Published online Aug 6, 2022. doi: 10.12998/wjcc.v10.i22.7686
Peer-review started: March 17, 2022
First decision: May 12, 2022
Revised: May 19, 2022
Accepted: June 22, 2022
Article in press: June 22, 2022
Published online: August 6, 2022
Processing time: 126 Days and 15.6 Hours
The carcinogenesis of colorectal cancer (CRC) involves many different molecules and multiple pathways, and the specific mechanism has not been elucidated until now. Existing studies on the proteomic signature profiles of CRC are relatively limited. Therefore, we herein aimed to provide a more comprehensive proteomic signature profile and discover new prognostic markers and therapeutic targets by performing proteomic analysis of CRC and paired normal tissues.
To investigate the proteomic signature and identify novel protein prognostic biomarkers of CRC.
Cancer tissues and paired normal tissues were collected from 48 patients who underwent surgical removal at the China-Japan Friendship Hospital from January 2020 to June 2021. Data independent acquisition (DIA) quantitative proteomic analysis was performed using high-performance liquid chromatography–mass spectrometry/mass spectrometry (nano-UHPLC–MS/MS) to identify differentially expressed proteins, among which those with a P adj value (t test, BH correction) < 0.05 and an absolute fold change (|log2FC|) > 2 were identified as potential markers. Differentially expressed proteins were selected by bioinformatics analysis and validated by immunohistochemical tissue microarrays, and their association with prognosis was further analyzed with the Gene Expression Profiling Interactive Analysis database to identify prognostic protein biomarkers of CRC.
Significantly differential protein expression was observed between cancer tissues and normal tissues. Compared with normal tissues, 1115 proteins were upregulated and 705 proteins were downregulated in CRC based on P adj < 0.05 and |log2FC| > 2, and bioinformatics analysis revealed that the differentially expressed proteins were involved in multiple biological processes associated with tumorigenesis, including ribosome biogenesis in eukaryotes, focal adhesion, extracellular matrix-receptor interactions and other tumor metabolism processes. Moreover, cyclin-dependent kinase inhibitor 2A (CDKN2A) expression was markedly upregulated in CRC, as validated by immunohistochemistry (0.228 vs 0.364, P = 0.0044), and was significantly enriched in tumor proliferation and signal transduction pathways such as the cell cycle and p53 signaling pathways. High CDKN2A expression was significantly correlated with poor prognosis (P = 0.021). These results demonstrated that CDKN2A functions as a driver of CRC.
Our study provides a comprehensive proteomic signature of CRC and highlights CDKN2A as a potential powerful prognostic marker and precision therapeutic target.
Core Tip: In this study, quantitative proteomic analysis of colorectal cancer (CRC) was comprehensively performed, revealing many differentially expressed proteins that may be useful for mining novel targets. The results revealed the overexpression of thousands of tumor proteins, among which cyclin-dependent kinase inhibitor 2A (CDKN2A) was the highlight of this study, and high CDKN2A expression in CRC was significantly associated with poor prognosis and could serve as a powerful prognostic marker and precision therapeutic target.
- Citation: Wang QQ, Zhou YC, Zhou Ge YJ, Qin G, Yin TF, Zhao DY, Tan C, Yao SK. Comprehensive proteomic signature and identification of CDKN2A as a promising prognostic biomarker and therapeutic target of colorectal cancer. World J Clin Cases 2022; 10(22): 7686-7697
- URL: https://www.wjgnet.com/2307-8960/full/v10/i22/7686.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i22.7686
According to the newest global cancer statistics, colorectal cancer (CRC) has become the second leading cause of cancer-related morbidity and mortality worldwide, and these rates continue to increase every year[1]. In the past few decades, the primary CRC treatment methods, including surgical resection, chemo- or chemoradiotherapy, immunotherapy and targeted therapy, have significantly improved[2]. However, the prognosis of CRC patients, especially those diagnosed at an advanced stage, remains poor. Colorectal carcinogenesis encompasses mechanisms of abnormal proliferation, differentiation, resistance to apoptosis and surrounding invasion of colonic epithelial cells[3]. A variety of genes and the interplay of multiple signaling pathways have been proposed to underlie the tumorigenesis of CRC, but the complex mechanism remains incompletely understood.
On the basis of the biological central dogma, gene mutation plays an important role in the occurrence and development of malignant tumors, including CRC. Previous studies have revealed possible gene mutations in patients with CRC; however, their effects are not completely consistent. For example, Pearlman et al[4] proposed that cyclin-dependent kinase inhibitor 2A (CDKN2A)mutation had nothing to do with CRC risk; in contrast, Lee et al[5] found that CDKN2A was a tumor suppressor, and Li et al[6] insisted that CDKN2A was an oncogenic gene that was significantly correlated with poor prognosis. Therefore, gene mutation alone could not fully explain the occurrence and metastasis of tumors: the perspective of proteins, the executors of function, were also needed.
Proteins play important physiological roles and have an enormous impact on cellular biology and human health, and proteomics has thus become an indispensable tool for mechanistic studies[7,8]. Although rapidly accumulating omics studies, such as genomic, transcriptomic and epigenetic studies, have been conducted, genomic and epigenetic analyses cannot be used to fully elucidate the large variance in cancer mechanisms[9]. Proteomics analysis has been gradually applied to a variety of cancer types, such as pancreatic ductal adenocarcinoma and breast cancer, to characterize the tumor stage, predict metastasis, identify candidate cancer biomarkers, and evaluate therapeutic effects[10,11]. Nevertheless, our knowledge of the proteomics mechanisms of CRC is still limited.
