Published online Aug 26, 2022. doi: 10.12998/wjcc.v10.i24.8634
Peer-review started: August 7, 2021
First decision: November 7, 2021
Revised: December 11, 2021
Accepted: June 24, 2022
Article in press: June 24, 2022
Published online: August 26, 2022
Processing time: 373 Days and 12.4 Hours
Cronkhite-Canada syndrome (CCS) is a rare, non-genetic disorder characterized by multiple gastrointestinal polyps, and ectodermal lesions such as alopecia, fingernail atrophy, and skin mucosal pigmentation. Unfortunately, the pathoge
Here, we describe the case of an elderly female with diarrhea, fatigue, and hair loss, who experienced abdominal pain for over half a year and was found to have multiple gastrointestinal polyps. She was diagnosed with CCS and was treated with albumin supplementation and prednisone, and her electrolyte imbalance was corrected. Following treatment, her symptoms significantly improved. To elucidate the role of potential genetic events in the pathogenesis of CCS, we performed exome sequencing using an extract of her colorectal adenoma.
Our data revealed multiple somatic mutations and copy number variations. Our findings provide a novel insight into the potential mechanisms of CCS etiology.
Core Tip: Cronkhite-Canada syndrome (CCS) is a rare, non-genetic disorder characterized by multiple gastrointestinal polyps, ectodermal lesions including alopecia, fingernail atrophy, and skin mucosal pigmentation. However, its pathogenesis is unclear. We performed exome sequencing in an elderly female patient and obtained somatic mutations in the hope that these data could provide a genetic perspective on the pathogenesis of CCS.
- Citation: Li ZD, Rong L, He YJ, Ji YZ, Li X, Song FZ, Li XA. Exome analysis for Cronkhite-Canada syndrome: A case report. World J Clin Cases 2022; 10(24): 8634-8640
- URL: https://www.wjgnet.com/2307-8960/full/v10/i24/8634.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i24.8634
Cronkite and Canada reported the first case of Cronkhite-Canada syndrome (CCS) in 1955, which was characterized by multiple gastrointestinal polyps, ectodermal lesions including alopecia, fingernail atrophy, and skin mucosal pigmentation[1]. Unfortunately, despite much research, the pathogenesis of CCS remains unknown. Some studies have reported an association between CCS and immune factors, especially infiltration of IgG4-positive plasma cells[2]. To date, there is no standard treatment for CCS; however, some studies have recommended glucocorticoid therapy for partial symptom relief[3]. Herein, we describe the case of an elderly female with the diagnosis of CCS. As part of her treatment regimen, she received albumin supplementation and prednisone, and her electrolyte disturbance was corrected. After 15 days, her symptoms, namely, hypokalemia and diarrhea, significantly improved. To elucidate the role of potential genetic events in the pathogenesis of CCS, we performed exome sequencing using DNA extracted from a colorectal adenoma. Our data revealed multiple somatic mutations and copy number variations. We are the first to identify 3 novel genetic mutations (USP24, KCNQ5, and FKBP10) in a CCS patient. Moreover, we demonstrated that the HPSE2, SPATA7, and ZC3H18 genes had markedly elevated copy numbers. Given this evidence, we hypothesized that these specific gene mutations and copy number variations are associated with the pathogenesis of CCS.
An elderly female patient sought treatment for diarrhea, fatigue, and hair loss, as well as abdominal pain for half a year at the Department of Gastroenterology on October 11, 2019.
