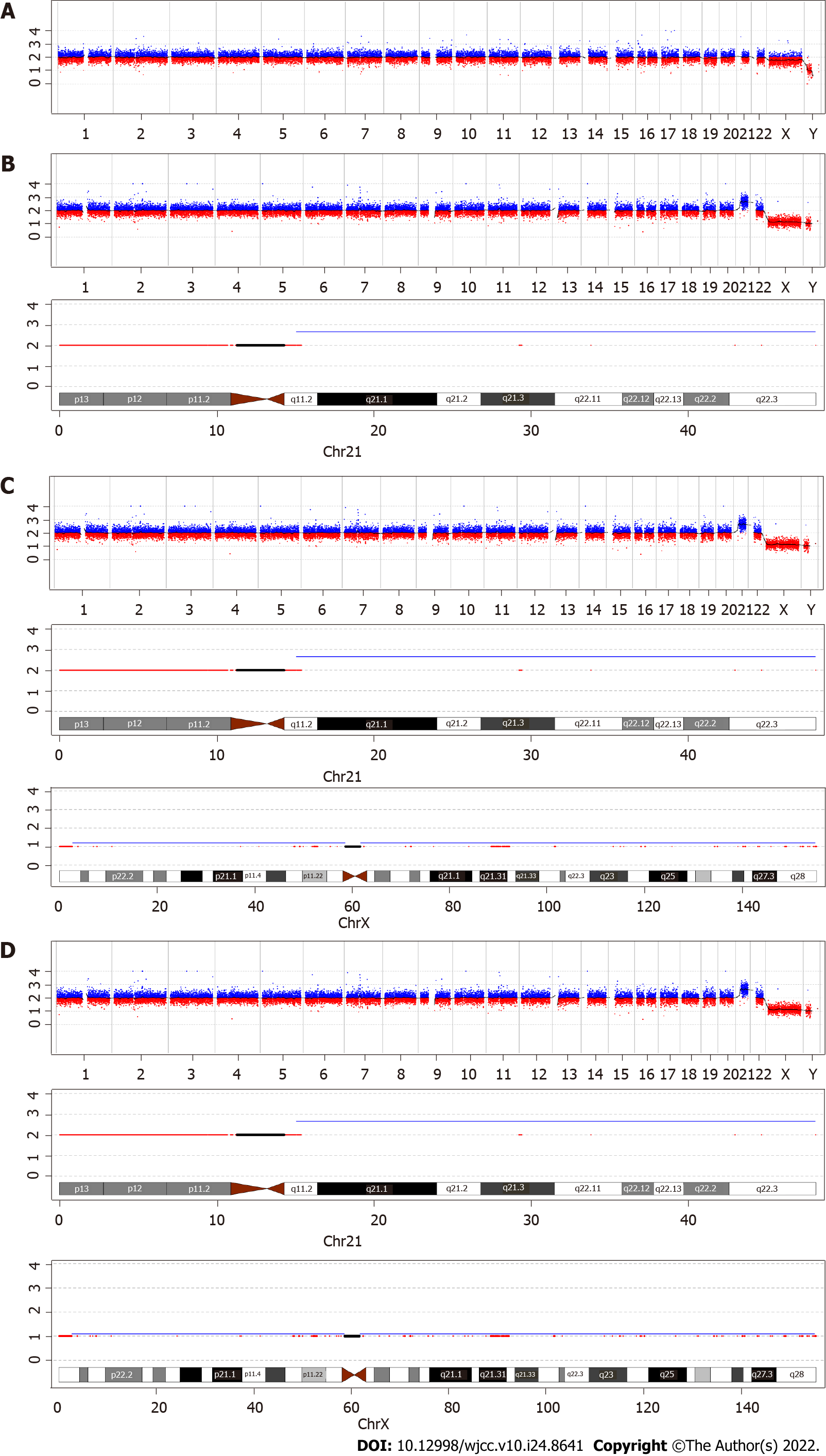
To confirm the positive NIPT results, amniocentesis was performed at 19 + 2 wk. Copy-number variation sequencing (CNV-seq) and karyotype analysis performed on amniotic fluid cells suggested that the fetal karyotype was XXY, as shown in Figures 1 and 2 and Table 1. The patient underwent genetic counseling and decided to terminate her pregnancy. After written informed consent for the procedure and further testing was obtained, she underwent an induced abortion at 22 + 5 wk. Samples from the fetus were collected after delivery - including fetal muscle tissue, the middle segment of the umbilical cord, and placental tissue - and sent for CNV-seq. The placental samples included a mid-thickness section from the center of the placenta and samples from the center and margin of the maternal face, and the center and margin of the fetal face. As shown in Table 1 and Figure 2, the fetal muscle tissue and umbilical cord tissue had a karyotype of 47,XXY - matching that of the amniotic fluid cells. However, the center and margin samples from the fetal face and the margin of the maternal face of the placenta had a mosaic karyotype of 47,XY,+21 (65%) and 46,XY (35%), respectively. The mid-thickness sample from the placental center and the sample from the center of the maternal face of the placenta demonstrated a mosaic of 47,XY,+21; 47,XXY; and 46,XY with different proportions in each sample. In brief, the placenta was a mosaic of 47, XY,+21; 47,XXY; and 46,XY.