Therefore, this study aimed to elucidate the proteomic profiles of CRC by performing proteomics analysis of CRC tissues and adjacent normal mucosal tissues. By performing tissue microarray validation, gene expression profiling interactive analysis (GEPIA) genomics and survival analyses in combination, we aimed to identify potential target proteins among numerous differentially expressed proteins and provide clinicians with more evidence for precision CRC treatment.
A total of 48 CRC patients underwent surgical resection of colorectal tumors between January 2020 and June 2021 at the China-Japan Friendship Hospital. The inclusion criteria were as follows: The diagnosis of CRC was confirmed via identification by the pathology department. The exclusion criteria were as follows: (1) Patients with familial adenomatous polyposis, hereditary nonpolyposis colorectal cancer, or synchronous multiple CRCs; (2) Patients with inflammatory bowel disease; (3) Patients who had received prior treatments, including chemoradiotherapy, targeted therapy, and immunosuppressive therapy, during the previous 6 mo before resection; and (4) Patients were lacking informed consent or had missing data. Medical records were reviewed to obtain clinicopathological information, such as the patient’s age, sex, differentiation status, and TNM stage. The study was approved by the Ethics Committee of China-Japan Friendship Hospital (No. 2018-116-K85) and was conducted in accordance with the Declaration of Helsinki.
After resection of fresh colorectal cancer tissues along with the adjacent noncancerous tissues (>5 cm away from the tumor), the collected samples were cleaned with cold normal saline, immediately frozen in liquid nitrogen and stored at -80 ℃ until use. Sample preparation includes multiple steps: washes, denaturation, reduction and alkylation, digestion by trypsin and extraction and cleanup of mixed peptides. Commercially available iST Sample Preparation kit (PreOmics, Germany) was used following the manufacturer’s recommendation. Briefly, 50 µL of Lyse buffer was added and heated at 95 °C for 10 min at 1000 rpm with agitation. After cooling the sample to room temperature, trypsin digestion buffer was added, and the sample incubated at 37 °C for 2 h at 500 rpm with shaking. The digestion process was stopped with a stop buffer. Sample clean-up and desalting was carried out in the iST cartridge using the recommended wash buffers. Peptides were eluted with elution buffer (2 × 100 µL), and then lyophilized by SpeedVac.
The peptides were redissolved in solvent A (A: 0.1% formic acid in water) and analyzed by an Orbitrap Fusion Lumos Tribrid coupled to an EASY-nanoLC 1200 system (Thermo Fisher Scientific, MA, USA). Next, 4 μL of the peptide sample was loaded onto a 25 cm analytical column (75 μm inner diameter, 1.9 μm resin (Dr Maisch)) and separated by gradient applied over 90 min as follows: 4% buffer B (80% ACN with 0.1% FA) for 3 min; a stepwise increase to 50% buffer B in 82 min; increase to 95% buffer B in 1 min; and a hold for 7 min. The column flow rate was maintained at 250 nL/min, and the column temperature was 55°C. The electrospray voltage was set to 2 kV. The mass spectrometer was run under data independent acquisition (DIA) mode with hybrid data strategy. A survey scan was acquired at 120,000 resolution, normalized AGC target of 250% and a maximum injection time of 100ms. In the DIA MS2 acquisition, variable Isolation window were performed. One full scan followed by 20 windows with resolution of 50,000, normalized AGC target of 200%, a maximum injection time of 86ms and normalized collision energy at 33.
Raw DIA data were processed and analyzed by Spectronaut 15.0 (Biognosys AG, Switzerland) with default settings. Spectronaut was set up to search the UniProt-Homo sapiens database (version 201907, 20428 entries) assuming that trypsin was used as the digestion enzyme. Carbamidomethyl (C) was specified as the fixed modification. Oxidation (M) was specified as the variable modification. The retention time prediction type was set to dynamic iRT. Data extraction was performed by Spectronaut based on extensive mass calibration. Spectronaut was used to determine the ideal extraction window dynamically depending on the iRT calibration and gradient stability. The Q value (FDR) cutoff on the precursor level was 1%, and the protein level was 1%. Decoy generation was set to mutated, which was similar to scrambled but only applied a random number of AA position swamps (min = 2, max = length/2). The normalization strategy was set to local normalization. The average top 3 filtered peptides that passed the 1% Q value cutoff were used to calculate the major group quantities. After application of the t test, differentially expressed proteins were identified by a P adj value < 0.05 and an absolute fold change > 2. The R package was used to visualize the differential expression for bioinformatics analysis. Gene set enrichment analysis (GSEA) was completed by using GSEA v4.2.3 (https://www.gsea-msigdb.org/gsea/index.jsp).
The Human Protein Atlas (HPA, http://www.proteinatlas.org/) contains images of histological sections from normal and cancer tissues obtained by immunohistochemistry and is publicly available at v20.proteinatlas.org. Antibodies were labeled with DAB (3, 3'-diaminobenzidine), and the resulting brown staining indicated where an antibody had bound to its corresponding antigen. The section was further counterstained with hematoxylin to enable visualization of microscopic features. Each sample was represented by 1 mm tissue cores. CRC pathology and normal colon tissue microarrays were downloaded to quantify protein expression. ImageJ software was used to assess the staining area and integrated optical density (IOD) to determine the average optical density (AOD) values. All images were manually evaluated by two independent observers. The Mann–Whitney U test was used for statistical analyses, and the statistical significance level was set to 0.05.
GEPIA (http://gepia.cancer-pku.cn/index.html) provides customizable functions such as tumor/ normal differential expression analysis and patient survival analysis[12]. A box plot was generated to compare gene expression in CRC. Genes with |log2FC| values >1 and q values < 0.01 were considered to be differentially expressed. GEPIA was also used to perform overall survival (OS) analysis based on gene expression. We selected the median gene expression as a group cutoff for splitting the high- and low-expression cohorts. Kaplan–Meier plots and the log-rank test were used to analyze differences in survival times between patients with high and low expression.