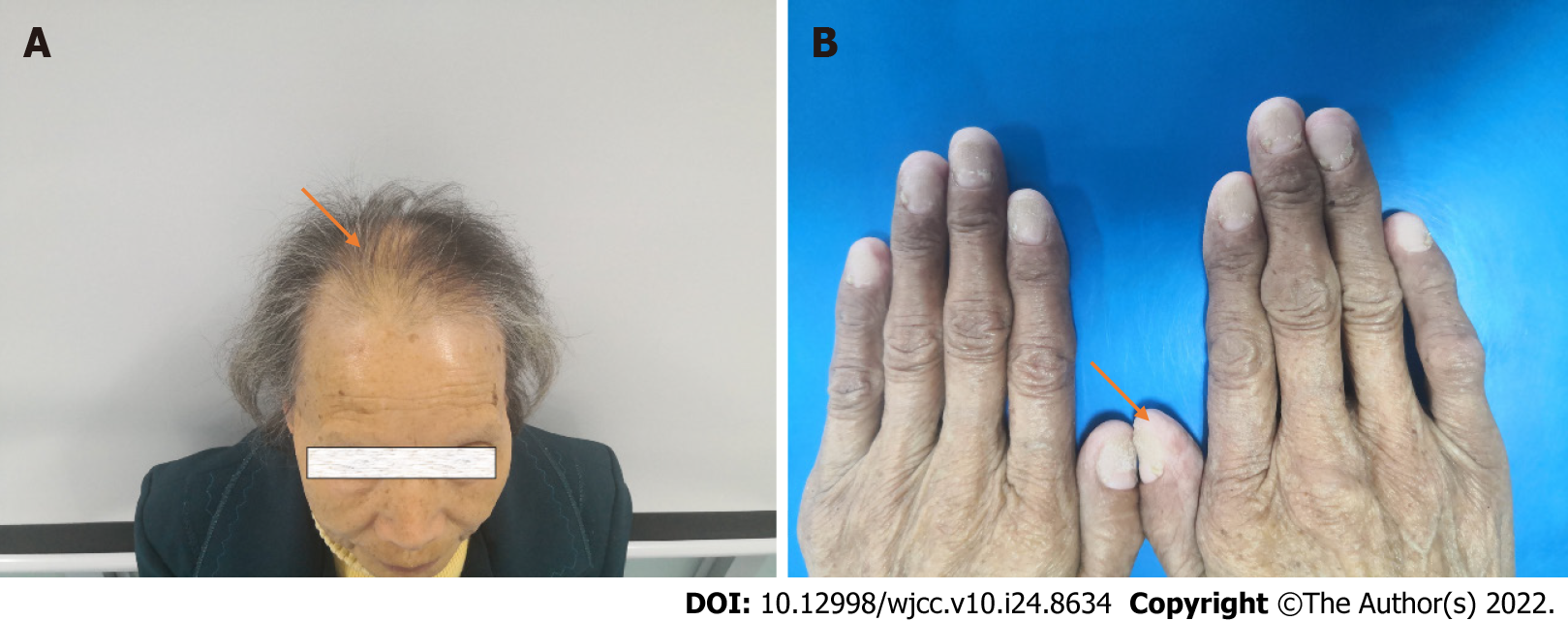
She reported diarrhea 4-5 times daily, watery stools, no blood, and abdominal pain under the xiphoid process, not related to eating. Physical examination revealed obvious emaciation, alopecia, and nail atrophy.
Her past medical history is unremarkable.
Her family history is unremarkable.
Physical examination revealed obvious emaciation, alopecia, and nail atrophy (Figure 1).
Laboratory tests revealed the following: (routine blood examination) white blood cells: 10.03 × 109/L, hemoglobin: 95 g/L, and platelets: 151 × 109/L; (liver function) albumin 25 g/L; (electrolytes) Na+: 128 mmol/L, K+: 2.8 mmol/L.