| Sample type | Sample | Result of CNV-seq |
| Amniotic fluid | Amniotic fluid cells | 47,XXY |
| Fetal tissue | Fetal muscle tissue | 47,XXY |
| Umbilical cord | Middle segment of umbilical cord | 47,XXY |
| Placenta | Center of fetal face | 47,XY,+21[65%]/46,XY[35%] |
| Margin of fetal face | 47,XY,+21[65%]/46,XY[35%] | |
| Margin of maternal face | 47,XY,+21[65%]/46,XY[35%] | |
| Center of maternal face | 47,XY,+21[60%]/47,XXY[20%]/46,XY[20%] | |
| Placental center | 47,XY,+21[65%]/47,XXY[10%]/46,XY[25%] |
No obvious abnormality was detected upon fetal ultrasonography.
The fetal karyotype was 47,XXY; whereas the placenta was a mosaic of 47,XY,+21; 47,XXY; and 46,XY.
Amniocentesis was used to determine the karyotype of the fetus. A placental sample was collected following induced abortion and was tested to determine the cause of the discrepancy between the NIPT results and the fetal karyotype.
The patient underwent an induced abortion after genetic counseling. The timeline is shown in Table 2.
| Gestational age (wk) | Examination items | Results |
| 15 + 1 | Serum Down’s screening | High risk for trisomy 21 |
| 15 + 3 | NIPT | High risk for trisomy 21, low risk for sex chromosome aneuploidy |
| 19 + 2 | Amniocentesis (CNV-seq and karyotype analysis) | 47,XXY |
| 22 + 5 | Abortion, collected fetal muscle tissue, umbilical cord and placental samples | Fetal muscle tissue and umbilical cord: 47,XXY placenta: A mosaic of 47,XY,+21; 47,XXY; and 46,XY |
The patient had a rare CPM consisting of 47,XY,+21; 47,XXY; and 46,XY, which caused a discrepancy between the results of NIPT and the actual fetal karyotype. The cffDNA in maternal blood has a dominant peak size of 143 base pairs, which is shorter than the free DNA fragments typically found in maternal plasma (around 166 base pairs)[8]. cffDNA can be detected as early as 4.5 wk of pregnancy[9], is present throughout pregnancy, and disappears from the maternal circulation within hours after delivery[10]. The proportion of cffDNA to total free DNA (fetal and maternal) is referred to as the fetal fraction, and it increases throughout pregnancy. At 10-20 wk of gestation, the average fetal fraction in maternal plasma is 10%-15%; however, it may range from less than 3% to over 30%[11].
The introduction of NIPT in the late 2000s was revolutionary for aneuploidy screening, and it is now a commonly used screening method. The sensitivity and positive predictive value of serologic screening for trisomy 21 is only about 80% and 5%, respectively[3]; while the sensitivity of NIPT can reach up to 99%, with a positive predictive value of 94.5%[1]. Thus, the expanded use of NIPT can greatly reduce the use of invasive diagnostic procedures, thereby avoiding the resulting complications of miscarriage or intrauterine infection. The sensitivity and specificity of NIPT for other common aneuploidies, including trisomy 18, trisomy 13, and sex chromosome aneuploidy, are as high as 99%[1]. However, false positive and false negative results for NIPT occur at a rate of 0.3% and 1.1%, respectively[1]. There are four factors that affect the results of NIPT: (1) A low fetal fraction, which can be present in overweight mothers, usually leading to a false negative result[12]; (2) Maternal conditions, such as the presence of a tumor, mosaicism, or chromosomal abnormalities, are often associated with false-positive results[13]; (3) Fetal chimerism and vanishing twin syndrome can affect the results[14]; and (4) CPM, which is also a very common cause of incorrect results[6,15]. In our patient with CPM, the results of NIPT were falsely positive for trisomy 21 and falsely negative for 47,XXY.
The mosaicism involved in CPM occurs only in the placenta, not in the fetus. In most situations, the fetal outcome is normal if the fetal chromosomes are normal[16]. However, 10% of pregnancies that involve a placenta with CPM are affected by fetal growth restriction, even after constitutional fetal chromosomal abnormalities are excluded[17,18]. According to a large-scale evaluation of chorionic villus sampling, the prevalence of CPM is about 0.6% to 1.0%[18,19]. Although the genetic makeup of placental and fetal tissue is usually identical, clinicians should be mindful of the possibility of CPM, especially as it accounts for a high proportion of incorrect results on NIPT[6]. Wu et al[20] found that CPM was present in 6 of 10 placentas from pregnancies in which there was a false-positive result on NIPT[20]. Our group identified three false negative NIPT results in a total of 34311 pregnancies, and all fetuses had structural abnormalities detected on follow-up ultrasound screening. Placental biopsies were collected from 2 of the 3 patients with false-negative NIPT results; both were confirmed to have CPM. One was the patient described in this report, and the other patient had a fetus with trisomy 21 and a placental mosaic of 47,XY,+21 and 46,XY.
There are two key elements that should be noted for NIPT. While its sensitivity and specificity are high, the positive predictive value varies from 94.5% for trisomy 21[21], to 82.1% for trisomy 18, 46.2% for trisomy 13, and 46.7% for sex chromosome aneuploidies[1]. A positive result on NIPT should always be confirmed with invasive testing (e.g., amniocentesis, umbilical cord blood sampling, chorionic villus sampling) before any irreversible procedure is performed, as the results on NIPT may not correlate with the true fetal genotype[16]. The other key element is that false-negative results on NIPT are associated with more serious consequences than false-positive results and cause more stress to pregnant women and their families. Majorly, the false-negative result can be proven when abnormalities are detected on routine follow-up ultrasound screening which is still necessary, even when the results of NIPT are normal. Attention should also be paid to low fetal fractions. The quality threshold for the fetal fraction is commonly accepted as 4%, and samples with values below this are often reported as having inconclusive results[11].
We describe our experience with a rare discrepancy between NIPT and karyotype testing. It is important to remember that NIPT is just a screening test, and any positive result should be confirmed with invasive testing. Patients with negative results on NIPT still require follow-up ultrasound examination.
Thanks for the patients’ family participation.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gislinge JIP, Denmark; Tolunay HE, Turkey S-Editor: Wang JJ L-Editor: A P-Editor: Wang JJ
| 1. | Liang D, Cram DS, Tan H, Linpeng S, Liu Y, Sun H, Zhang Y, Tian F, Zhu H, Xu M, Wang H, Yu F, Wu L. Clinical utility of noninvasive prenatal screening for expanded chromosome disease syndromes. Genet Med. 2019;21:1998-2006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 170] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 2. | Liang D, Lv W, Wang H, Xu L, Liu J, Li H, Hu L, Peng Y, Wu L. Non-invasive prenatal testing of fetal whole chromosome aneuploidy by massively parallel sequencing. Prenat Diagn. 2013;33:409-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 121] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 3. | Canick J. Prenatal screening for trisomy 21: recent advances and guidelines. Clin Chem Lab Med. 2012;50:1003-1008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 4. | Tjoa ML, Cindrova-Davies T, Spasic-Boskovic O, Bianchi DW, Burton GJ. Trophoblastic oxidative stress and the release of cell-free feto-placental DNA. Am J Pathol. 2006;169:400-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 162] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 5. | Faas BH, de Ligt J, Janssen I, Eggink AJ, Wijnberger LD, van Vugt JM, Vissers L, Geurts van Kessel A. Non-invasive prenatal diagnosis of fetal aneuploidies using massively parallel sequencing-by-ligation and evidence that cell-free fetal DNA in the maternal plasma originates from cytotrophoblastic cells. Expert Opin Biol Ther. 2012;12 Suppl 1:S19-S26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 108] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 6. | Suzumori N, Sekizawa A, Takeda E, Samura O, Sasaki A, Akaishi R, Wada S, Hamanoue H, Hirahara F, Sawai H, Nakamura H, Yamada T, Miura K, Masuzaki H, Nakayama S, Kamei Y, Namba A, Murotsuki J, Yamaguchi M, Tairaku S, Maeda K, Kaji T, Okamoto Y, Endo M, Ogawa M, Kasai Y, Ichizuka K, Yamada N, Ida A, Miharu N, Kawaguchi S, Hasuo Y, Okazaki T, Ichikawa M, Izumi S, Kuno N, Yotsumoto J, Nishiyama M, Shirato N, Hirose T, Sago H. Retrospective details of false-positive and false-negative results in non-invasive prenatal testing for fetal trisomies 21, 18 and 13. Eur J Obstet Gynecol Reprod Biol. 2021;256:75-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 7. | Cui W, Liu X, Zhang Y, Wang Y, Chu G, He R, Zhao Y. Evaluation of non-invasive prenatal testing to detect chromosomal aberrations in a Chinese cohort. J Cell Mol Med. 2019;23:7873-7878. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 8. | Lo YM, Chan KC, Sun H, Chen EZ, Jiang P, Lun FM, Zheng YW, Leung TY, Lau TK, Cantor CR, Chiu RW. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med. 2010;2:61ra91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 634] [Cited by in RCA: 748] [Article Influence: 53.4] [Reference Citation Analysis (0)] |
| 9. | D’Aversa E, Breveglieri G, Pellegatti P, Guerra G, Gambari R, Borgatti M. Non-invasive fetal sex diagnosis in plasma of early weeks pregnants using droplet digital PCR. Mol Med. 2018;24:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 10. | Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet. 1999;64:218-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 812] [Cited by in RCA: 811] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 11. | Shaw J, Scotchman E, Chandler N, Chitty LS. PREIMPLANTATION GENETIC TESTING: Non-invasive prenatal testing for aneuploidy, copy-number variants and single-gene disorders. Reproduction. 2020;160:A1-A11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 12. | Canick JA, Palomaki GE, Kloza EM, Lambert-Messerlian GM, Haddow JE. The impact of maternal plasma DNA fetal fraction on next generation sequencing tests for common fetal aneuploidies. Prenat Diagn. 2013;33:667-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 258] [Cited by in RCA: 266] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 13. | Bianchi DW, Chudova D, Sehnert AJ, Bhatt S, Murray K, Prosen TL, Garber JE, Wilkins-Haug L, Vora NL, Warsof S, Goldberg J, Ziainia T, Halks-Miller M. Noninvasive Prenatal Testing and Incidental Detection of Occult Maternal Malignancies. JAMA. 2015;314:162-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 305] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 14. | Curnow KJ, Wilkins-Haug L, Ryan A, Kırkızlar E, Stosic M, Hall MP, Sigurjonsson S, Demko Z, Rabinowitz M, Gross SJ. Detection of triploid, molar, and vanishing twin pregnancies by a single-nucleotide polymorphism-based noninvasive prenatal test. Am J Obstet Gynecol. 2015;212:79.e1-79.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 116] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 15. | Li J, Xie M, Wang F, Ma J, Li J, Chen C, Li Z, Wang J, Zhang Y, Li Y. A rare case of NIPT discrepancy caused by the placental mosaicism of three different karyotypes, 47,XXX, 47,XX,+21, and 48,XXX,+21. Mol Genet Genomic Med. 2020;8:e1279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 16. | Mardy A, Wapner RJ. Confined placental mosaicism and its impact on confirmation of NIPT results. Am J Med Genet C Semin Med Genet. 2016;172:118-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 17. | Hayata K, Hiramatsu Y, Masuyama H, Eto E, Mitsui T, Tamada S. Discrepancy between Non-invasive Prenatal Genetic Testing (NIPT) and Amniotic Chromosomal Test due to Placental Mosaicism: A Case Report and Literature Review. Acta Med Okayama. 2017;71:181-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 18. | Toutain J, Goutte-Gattat D, Horovitz J, Saura R. Confined placental mosaicism revisited: Impact on pregnancy characteristics and outcome. PloS One. 2018;13:e0195905. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 19. | West JD, Everett CA. Preimplantation chromosomal mosaics, chimaeras and confined placental mosaicism. Reprod Fertil. 2022;3:R66-R90. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 20. | Wu X, Li Y, Xie X, Su L, Cai M, Lin N, Du S, Xu L, Huang H. Clinical Review of Noninvasive Prenatal Testing: Experience from 551 Pregnancies with Noninvasive Prenatal Testing-Positive Results in a Tertiary Referral Center. J Mol Diagn. 2020;22:1469-1475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 21. | Petersen AK, Cheung SW, Smith JL, Bi W, Ward PA, Peacock S, Braxton A, Van Den Veyver IB, Breman AM. Positive predictive value estimates for cell-free noninvasive prenatal screening from data of a large referral genetic diagnostic laboratory. Am J Obstet Gynecol. 2017;217:691.e1-691.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 128] [Article Influence: 16.0] [Reference Citation Analysis (0)] |