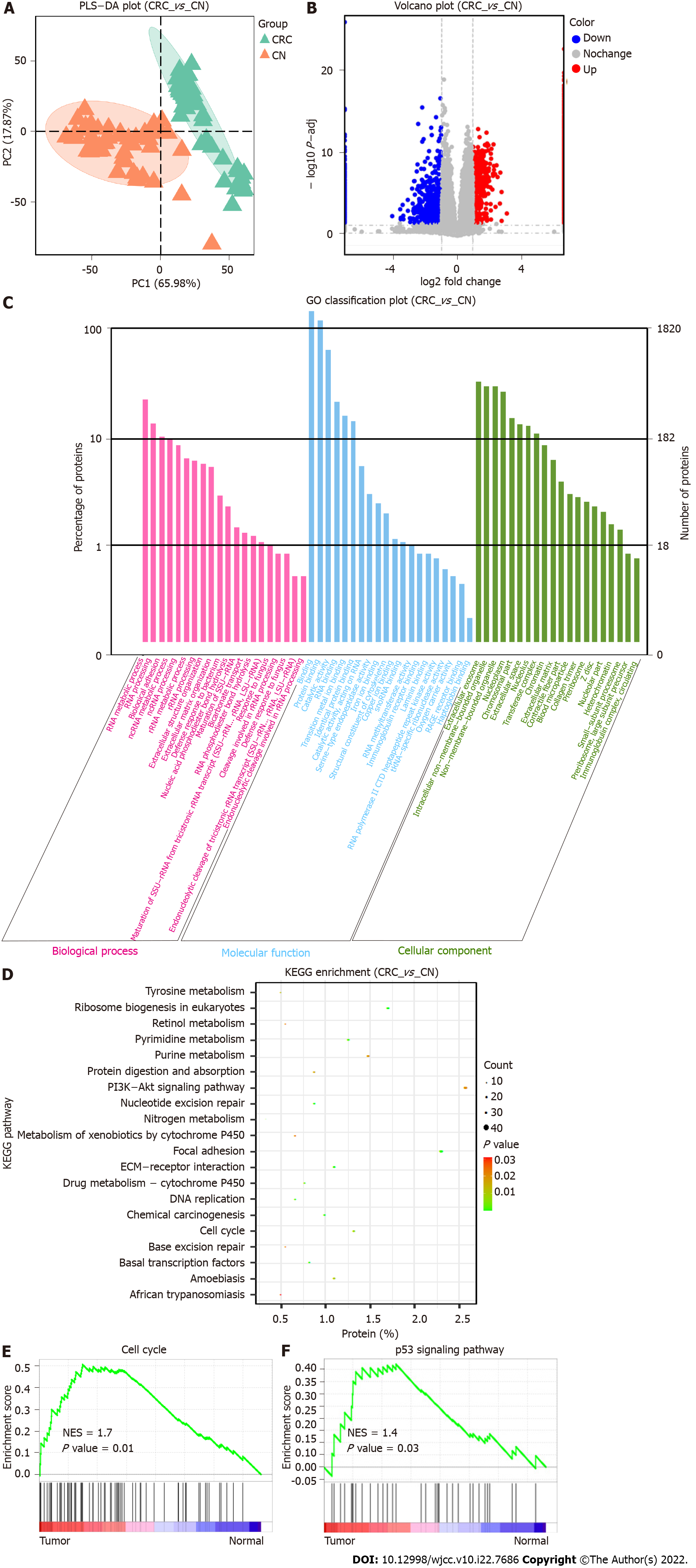
We collected a total of 48 pairs of cancer tissues and matched adjacent normal tissues after surgical treatment of CRC for quantitative proteomic detection. We found differences in the protein signatures between the CRC group and the normal group, and PLS-DA could clearly distinguish the two groups (Figure 1A). In total, we identified 7643 proteins as well as 1820 differentially expressed proteins with an FDR < 1% in the CRC group vs the normal group, which were used for subsequent enrichment analysis and tumor target mining. There were 1115 proteins upregulated in the tumor group, including CEAM6/5, IFM1, LAT1, RAI3, VISL1, TACC3, HS71L, IMA1, DIEXF, and UPAR, while 705 proteins, including SYUG, NOS1, RT26, GEMI4, HAUS5, TET1, NB5R2, PERI, LIPS and PPLA, were downregulated in the tumor group compared with the normal group (Figure 1B, Supplementary Figure 1).
GO analysis of the differentially expressed proteins was performed, and the differentially expressed proteins were significantly enriched in biological processes (BPs; RNA metabolic processing, RNA processing, biological adhesion, and other processes), cellular components (CCs, binding, protein binding, catalytic activity, RNA binding, etc.), and molecular functions (MFs, extracellular exosome, intracellular nonmembrane-bounded organelle, nucleoplasm, etc.)(Figure 1C). The differentially expressed proteins were further subjected to subsequent Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis, which identified the PI3K-Akt signaling, focal adhesion, ribosome biogenesis in eukaryotes, cell cycle, extracellular matrix-receptor interaction and DNA replication pathways as being significantly enriched (Figure 1D). To explore the mechanisms of the differentially expressed proteins in cancer development to the greatest extent possible, we performed further GSEA, which showed that transcription factors, the cell cycle, the p53 signaling pathway and other processes were significantly enriched in CRC tissues (Figure 1E and F, Supplementary Figures 2 and 3).