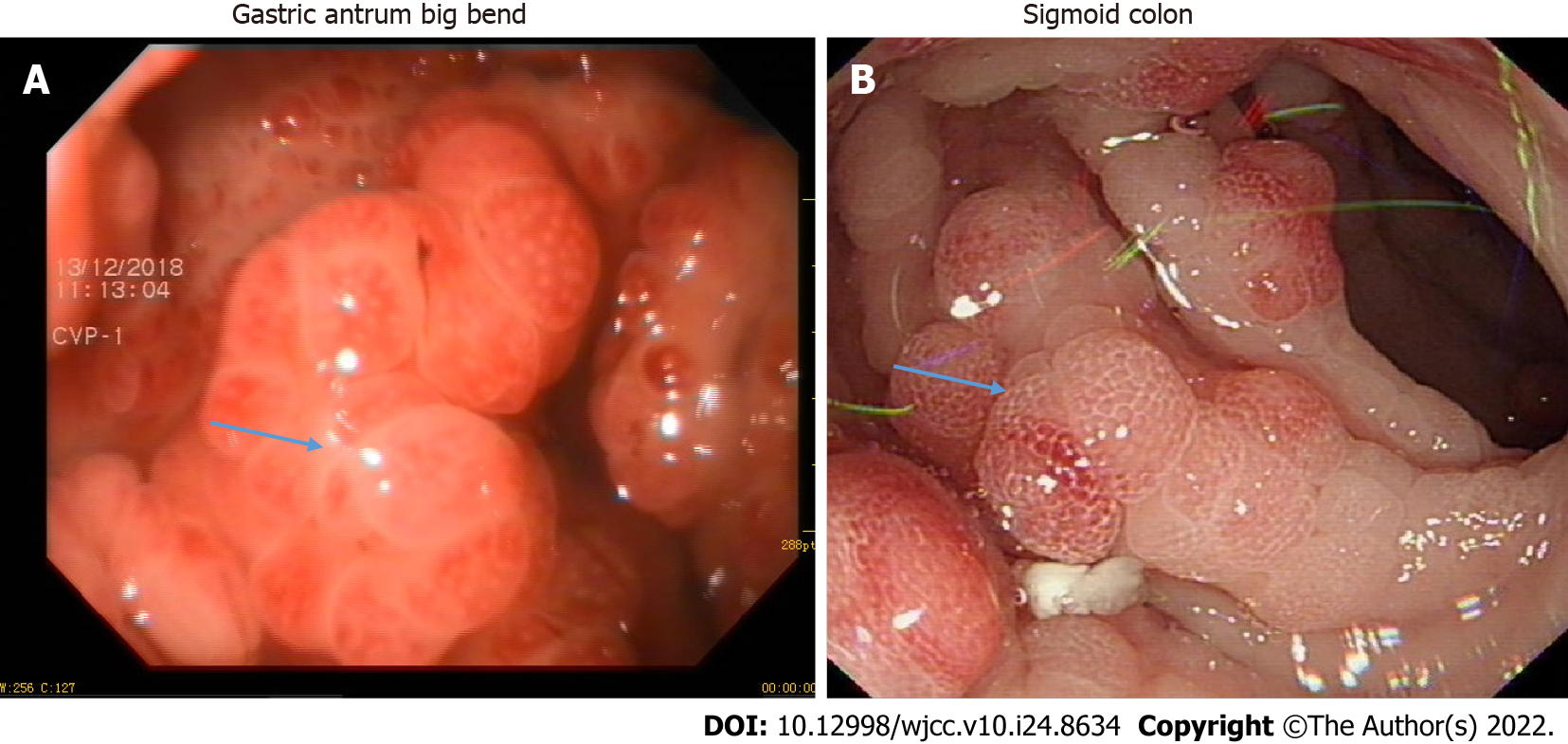
She underwent gastroenteroscopy, which revealed multiple small polyps in the gastrointestinal tract (Figure 2).
DNA samples: This study was performed in accordance with the guidelines of the Ethics Committee of the Mianyang Central Hospital. DNA was extracted from a sample of the patient's colonic adenomas and normal colon tissue. The samples were obtained via endoscopic mucosal resection and were preserved in liquid nitrogen.
Whole exome sequencing: Genomic DNA ≥ 1.5 μg from colonic adenomas and normal colon tissue was used for whole exome sequencing library construction. The Agilent liquid chip capture system was employed for the efficient enrichment of human DNA in all exon regions. Upon library construction, Qubit2.0 was used for preliminary quantification, and the library was diluted to 1 ng/μL. Subsequently, Agilent 2100 was utilized for detection of the library insert size to ensure library quality (the effective concentration of the library was > 2 nmol/L). Next, PE150 high-throughput sequencing was performed based on the Illumina Hiseq platform. Finally, the library and capture experiment were constructed using the Agilent SureSelect v5 kit, according to the manufacturer’s instructions.
Quality control: Upon collection of the original sequenced reads (Sequenced Reads), Cutadapt software was used to remove the adapter and reads with N (N denotes that the base information cannot be determined) ratio ≥ 10%. In addition, the read pairs were discarded if the low-quality bases (quality score ≤ 5) accounted for 50% of the entire single read. Furthermore, quality controls (including, sequencing error rate distribution, base quality distribution, and GC content distribution) were performed, based on the selected filtered reads using the above methods.