Visualizing and displaying the bioinformatics analysis results led to the identification of a special protein, CDKN2A, which was significantly upregulated in tumors compared with the normal tissues. KEGG pathway enrichment analysis showed that CDKN2A was involved in the cell cycle, PI3K-Akt signaling pathway, and p53 signaling pathway. GSEA further confirmed that CDKN2A was involved in regulating the cell cycle and p53 signaling pathway and functioned as a core gene in the gene set. Therefore, we speculated that CDKN2A plays a pivotal role in colorectal carcinogenesis and has potential as a prognostic biomarker.
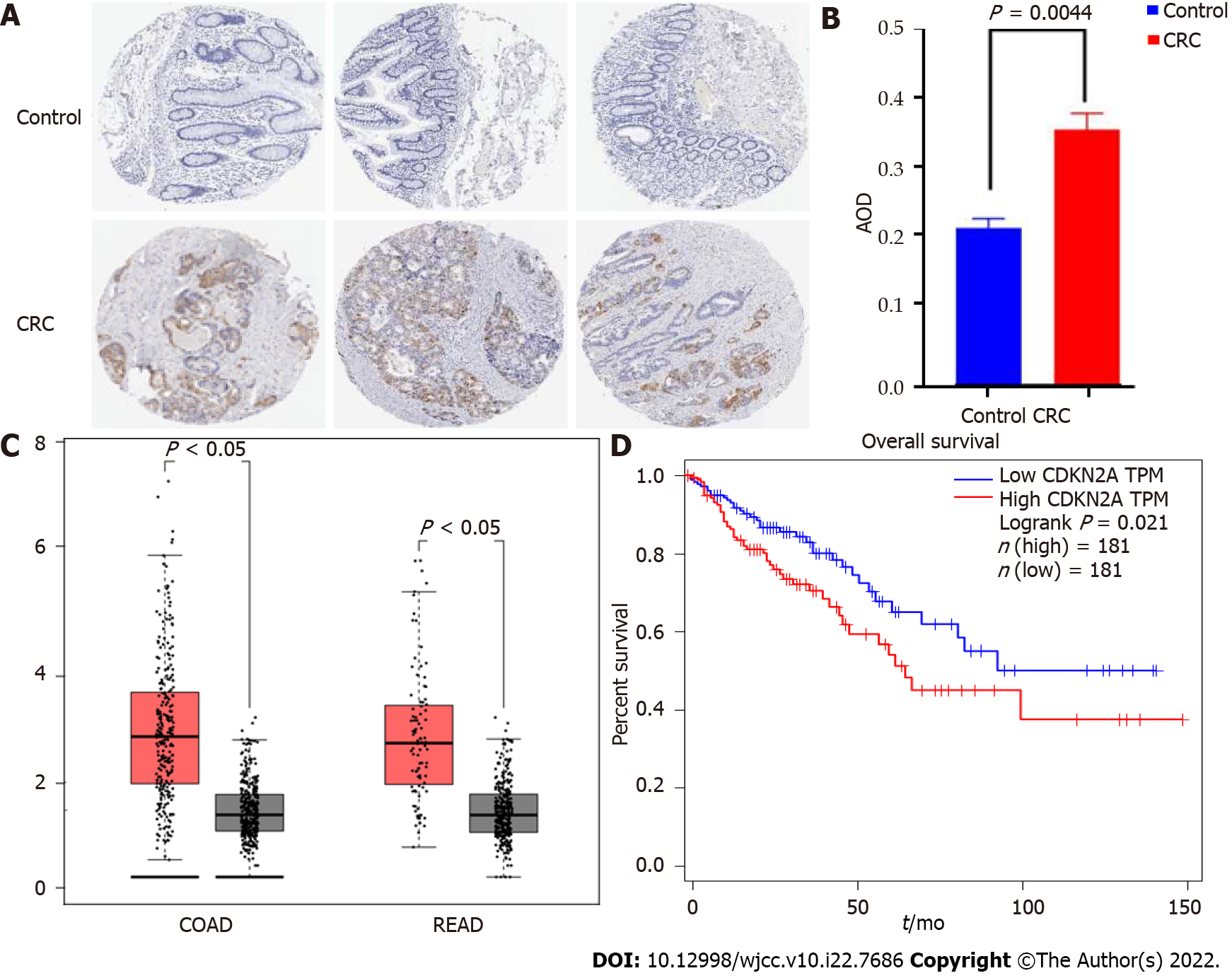
We collected protein microarrays from a total of 3 normal colon tissues and 12 CRC tissues from the HPA database that had been subjected to antigen antibody labeling reactions using CAB00093 antibodies (Supplementary Figure 4). The AOD value of each chip was calculated using ImageJ (Supplementary Table 1). The protein expression of CDKN2A was significantly different between CRC and normal samples (P < 0.05) (Figure 2A and B). Low or no protein expression of CDKN2A was observed in the normal samples, while the staining intensity and area were significantly increased in CRC samples, implying that CDKN2A protein expression was significantly upregulated (P = 0.0044). These results were consistent with those of proteomic analysis and further confirmed that the protein expression of CDKN2A was significantly increased in CRC. The results are presented as a bar graph (Figure 2B).
To explore the expression profile of the CDKN2A gene in normal mucosa and CRC tissues as well as its association with patient prognosis, we performed differential gene expression analysis and survival analysis using the GEPIA database. We found that CDKN2A gene expression differed significantly between CRC and normal tissues, with significantly higher expression in colon cancer (P < 0.05), and the same trend was observed in rectal cancer compared with normal tissue (P < 0.05) (Figure 2C). Further overall survival analysis using GEPIA showed that a total of 362 colorectal cancer patients were matched to normal tissues from the TCGA and GTEx databases and that patients with high expression of the CDKN2A gene had a significantly decreased overall survival (P = 0.021), while patients with low expression of the CDKN2A gene had a relatively good prognosis (Figure 2D).