Data analysis: The BWA (v0.7.15) software was used to map the sequenced reads to the human reference genome GRCh37 (hg19). Picard software was used to remove the sequence generated by PCR-duplication. The somatic single nucleotide variations (SNVs) and InDel detection were performed using the GATK (v4.1.0.0) and muTect2 software, respectively. The ANNOVAR software (Mon, 17 Jul 2017) was used to annotate gene mutations. Lastly, the control-free software (v11.4) was used for copy number variation analysis.
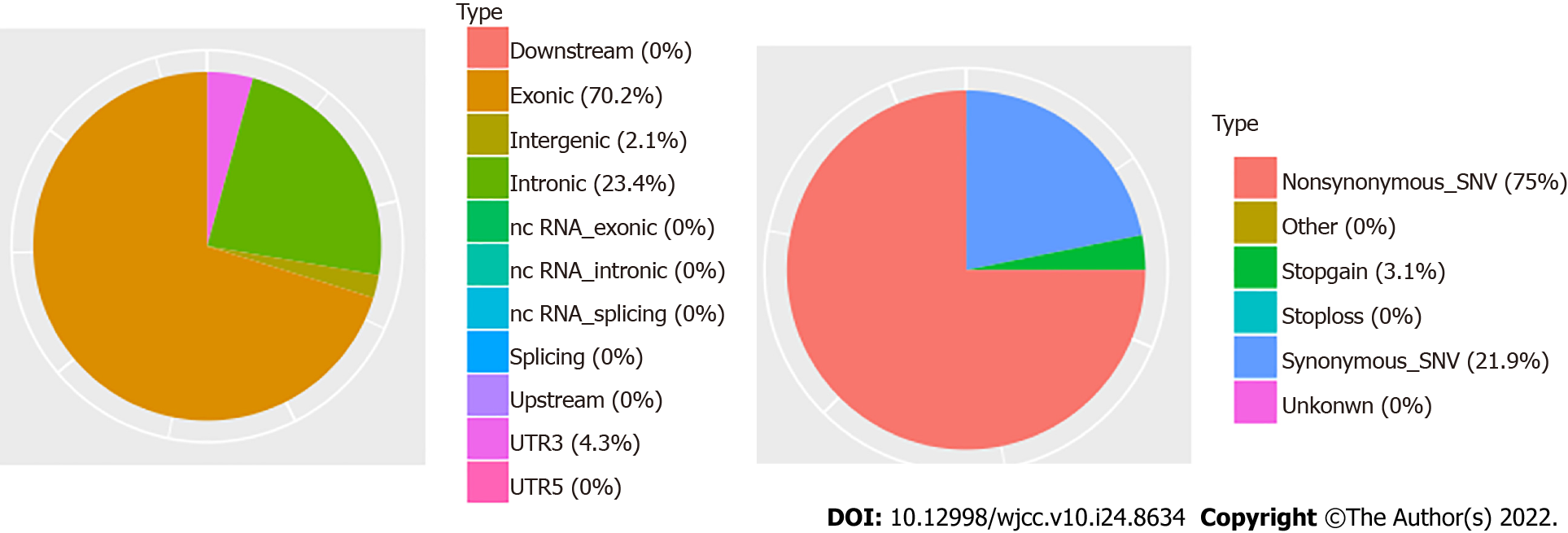
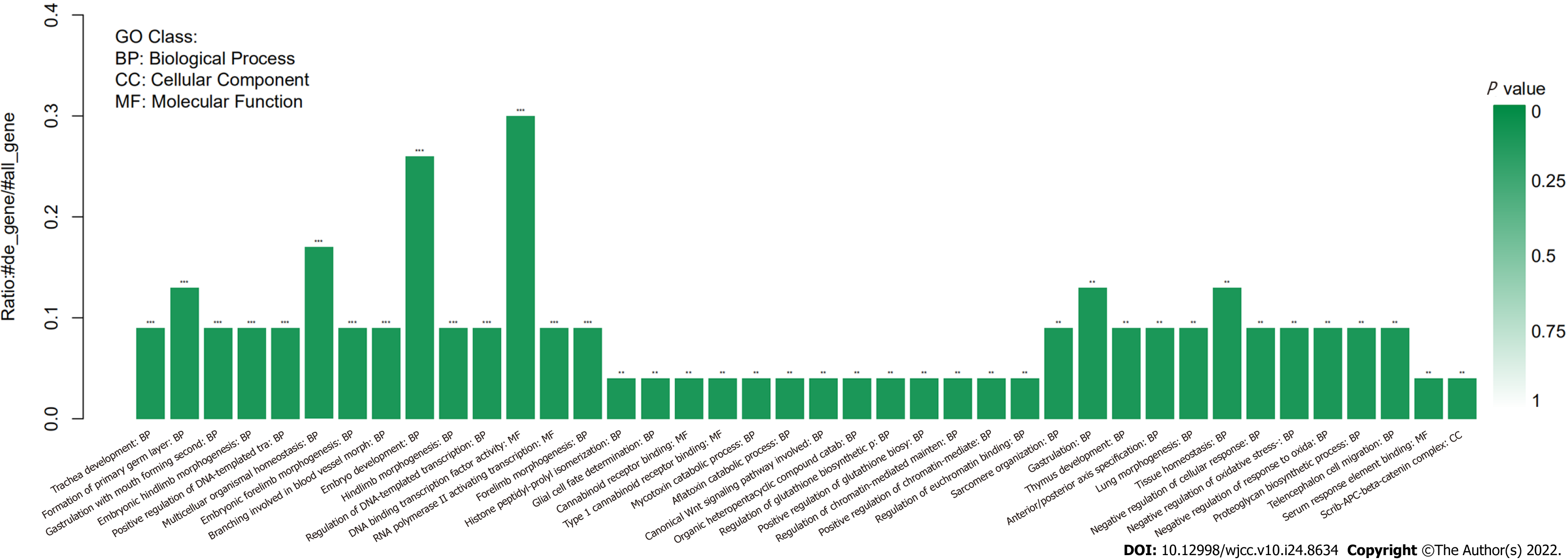
Somatic mutation analysis, based on samples from the CCS patient, identified 47 SNVs. 75% of them were nonsynonymous SNVs. In addition, approximately 70% of them occurred in exonic regions (Figure 3). Mutations of the USP24, KCNQ5, and FKBP10 genes were identified as deleterious via SIFT, LRT, Polyphen2, and Mutation Taster software (Supplementary Table 1). Based on the COSMIC data, the three mutations in genes USP24, KCNQ5, and FKBP10 may be novel in CCS. The mutated genes are associated with the regulation of DNA-templated transcription (Figure 4). Our analysis showed that the HPSE2, SPATA7, and ZC3H18 genes had markedly elevated copy numbers, while other significant altered fragments were located in the intergenic regions (Table 1).
| Chr | Start | End | Gene | Copy number | Status | Wilcoxon rank sum test P value | Kolmogorov-Smirnov test P value |
| 2 | 1.32E+08 | 1.32E+08 | 3 | Gain | 0.02315 | 0.14375 | |
| 3 | 37095399 | 37000000 | 3 | Gain | 0.00756 | 0.02719 | |
| 6 | 29910395 | 30000000 | 3 | Gain | 0.01482 | 0.03778 | |
| 6 | 1.37E+08 | 1.37E+08 | 3 | Gain | 0.03443 | 0.10684 | |
| 10 | 98711799 | 99000000 | HPSE2 | 3 | Gain | 0.0104 | 0.04592 |
| 11 | 73534113 | 74000000 | 1 | Loss | 0.00139 | 0.0043 | |
| 14 | 88442644 | 88000000 | SPATA7 | 3 | Gain | 0.02368 | 0.06465 |
| 16 | 3074297 | 3078522 | 3 | Gain | 0.00136 | 0.0099 | |
| 16 | 88600346 | 89000000 | ZC3H18 | 3 | Gain | 0.00311 | 0.00561 |
| 17 | 2274380 | 2276393 | 3 | Gain | 0.0146 | 0.03707 | |
| 19 | 38702846 | 39000000 | 3 | Gain | 0.00017 | 0.00075 | |
| 19 | 47287593 | 47000000 | 3 | Gain | 0.01445 | 0.03707 | |
| 22 | 28194746 | 28000000 | 3 | Gain | 0.0032 | 0.00641 | |
| Y | 1456244 | 1486988 | 3 | Gain | 0.00126 | 0.0036 | |
| Y | 13309548 | 14000000 | 1 | Loss | 0.00059 | 0.00437 | |
| Y | 59359925 | 59000000 | 3 | Gain | 0.01688 | 0.04401 |
The patient was diagnosed with CCS.