Signaling proteins are very attractive for the precise treatment of cancer. However, research on the proteomic spectrum of CRC is still limited. In this study, 48 pairs of cancer and adjacent normal tissues were collected for proteomics analysis, and 1115 upregulated proteins and 705 downregulated proteins were observed in the CRC group. First, we made an enormous effort to provide more comprehensive proteomic signatures for CRC. Second, we identified many proteins with abnormal expression abundance in CRC and validated their potential as tumor targets. Interestingly, among all the differentially expressed proteins, we highlighted one upregulated protein, cyclin-dependent kinase inhibitor 2A (CDKN2A), which is involved in both the cell cycle and the p53 signaling pathway, and a tissue microarray confirmed that the CDKN2A protein was highly expressed in CRC and was associated with low OS survival and poor prognosis. This suggests that CDKN2A plays a cancer-driving role in CRC and is predictive of poor survival.
CDKN2A, is a well-known tumor suppressor located at chromosome 9p21[13,14]. Two proteins are encoded, p16INK4a and p14ARF, which exert different regulatory effects on the cell cycle: p14ARF interacts with and degrades MDM2, preventing p53 inactivation by ubiquitin-mediated proteolysis or transcriptional silencing, and P16INK4a binds CDK4 and CDK6 to prevent phosphorylation of the Rb protein[13,15]. Loss-of-function mutations or homozygous deletions of CDKN2A resulted in the loss of both proteins, releasing the G1–S and G2–M cell cycle checkpoints and resulting in uninhibited cell proliferation and tumor formation[16]. A literature database search revealed that CDKN2A has been extensively researched in the context of melanoma, pancreatic cancer and other tumor types, but the findings are controversial[15,17].
CDKN2A plays a role in CRC initiation and progression via multiple mechanisms. Ferroptosis, a newly defined form of cell death that differs from apoptosis and autophagy and is characterized by iron overload, lipid reactive oxygen species and lipid peroxidation, is involved in the carcinogenesis, progression, and treatment of CRC[18]. A study showed that CDKN2A is a ferroptosis-associated gene that is involved in the iron metabolism and tumorigenesis of CRC by enhancing p53-dependent transactivation and ferroptosis. The upregulation of this gene in CRC correlated with poor prognosis and could be considered part of a predictive model[19]. However, our differential protein enrichment analysis did not reveal the involvement of CDKN2A in this process. In addition, CDKN2A is a prominent hallmark of cellular senescence, whereas reactive oxygen species, DNA damage, and chronic inflammation all induce cellular senescence[20-22]. In mice, p53 genes in senescent cells might transform these cells into highly aggressive, cancer-initiating cells with long-term accumulation progressing to a cancerous state[23]. Another recent study on the inflammatory microenvironment in CRC found significant differences in CDKN2A expression between the epithelium and stroma, with a 10-fold decrease in the epithelium and a 17-fold increase in the stroma[24,25]. This study speculated that the activation of CDKN2A expression in the stroma was influenced by cellular senescence and oncogene activation, and senescent fibroblasts prematurely accumulated in the stroma as a result of chronic inflammation and oxidative stress[24]. Moreover, CDKN2A participates in the development of CRC through the Wnt/β-catenin signaling pathway[13,26].
The role of CDKN2A in CRC is also controversial. In 2017, a comprehensive cancer center conducted exploratory research on CRC cancer susceptibility genes and found that CDKN2A had high- or moderate-level gene mutations. This study pointed out that CDKN2A gene mutation was not traditionally associated with CRC risk[4,27]. Some studies have shown that CDKN2A usually acts as a tumor suppressor. On the one hand, CRC patients have a poor prognosis due to the occurrence of promoter region hypermethylation, CDKN2A gene silencing or dysfunction, which causes uncontrolled cell proliferation and carcinogenesis[5,28]. On the other hand, CDKN2A participates in the ILF3-AS1/EZH2/CDKN2A/H3K27me3 axis, and downregulation of CDKN2A accelerates CRC proliferation and metastasis[29]. However, the results of some other studies were opposite but consistent with our findings. A recent convincing study demonstrated that CDKN2A was a cancer-driver gene that could effectively predict the poor prognosis and clinical status of CRC patients[6]. Concordant with this, several studies found that CDKN2A was a risk gene for overexpression in CRC and highlighted that increased CDKN2A protein expression, rather than loss of protein expression, was associated with poor prognosis in CRC[3,19,25,30].
Possible explanations for these contradictory results are as follows: (1) It is well known that CDKN2A protein expression is affected by genomic deletions, point mutations, and promoter region hypermethylation[28,31]. The promoter region of the CDKN2A gene was hypermethylated, combined with methylase, and thus could not bind to the RNA binding enzyme, resulting in decreased or silent protein expression. Therefore, we speculated that this result was due to hypermethylation of gene body regions rather than promoter regions, and CDKN2A protein expression was thus positively altered in the tumor with the help of histone modification[32]; (2) An additional speculation was that CDKN2A protein expression was influenced by the stage of tumor development. CDKN2A is known to be involved in the epithelial-stromal microenvironment during CRC development, with different stages of tumor progression and diverse states of the tumor microenvironment, and CDKN2A protein expression thus varies substantially in multiple studies[24]. It has also been proposed that the upregulation of CDKN2A protein expression is a consequence of tumor metastasis[30]; and (3) The degree of cellular senescence varies, leading to different trends in the expression of CDKN2A and therefore to conflicting inferences about its role in tumors[22]. Additionally, since the mRNA expression of CDKN2A could be promoted by nontumor cells present in the tumor microenvironment, interpreting of bulk transcriptional data remains challenging, requiring single-cell RNA-seq analysis to analyze population-specific transcription of this gene[33]. Because of the limitations of the study design, small sample size, and publication bias, the mechanism of CDKN2A in CRC has not been fully elucidated despite multiple investigations being conducted. This problem needs to be resolved in the future to fully construct the genomic, transcriptomic, epigenetic and proteomic landscape of CDKN2A in CRC.
Our study more comprehensively revealed the proteomic signature of CRC, although not for the first time, and provided important biological and clinical insights. Of course, our study has some limitations. First, similar to the shortcomings of previous studies, this was a single-center study, and the findings may therefore not have general applicability. Second, the lack of genomic and epigenetic data from our own tissues prevented the complete elucidation of the gene-mRNA-protein chain. To some extent, it is possible to complement the understanding of the CRC mechanism at the protein level. Finally, to fully substantiate the tumor suppressor or cancer-promoting role of the CDKN2A gene, multicenter studies are needed.
In conclusion, this study more comprehensively revealed the proteomic signature of CRC. CDKN2A was identified among all the differentially expressed proteins and shown to be involved in the cell cycle, p53 signaling pathway and other mechanisms. Moreover, CDKN2A is associated with poor overall survival and may serve as a prognostic biomarker and treatment target for CRC.
Colorectal cancer (CRC) has become the second leading cause of cancer-related deaths worldwide; however, its specific pathogenic mechanism has not been elucidated until now. Exploring the proteomic features of CRC, mining protein prognostic biomarkers and identifying precise therapeutic targets are important for improving the prognosis of CRC patients.
Despite improvements in diagnosis and treatment, the overall survival of CRC patients is still not very satisfactory. In particular, the 5-year survival rate of patients with advanced CRC remains less than 20%. Therefore, there is an urgent need for the clinical discovery of novel biomarkers and therapeutic targets. With the development of proteomic technology, proteins are considered potential biomarkers and precision therapy targets in CRC.
To comprehensively characterize the proteomic features and identify novel prognostic biomarkers and precise therapeutic targets of CRC.
The differentially expressed proteins (DEPs) were obtained by performing liquid chromatography–mass spectrometry detection of clinical samples, including colorectal cancer tissues and paired paracancerous tissues. Through further bioinformatic analysis, immunohistochemical (IHC) verification, and the correlation between DEPs and overall survival, protein prognostic biomarkers and therapeutic targets for CRC were identified.
The authors first provide a comprehensive characterization of the proteomic signature of CRC. Compared with normal tissues, 1115 proteins were upregulated and 705 proteins downregulated in CRC based on a P adj < 0.05 and |log2FC| > 2 criteria, and the DEPs were involved in a variety of different molecular functions and signal transduction pathways. In addition, through IHC verification and survival analysis, we found that high expression of cyclin-dependent kinase inhibitor 2A (CDKN2A) protein was significantly correlated with poor prognosis in CRC. This demonstrated that CDKN2A could be used as a prognostic biomarker and a target for precision therapy in CRC.
The proteomic signature of CRC has been comprehensively characterized, and CDKN2A has strong potential as a prognostic biomarker and a target for precision therapy in CRC.
Novel protein prognostic biomarkers and precise therapeutic targets provide new opportunities to improve the prognosis of CRC patients.
The authors thank The Human Protein Atlas for providing invaluable datasets for statistical analyses.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Batyrbekov K, Kazakhstan; Koustas E, Greece S-Editor: Ma YJ L-Editor: A P-Editor: Ma YJ
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64104] [Article Influence: 16026.0] [Reference Citation Analysis (174)] |
| 2. | Benson AB, Venook AP, Al-Hawary MM, Arain MA, Chen YJ, Ciombor KK, Cohen S, Cooper HS, Deming D, Farkas L, Garrido-Laguna I, Grem JL, Gunn A, Hecht JR, Hoffe S, Hubbard J, Hunt S, Johung KL, Kirilcuk N, Krishnamurthi S, Messersmith WA, Meyerhardt J, Miller ED, Mulcahy MF, Nurkin S, Overman MJ, Parikh A, Patel H, Pedersen K, Saltz L, Schneider C, Shibata D, Skibber JM, Sofocleous CT, Stoffel EM, Stotsky-Himelfarb E, Willett CG, Gregory KM, Gurski LA. Colon Cancer, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2021;19:329-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1054] [Cited by in RCA: 944] [Article Influence: 236.0] [Reference Citation Analysis (16)] |
| 3. | Poursheikhani A, Abbaszadegan MR, Nokhandani N, Kerachian MA. Integration analysis of long non-coding RNA (lncRNA) role in tumorigenesis of colon adenocarcinoma. BMC Med Genomics. 2020;13:108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 4. | Pearlman R, Frankel WL, Swanson B, Zhao W, Yilmaz A, Miller K, Bacher J, Bigley C, Nelsen L, Goodfellow PJ, Goldberg RM, Paskett E, Shields PG, Freudenheim JL, Stanich PP, Lattimer I, Arnold M, Liyanarachchi S, Kalady M, Heald B, Greenwood C, Paquette I, Prues M, Draper DJ, Lindeman C, Kuebler JP, Reynolds K, Brell JM, Shaper AA, Mahesh S, Buie N, Weeman K, Shine K, Haut M, Edwards J, Bastola S, Wickham K, Khanduja KS, Zacks R, Pritchard CC, Shirts BH, Jacobson A, Allen B, de la Chapelle A, Hampel H; Ohio Colorectal Cancer Prevention Initiative Study Group. Prevalence and Spectrum of Germline Cancer Susceptibility Gene Mutations Among Patients With Early-Onset Colorectal Cancer. JAMA Oncol. 2017;3:464-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 369] [Cited by in RCA: 514] [Article Influence: 64.3] [Reference Citation Analysis (0)] |
| 5. | Lee CC, Kuo YC, Hu JM, Chang PK, Sun CA, Yang T, Li CW, Chen CY, Lin FH, Hsu CH, Chou YC. MTNR1B polymorphisms with CDKN2A and MGMT methylation status are associated with poor prognosis of colorectal cancer in Taiwan. World J Gastroenterol. 2021;27:5737-5752. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 6] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 6. | Li CH, Haider S, Boutros PC. Age influences on the molecular presentation of tumours. Nat Commun. 2022;13:208. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 47] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 7. | Xiao Q, Zhang F, Xu L, Yue L, Kon OL, Zhu Y, Guo T. High-throughput proteomics and AI for cancer biomarker discovery. Adv Drug Deliv Rev. 2021;176:113844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 79] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 8. | Gong Y, Liu Y, Wang T, Li Z, Gao L, Chen H, Shu Y, Li Y, Xu H, Zhou Z, Dai L. Age-Associated Proteomic Signatures and Potential Clinically Actionable Targets of Colorectal Cancer. Mol Cell Proteomics. 2021;20:100115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 9. | Vasaikar S, Huang C, Wang X, Petyuk VA, Savage SR, Wen B, Dou Y, Zhang Y, Shi Z, Arshad OA, Gritsenko MA, Zimmerman LJ, McDermott JE, Clauss TR, Moore RJ, Zhao R, Monroe ME, Wang YT, Chambers MC, Slebos RJC, Lau KS, Mo Q, Ding L, Ellis M, Thiagarajan M, Kinsinger CR, Rodriguez H, Smith RD, Rodland KD, Liebler DC, Liu T, Zhang B; Clinical Proteomic Tumor Analysis Consortium. Proteogenomic Analysis of Human Colon Cancer Reveals New Therapeutic Opportunities. Cell. 2019;177:1035-1049.e19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 527] [Article Influence: 87.8] [Reference Citation Analysis (0)] |
| 10. | Cao L, Huang C, Cui Zhou D, Hu Y, Lih TM, Savage SR, Krug K, Clark DJ, Schnaubelt M, Chen L, da Veiga Leprevost F, Eguez RV, Yang W, Pan J, Wen B, Dou Y, Jiang W, Liao Y, Shi Z, Terekhanova NV, Cao S, Lu RJ, Li Y, Liu R, Zhu H, Ronning P, Wu Y, Wyczalkowski MA, Easwaran H, Danilova L, Mer AS, Yoo S, Wang JM, Liu W, Haibe-Kains B, Thiagarajan M, Jewell SD, Hostetter G, Newton CJ, Li QK, Roehrl MH, Fenyö D, Wang P, Nesvizhskii AI, Mani DR, Omenn GS, Boja ES, Mesri M, Robles AI, Rodriguez H, Bathe OF, Chan DW, Hruban RH, Ding L, Zhang B, Zhang H; Clinical Proteomic Tumor Analysis Consortium. Proteogenomic characterization of pancreatic ductal adenocarcinoma. Cell. 2021;184:5031-5052.e26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 285] [Cited by in RCA: 335] [Article Influence: 83.8] [Reference Citation Analysis (0)] |
| 11. | Terkelsen T, Pernemalm M, Gromov P, Børresen-Dale AL, Krogh A, Haakensen VD, Lethiö J, Papaleo E, Gromova I. High-throughput proteomics of breast cancer interstitial fluid: identification of tumor subtype-specific serologically relevant biomarkers. Mol Oncol. 2021;15:429-461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 12. | Tang Z, Li C, Kang B, Gao G, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5550] [Cited by in RCA: 7036] [Article Influence: 879.5] [Reference Citation Analysis (0)] |
| 13. | Adib E, Nassar AH, Akl EW, Abou Alaiwi S, Nuzzo PV, Mouhieddine TH, Sonpavde G, Haddad RI, Mouw KW, Giannakis M, Hodi FS, Shukla SA, Gusev A, Braun DA, Choueiri TK, Kwiatkowski DJ. CDKN2A Alterations and Response to Immunotherapy in Solid Tumors. Clin Cancer Res. 2021;27:4025-4035. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 75] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 14. | Zhao R, Choi BY, Lee MH, Bode AM, Dong Z. Implications of Genetic and Epigenetic Alterations of CDKN2A (p16(INK4a)) in Cancer. EBioMedicine. 2016;8:30-39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 172] [Cited by in RCA: 285] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 15. | Chan SH, Chiang J, Ngeow J. CDKN2A germline alterations and the relevance of genotype-phenotype associations in cancer predisposition. Hered Cancer Clin Pract. 2021;19:21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 50] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 16. | Bihl MP, Foerster A, Lugli A, Zlobec I. Characterization of CDKN2A(p16) methylation and impact in colorectal cancer: systematic analysis using pyrosequencing. J Transl Med. 2012;10:173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 17. | Ipenburg NA, El Sharouni MA, van Doorn R, van Diest PJ, van Leerdam ME, van der Rhee JI, Goeman J, Kukutsch NA; Netherlands Foundation for Detection of Hereditary Tumors collaborative investigators. Lack of association between CDKN2A germline mutations and survival in patients with melanoma: A retrospective cohort study. J Am Acad Dermatol. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 18. | Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, Morrison B 3rd, Stockwell BR. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060-1072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4711] [Cited by in RCA: 11531] [Article Influence: 887.0] [Reference Citation Analysis (1)] |
| 19. | Shao Y, Jia H, Huang L, Li S, Wang C, Aikemu B, Yang G, Hong H, Yang X, Zhang S, Sun J, Zheng M. An Original Ferroptosis-Related Gene Signature Effectively Predicts the Prognosis and Clinical Status for Colorectal Cancer Patients. Front Oncol. 2021;11:711776. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 66] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 20. | Uyar B, Palmer D, Kowald A, Murua Escobar H, Barrantes I, Möller S, Akalin A, Fuellen G. Single-cell analyses of aging, inflammation and senescence. Ageing Res Rev. 2020;64:101156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 116] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 21. | Avelar RA, Ortega JG, Tacutu R, Tyler EJ, Bennett D, Binetti P, Budovsky A, Chatsirisupachai K, Johnson E, Murray A, Shields S, Tejada-Martinez D, Thornton D, Fraifeld VE, Bishop CL, de Magalhães JP. A multidimensional systems biology analysis of cellular senescence in aging and disease. Genome Biol. 2020;21:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 172] [Cited by in RCA: 228] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 22. | Liu JY, Souroullas GP, Diekman BO, Krishnamurthy J, Hall BM, Sorrentino JA, Parker JS, Sessions GA, Gudkov AV, Sharpless NE. Cells exhibiting strong p16 INK4a promoter activation in vivo display features of senescence. Proc Natl Acad Sci U S A. 2019;116:2603-2611. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 138] [Cited by in RCA: 238] [Article Influence: 39.7] [Reference Citation Analysis (0)] |
| 23. | Brenner E, Schörg BF, Ahmetlić F, Wieder T, Hilke FJ, Simon N, Schroeder C, Demidov G, Riedel T, Fehrenbacher B, Schaller M, Forschner A, Eigentler T, Niessner H, Sinnberg T, Böhm KS, Hömberg N, Braumüller H, Dauch D, Zwirner S, Zender L, Sonanini D, Geishauser A, Bauer J, Eichner M, Jarick KJ, Beilhack A, Biskup S, Döcker D, Schadendorf D, Quintanilla-Martinez L, Pichler BJ, Kneilling M, Mocikat R, Röcken M. Cancer immune control needs senescence induction by interferon-dependent cell cycle regulator pathways in tumours. Nat Commun. 2020;11:1335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 91] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 24. | Ideta T, Li B, Flynn C, Igarashi Y, Lowman G, Looney T, Devers TJ, Birk J, Forouhar F, Giardina C, Rosenberg DW. The Epithelial-Stromal Microenvironment in Early Colonic Neoplasia. Mol Cancer Res. 2022;20:56-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 25. | Chen Z, Guo Y, Zhao D, Zou Q, Yu F, Zhang L, Xu L. Comprehensive Analysis Revealed that CDKN2A is a Biomarker for Immune Infiltrates in Multiple Cancers. Front Cell Dev Biol. 2021;9:808208. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 54] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 26. | Ewing RM, Song J, Gokulrangan G, Bai S, Bowler EH, Bolton R, Skipp P, Wang Y, Wang Z. Multiproteomic and Transcriptomic Analysis of Oncogenic β-Catenin Molecular Networks. J Proteome Res. 2018;17:2216-2225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 27. | Ohuchi M, Sakamoto Y, Tokunaga R, Kiyozumi Y, Nakamura K, Izumi D, Kosumi K, Harada K, Kurashige J, Iwatsuki M, Baba Y, Miyamoto Y, Yoshida N, Shono T, Naoe H, Sasaki Y, Baba H. Increased EZH2 expression during the adenoma-carcinoma sequence in colorectal cancer. Oncol Lett. 2018;16:5275-5281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 28. | Xing X, Cai W, Shi H, Wang Y, Li M, Jiao J, Chen M. The prognostic value of CDKN2A hypermethylation in colorectal cancer: a meta-analysis. Br J Cancer. 2013;108:2542-2548. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 89] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 29. | Hong S, Li S, Bi M, Yu H, Yan Z, Liu T, Wang H. lncRNA ILF3-AS1 promotes proliferation and metastasis of colorectal cancer cells by recruiting histone methylase EZH2. Mol Ther Nucleic Acids. 2021;24:1012-1023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 30. | Dasgupta N, Kumar Thakur B, Chakraborty A, Das S. Butyrate-Induced In Vitro Colonocyte Differentiation Network Model Identifies ITGB1, SYK, CDKN2A, CHAF1A, and LRP1 as the Prognostic Markers for Colorectal Cancer Recurrence. Nutr Cancer. 2019;71:257-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 31. | Sugai T, Yoshida M, Eizuka M, Uesugii N, Habano W, Otsuka K, Sasaki A, Yamamoto E, Matsumoto T, Suzuki H. Analysis of the DNA methylation level of cancer-related genes in colorectal cancer and the surrounding normal mucosa. Clin Epigenetics. 2017;9:55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 32. | Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577-590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 763] [Cited by in RCA: 858] [Article Influence: 78.0] [Reference Citation Analysis (0)] |
| 33. | Banchereau R, Leng N, Zill O, Sokol E, Liu G, Pavlick D, Maund S, Liu LF, Kadel E, Baldwin N, Jhunjhunwala S, Nickles D, Assaf ZJ, Bower D, Patil N, McCleland M, Shames D, Molinero L, Huseni M, Sanjabi S, Cummings C, Mellman I, Mariathasan S, Hegde P, Powles T. Molecular determinants of response to PD-L1 blockade across tumor types. Nat Commun. 2021;12:3969. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 106] [Article Influence: 26.5] [Reference Citation Analysis (0)] |