As part of her treatment regimen, she received albumin supplementation 10 g/d and prednisone 10 mg/d, and her electrolyte imbalance was corrected.
Treatment lasted for 15 d and her diarrhea, hypokalemia, and abdominal pain significantly improved.
Approximately 500 cases of CCS are currently described in the literature, with an estimated incidence of 1 in 1 million per year. The average age at CCS diagnosis is early 60s and it is predominantly diagnosed in males[4]. At present, there is no definite treatment plan for CCS. Previous literature reported that glucocorticoids and immunosuppression have certain benefits, and some studies reported that anti-tumor necrosis factor therapy is effective in CCS patients[5]. Our treatment plan and effect are consistent with these published works. However, there are limited studies on the pathogenesis of CCS. Therefore, based on the fact that CCS is an acquired non-genetic disease, we performed exon sequencing of excised diseased and non-diseased tissues from our CCS patient, in an attempt to elucidate CCS pathogenesis. Based on our exon sequencing results, three genes, namely, USP24, KCNQ5, and FKBP10 may be related to CCS pathogenesis.
We searched Human Gene Mutation Database (HGMD) and found that USP24 belongs to a large family of cysteine proteases that function as deubiquitinating enzymes. USP24 stabilizes the bromine domain protein and promotes malignant lung cancer[6]. In addition, Zhang L et al[7] found that USP24 deubiquitinase regulates DNA damage by directly targeting the tumor suppressor gene p53. Also, the USP24-Mcl-1 axis may represent a novel strategy in treating acute T cell lymphoma[8], whereas functional studies of the FKBP10 mutation reported an association with osteogenesis imperfecta[9]. The above two genes are both related to tumor formation and body development. Combined with the clinicopathological characteristics of CCS, we speculated that gene mutations are involved in the formation of multiple intestinal adenomas. The KCNQ family protein activates slowly during depolarization and forms heterogeneous channels with the protein encoded by KCNQ5 gene. KCN5 dependent potassium channels play an important role in airway smooth muscle relaxation[10]. Given the symptoms of diarrhea and difficult-to-correct hypokalemia of CCS, the KNQ3 mutation seems to suggest an association.
In terms of copy variation, ZC3H18 copy number losses are known to contribute to homologous recombination defects in high-grade serous ovarian cancers[11]. HPSE2 was reported to play an inhibitory role in bladder cancer[12]. Mutations in SPATA7 are associated with fundus macular degeneration[13]. CCS patients have multiple clinicopathological manifestations, such as, multiple gastrointestinal adenomas, nail atrophy, skin pigmentation, and alopecia, which may be related to the increased copy number of the three genes mentioned above.
However, due to the isolation of individual cases, the sample size of phenotypic alterations, caused by the above gene mutations, needs to be further expanded.
CCS is a rare disease and its etiology is unclear. The autoimmune etiology of CCS was previously proposed, and case reports described beneficial responses to immunosuppressive therapies such as azathioprine, anti-tumor necrosis factor antibodies, cyclosporine, and sirolimus[14,15]. Interestingly, Brigid S. Boland also conducted exome sequencing on tissue from a CCS patient who responded effectively to infliximab, and found that PRKDC mutations may be involved. However, this gene was not included in our analysis[14]. This suggests that the data on these mutations require further validation using tissues from a large CCS patient population.
In conclusion, we report a classic case of CCS, which was effectively treated with parenteral nutritional support and glucocorticoids, and we explored the pathogenesis of CCS from the perspective of gene mutation. Based on our analysis, we identified several gene mutations and alterations in gene copy numbers. However, we acknowledge that more genetic and epidemiological research is necessary to understand the complex pathogenesis of this rare but highly fatal disease.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Azarbakhsh H, Iran; VV R, India S-Editor: Ma YJ L-Editor: Webster JR P-Editor: Ma YJ
| 1. | Cronkhite Lw Jr, CANADA WJ. Generalized gastrointestinal polyposis; an unusual syndrome of polyposis, pigmentation, alopecia and onychotrophia. N Engl J Med. 1955;252:1011-1015. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 247] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 2. | Fan RY, Wang XW, Xue LJ, An R, Sheng JQ. Cronkhite-Canada syndrome polyps infiltrated with IgG4-positive plasma cells. World J Clin Cases. 2016;4:248-252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 15] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 3. | Yamakawa K, Yoshino T, Watanabe K, Kawano K, Kurita A, Matsuzaki N, Yuba Y, Yazumi S. Effectiveness of cyclosporine as a treatment for steroid-resistant Cronkhite-Canada syndrome; two case reports. BMC Gastroenterol. 2016;16:123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 4. | Dore MP, Satta R, Murino A, Pes GM. Long-lasting remission in a case of Cronkhite-Canada syndrome. BMJ Case Rep. 2018;2018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 5. | Taylor SA, Kelly J, Loomes DE. Cronkhite-Canada Syndrome: Sustained Clinical Response with Anti-TNF Therapy. Case Rep Med. 2018;2018:9409732. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 6. | Wang SA, Young MJ, Jeng WY, Liu CY, Hung JJ. USP24 stabilizes bromodomain containing proteins to promote lung cancer malignancy. Sci Rep. 2020;10:20870. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 7. | Zhang L, Nemzow L, Chen H, Lubin A, Rong X, Sun Z, Harris TK, Gong F. The deubiquitinating enzyme USP24 is a regulator of the UV damage response. Cell Rep. 2015;10:140-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 8. | Luo H, Jing B, Xia Y, Zhang Y, Hu M, Cai H, Tong Y, Zhou L, Yang L, Yang J, Lei H, Xu H, Liu C, Wu Y. WP1130 reveals USP24 as a novel target in T-cell acute lymphoblastic leukemia. Cancer Cell Int. 2019;19:56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 9. | Essawi OH, Tapaneeyaphan P, Symoens S, Gistelinck C C, Malfait F, Eyre DR, Essawi T, Callewaert B, Coucke PJ. New insights on the clinical variability of FKBP10 mutations. Eur J Med Genet. 2020;63:103980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 10. | Brueggemann LI, Haick JM, Cribbs LL, Byron KL. Differential activation of vascular smooth muscle Kv7.4, Kv7.5, and Kv7.4/7.5 channels by ML213 and ICA-069673. Mol Pharmacol. 2014;86:330-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 41] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 11. | Kanakkanthara A, Huntoon CJ, Hou X, Zhang M, Heinzen EP, O'Brien DR, Oberg AL, John Weroha S, Kaufmann SH, Karnitz LM. ZC3H18 specifically binds and activates the BRCA1 promoter to facilitate homologous recombination in ovarian cancer. Nat Commun. 2019;10:4632. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 12. | Gross-Cohen M, Feld S, Naroditsky I, Nativ O, Ilan N, Vlodavsky I. Heparanase 2 expression inversely correlates with bladder carcinoma grade and stage. Oncotarget. 2016;7:22556-22565. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 13. | Xiao X, Sun W, Li S, Jia X, Zhang Q. Spectrum, frequency, and genotype-phenotype of mutations in SPATA7. Mol Vis. 2019;25:821-833. [PubMed] |
| 14. | Boland BS, Bagi P, Valasek MA, Chang JT, Bustamante R, Madlensky L, Sandborn WJ, Harismendy O, Gupta S. Cronkhite Canada Syndrome: Significant Response to Infliximab and a Possible Clue to Pathogenesis. Am J Gastroenterol. 2016;111:746-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 15. | Ohmiya N, Nakamura M, Yamamura T, Yamada K, Nagura A, Yoshimura T, Hirooka Y, Matsumoto T, Hirata I, Goto H. Steroid-resistant Cronkhite-Canada syndrome successfully treated by cyclosporine and azathioprine. J Clin Gastroenterol. 2014;48:463-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |