Published online Jul 16, 2022. doi: 10.12998/wjcc.v10.i20.6944
Peer-review started: September 20, 2021
First decision: December 27, 2021
Revised: January 7, 2022
Accepted: May 13, 2022
Article in press: May 13, 2022
Published online: July 16, 2022
Processing time: 287 Days and 3.5 Hours
This study aimed to explore clinical and molecular factors that cause discordance for clinical expression of Leber’s hereditary optic neuropathy (LHON) in a pair of identical twins with the 14484 point mutation.
Twin patients with the 14484 point mutation were studied for zygosity by using the Short Tandem Repeats Typing system. For the monozygotic twins, the radioactive restriction and densitometric analyses were used to quantitate the heteroplasmy level for the 14484 point mutation. The mitochondrial genome was analyzed to determine influential factors by mitochondrial deoxyribonucleic acid (DNA) sequencing, denaturing high-performance liquid chromatography and next generation sequencing. For the dizygotic twins, the nuclear DNA was analyzed. The twins with 14484 LHON were monozygotic with homoplasmy. No difference in the point mutation in mitochondrial DNA was found. No modifying genes that potentially influenced the disparity in phenotypic expression of LHON were detected in these twins.
This 11-year follow-up of monozygotic twins showed additional genetic modifications and epigenetic factors are possibly associated with discordance for LHON.
Core Tip: This is the first report of a pair of monozygotic twins homoplasmic for 14484T>C mutation discordant for Leber’s hereditary optic neuropathy during an 11-year follow-up. This interval between the discordant pair of twins is the longest reported to date.
- Citation: Chuenkongkaew WL, Chinkulkitnivat B, Lertrit P, Chirapapaisan N, Kaewsutthi S, Suktitipat B, Mitrpant C. Clinical expression and mitochondrial deoxyribonucleic acid study in twins with 14484 Leber’s hereditary optic neuropathy: A case report . World J Clin Cases 2022; 10(20): 6944-6953
- URL: https://www.wjgnet.com/2307-8960/full/v10/i20/6944.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i20.6944
Leber’s hereditary optic neuropathy (LHON) is the most common mitochondrial disease characterized by painless subacute asymmetrical bilateral optic neuropathy and impairment of central vision and predominantly presents in young adult males. Typical features are swelling and hyperemic optic disc, which ultimately becomes pale. The principal mechanism in LHON is mitochondrial dysfunction, which affects complex I subunit genes in the respiratory chain and causes selective degeneration of retinal ganglion cells. It eventually results in optic atrophy.
Three primary point mutations, including 11778G>A, 14484T>C and G3460G>A, contribute to 90% of clinical cases. The most prevalent 11778 mutation has the poorest visual outcome with a spontaneous partial recovery rate of 4%, while the best visual outcome is found in the 14484 point mutation, with a recovery rate of 37%-50%[1]. It has been long established that LHON causes a reduction of adenosine triphosphate synthesis at the optic nerve, leading to eventual vision loss. However, no scientific explanation has previously been provided for the variable clinical expression in certain families nor for the reason that explains why the disease is found predominantly in males as opposed to females.
However, scientists have proposed several hypotheses, such as the significance of other genetic factors (rs3749446 and rs1402000 in the PARL gene[2]) and/or epigenetic modifying factors (cyanide poisoning from cigarette smoking, alcohol, head injury, medically used drugs such as anti-retroviral drugs and anti-tuberculosis drugs, and environment toxins[3]). None of these have been determined to forecast the acute visual deterioration in affected patients, but it is possible that they can modify genes to elicit programmed cell death of retinal ganglion cells.
This study aimed to explore the molecular factors that cause the discordance for clinical expression of LHON in a pair of identical twins with the 14484 point mutation.
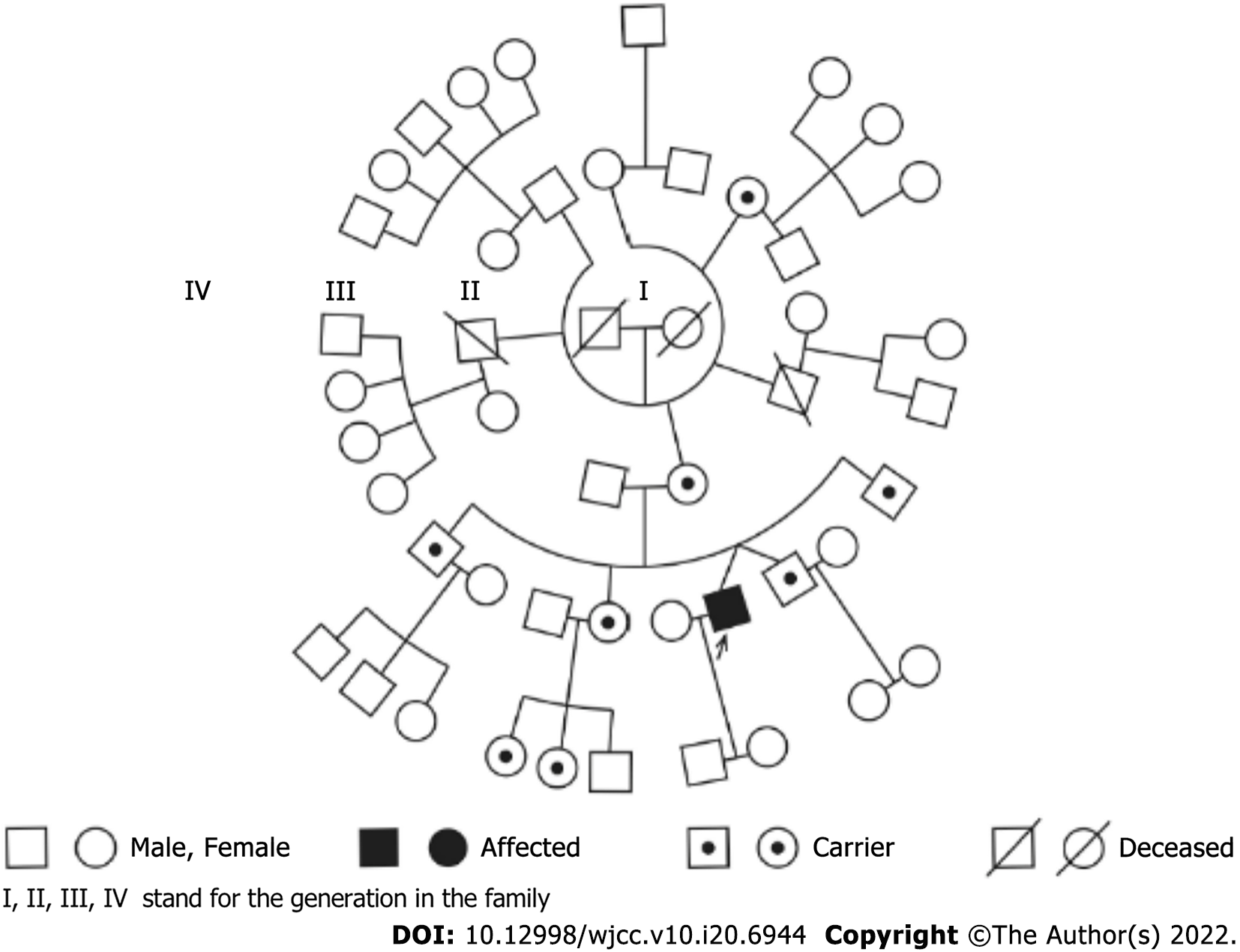
Twin A was a 33-year-old healthy man who displayed signs of bilateral acute painless visual loss for 6 wk. Twin B has remained visually asymptomatic for 11 years (Figure 1).
There was no relevant ocular or medical history.
There was no relevant ocular or medical history.
Twin A had a smoking history of one pack of cigarettes daily for 3 years and alcohol intake of one bottle daily for 5 years. Twin B had a smoking history of five cigarettes daily for 17 years and alcohol intake of half a bottle daily for 12 years; he also had a head trauma when he was 24-years-old.
Twin A’s initial visual acuity was counting fingers, and his color vision testing was abnormal in both eyes, with the absence of relative afferent pupillary defect. Both optic discs were pale with a cup-disc ratio of 0.5.
Genetic testing: Venous blood samples were obtained from the twin patients with the 14484 point mutation to identify zygosity (whether they are monozygotic or dizygotic) by using the short tandem repeats (STR) typing system.
STR typing: This study applied the AmpFLSTR® IdentifilerTM Plus polymerase chain reaction (PCR) Amplification kit, which was an STR multiplex assay that amplified 15 loci and amelogenin in a single tube. Procedures for PCR amplification, sample preparation and electrophoresis for the AmpFLSTR® IdentifilerTM Plus kit, which is composed of 15 loci (D8S1179, D21S11, D7S820, CSF1PO, D3S1358, TH01, D13S317, D16S539, D2S1338, D19S433, vWA, TPOX, D18S51, FGA and D5S818) were carried out based on the most recent instruction manual. Briefly, 10 mL of the AmpFLSTR® IdentifilerTM Master Mix [mixing AmpFLSTR® PCR Reaction Mix, AmpliTaq GoldTM deoxyribonucleic acid (DNA) polymerase] and 5 mL AmpFLSTR® IdentifilerTM Primer were distributed. The final volume of 25 mL was made by adding template DNA (1 ng). The PCR cycle time was 11 min at 95 °C followed by 28 cycles of 94 °C for 1 min, 59 °C for 1 min and 72 °C for 1 min, and a final extension at 60 °C for 60 min. The samples were held at 4 °C in the thermocycler (GeneAmp PCR System 9700; Perkin-Elmer Applied Biosystems). A 0.5 mL aliquot of PCR product was dissolved in 24.5 mL of formamide. These preparations and the fifth dye-labeled (LIZ) Size Standard (GeneScan-500; Perkin-Elmer Applied Biosystems) for sizing fragments were analyzed by using an Applied Biosystems ABI PrismTM 3130xl genetic analyzer.
The electrokinetic injection of the sample was for 3 kV/10 s. The raw data were analyzed after they were collected by the resident software (GeneMapper Analysis Software, version 2.1) using LIZ DNA size standards (range: 75-450 base pairs), and electrophoresis was conducted according to the internal size standard fragment. Genotypes were decisively made by matching the sizes[3].
Mitochondrial (mt) DNA sequencing: The mtDNA was amplified at position 14191 to 14873 using oligonucleotide primers L14191: 5’-AAACAATGGTCAACCAGGAAC-3’ and H14873: 5’-GGATCAGGCAGGGGCCAAGGAGTG-3’. The PCR reaction mixture was set up consisting of 10Xbuffer, 25 mmol/L MgCl2, 10 mmol/L deoxynucleotide triphosphates (dNTP), 20 pmol of each primer and 2.5 units of Thermus aquaticus (Taq) DNA polymerase. The cycling conditions were as follows: 94 °C for 5 min, 30 cycles of 94 °C for 1 min, 55 °C for 1 min and 72 °C for 1 min, and a final extension at 72 °C for 8 min. The PCR product was verified by agarose gel electrophoresis. The PCR product was purified by Exosap-IT® (USB Corporation).
The purification of PCR products for sequencing was obtained using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) and analyzed by 3730XL DNA Sequencer (Applied Biosystems).
Denaturing high-performance liquid chromatography (DHPLC) analysis to determine the optimum DHPLC temperature for detection of the mtDNA 14484T>C mutation: A mitochondrial PCR amp
To induce the heteroduplex formation suitable for DHPLC analysis, the putatively homoplasmic mutant (14484C) PCR products from each of the identical twins were separately mixed in equal amounts with the homoplasmic wild-type (14484T) PCR products and subjected to denaturation at 95 °C for 5 min, then slowly cooled down to 25 °C over a period of 70 min. Approximately 5 mL of both wild-type and mutant PCR products were injected into the DHPLC system followed by elution with a linear acetonitrile gradient (62.3% to 72.3%) in 0.1 M-triethyl ammonium acetate buffer at pondus hydrogenii 7.0 at the three different temperatures. The eluted DNA was inspected at 260 nm. The extraction of samples was detected as a peak of ultraviolet (UV) absorbance at 260 nm by plotting a graph of absorbance vs time.
The oven temperature at which the chromatographic peak pattern of the 50% mutant plus wild-type mixture was most prominently changed from the 100% wild-type PCR product was chosen (i.e. 55.5 °C) for further analysis.
Determination of the percentages of the putatively homoplasmic mutant (14484C) in each identical twin: The putatively homoplasmic mutant (14484C) PCR products from each of the identical twins were separately mixed with the homoplasmic wild-type (14484T) PCR products in equal amounts (1:1) to obtain a 50% mixture exactly. Subsequently, the 50% mixture of each twin was used as a precursor to prepare a serial dilution of 20%, 10%, 5%, 2.5%, 1.25%, 0.625% and 0.313%. Each dilution (eight total) from both twins was analyzed by DHPLC at 55.5 °C compared to the homoplasmic wild-type (100% of 14484T) after the induction of heteroduplex formation as mentioned above.
Next generation sequencing (NGS): An amplification of mtDNA was performed using two channels; one was the increase in mtDNA at 550-9838 using the oligonucleotide primers 5’-AACCAAACCCCAAAGACACC-3’ and 5’-GCCAATAATGACGTGAAGTCC-3’, and the other mtDNA amplification was performed at 9592-645 using the oligonucleotide primers 5’-TCCCACTCCTAAACACATCC-3’ and 5’-TTTATGGGGTGATGTGAGCC-3’, which were 9289 bp and 7626 bp in length, respectively. The PCR reaction consisted of 10Xbuffer, 25 mmol/L MgCl2, 10 mmol/L dNTP, 20 pmol of each primer and 2.5 units of Taq. The cycling conditions were as follows: 98 °C for 2 min, 30 cycles of 98 °C for 10 s, 59 °C for 5 s and 72 °C for 2.15 min, and a final extension at 72 °C for 2 min. Subsequently, an examination of the size and number of desired DNA produces was performed by separating the DNA on an agarose gel using electrophoresis and later purified by QIAquick gel extraction kit (Qiagen). After the purification, mtDNA library sequences were constructed and sequenced using GS junior: 454 Sequencing System (Roche).
A reduction of macular thickness was demonstrated on spectral domain optical coherence. Magnetic resonance imaging of the brain revealed mildly increased signal intensity on T2-weighted and fluid-attenuated inversion-recovery images at both optic nerves.
The first step was identification of twin zygosity by the STR typing system. The twin patients were monozygotic twins from the identical 16 unlinked portions on the chromosome. The study was compared with their relatives’ STR products (Tables 1 and 2, and Figure 2). Since the twins were monozygotic, we assumed that their nuclear DNA was identical.
| DNA (STRs) | Twin A | Twin B |
| D8S1179 | 10, 11 | 10, 11 |
| D21S11 | 30, 32.2 | 30, 32.2 |
| D7S820 | 11, 12 | 11, 12 |
| CSF1PO | 10, 12 | 10, 12 |
| D3S1358 | 17, 17 | 17, 17 |
| TH01 | 9, 10 | 9, 10 |
| D13S317 | 10, 11 | 10, 11 |
| D16S539 | 9, 10 | 9, 10 |
| D2S1338 | 23, 23 | 23, 23 |
| D19S433 | 13, 13.2 | 13, 13.2 |
| vWA | 14,18 | 14, 18 |
| TPOX | 8, 8 | 8, 8 |
| D18S51 | 11, 16 | 11, 16 |
| Amelogenin | X, Y | X, Y |
| D5S818 | 11, 13 | 11, 13 |
| FGA | 23, 25.2 | 23, 25.2 |
| DNA (STRs) | Twin A, B | Mother | Father | 1st Brother | 2nd Sister | 5th brother |
| D8S1179 | 10, 11 | 11, 13 | 10, 14 | 10, 11 | 10, 13 | 10, 11 |
| D21S11 | 30, 32.2 | 30, 32.2 | 30, 30 | 30, 32.2 | 30, 32.2 | 30, 30 |
| D7S820 | 11, 12 | 8, 12 | 8, 11 | 8, 8 | 8, 8 | 8, 12 |
| CSF1PO | 10, 12 | 10, 10 | 11, 12 | 10, 11 | 10, 11 | 10, 12 |
| D3S1358 | 17, 17 | 16, 17 | 16, 17 | 16, 17 | 16, 16 | 16, 17 |
| TH01 | 9, 10 | 7, 9 | 7, 10 | 7, 7 | 9, 10 | 7, 10 |
| D13S317 | 10, 11 | 10, 10 | 9, 11 | 9, 10 | 10, 11 | 10, 11 |
| D16S539 | 9, 10 | 9, 12 | 10, 12 | 12, 12 | 9, 12 | 12, 12 |
| D2S1338 | 23, 23 | 19, 23 | 21, 23 | 23, 23 | 21, 23 | 19, 23 |
| D19S433 | 13, 13.2 | 13, 14.2 | 13.2, 14.2 | 13, 13.2 | 13, 14.2 | 14.2, 14.2 |
| vWA | 14, 18 | 16, 18 | 14, 17 | 14, 18 | 14, 18 | 16, 17 |
| TPOX | 8, 8 | 8, 9 | 8, 8 | 8, 8 | 8, 9 | 8, 8 |
| D18S51 | 11, 16 | 16, 16 | 11, 13 | 13, 16 | 11, 16 | 11, 16 |
| Amelogenin | X, Y | X, X | X, Y | X, Y | X, X | X, Y |
| D5S818 | 11, 13 | 11, 11 | 12, 13 | 11, 12 | 11, 12 | 11, 13 |
| FGA | 23, 25.2 | 22, 23 | 22, 25.2 | 23, 25.2 | 22, 23 | 23, 25.2 |
The second step was aimed at the study of the differentiation of the mtDNA of the twins by mtDNA sequencing, DHPLC and NGS.
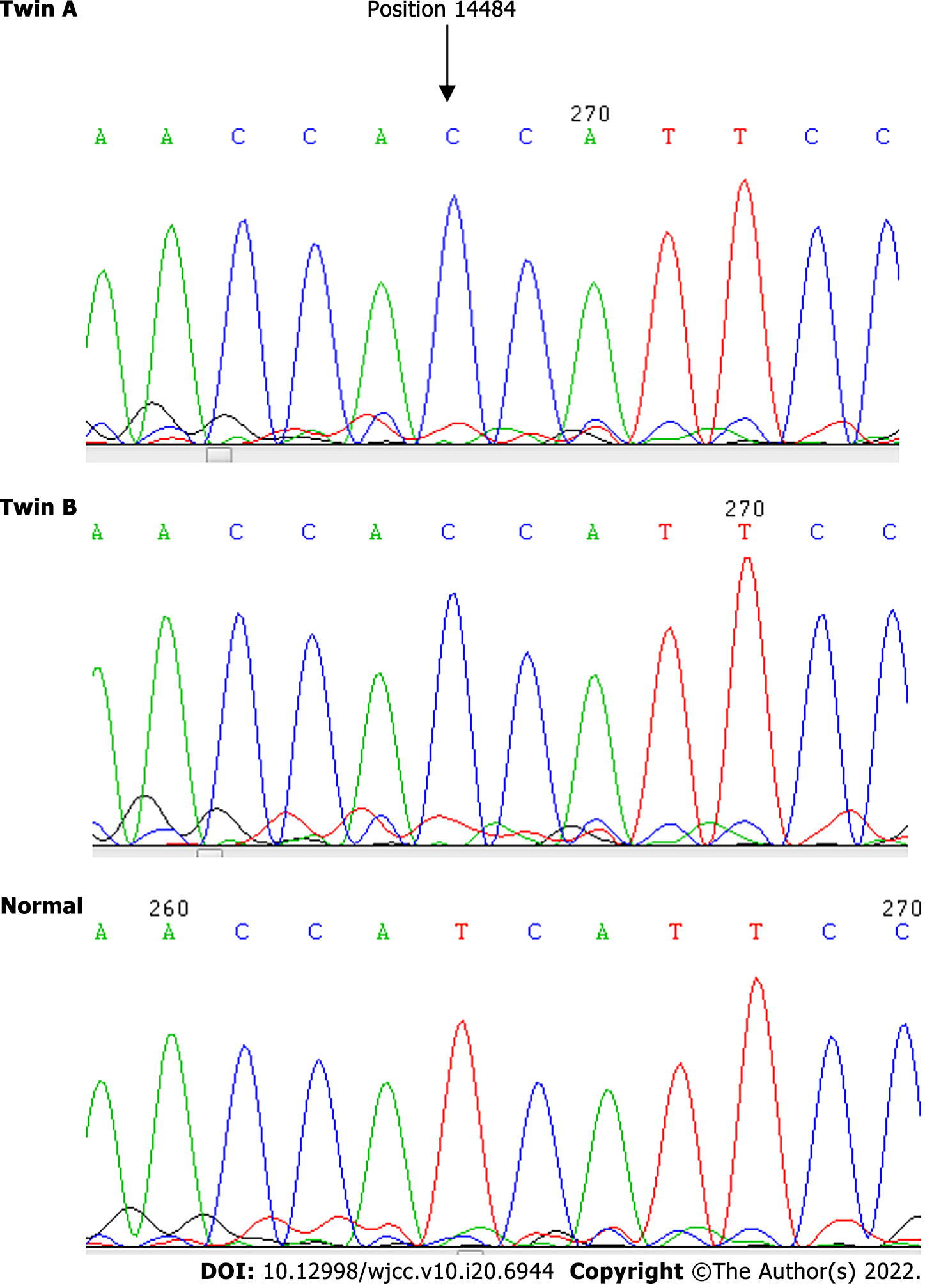
We analyzed the blood DNA of the twins by using the standard protocols to determine the degree of mutations. The result showed a point mutation at 14484T>C, but no differentiation was identified with respect to other point mutations on the mtDNA at 3115–3728, 11728–11942 and 14191–14873.
The oven temperature at 55.5 °C was found to be the optimum DHPLC temperature for detection of the mtDNA14484T>C mutation because the chromatographic peak pattern of the 50% mutant plus wild-type mixture (50% heteroplasmy) was prominently changed from the 100% wild-type (homoplasmy) PCR product. This DHPLC condition was thus used to detect the 14484T>C mutation in different heteroplasmic percentages from both twins for comparison to each other. The least heteroplasmic percentage of which the DHPLC peak pattern of the twin still showed a different profile compared to the 100% wild-type was 2.5% in both cases. It has been frequently reported that the sensitivity of DHPLC analysis for mutation detection is around 1%-5%[4-6]. Therefore, it can be inferred from the results that the starting template of the mutant (14484C) from both twins was 100% homoplasmy. This means that there is no difference between the heteroplasmic/homoplasmic status of the mtDNA mutation, 14484T>C, in these identical twin patients.
From the study, the twins had 39 portions of mutated points that were different from the wild-type, as shown in Table 3. However, the twins had similar mutations at every location in the mitochondria.
| Reference bases | Twin A | Twin B | |||
| Variation bases | Total variation percent | Variation bases | Total variation percent | ||
| 73 | A | G | 100 | G | 97.6 |
| 150 | C | T | 100 | T | 98.6 |
| 263 | A | G | 100 | G | 98 |
| 489 | T | C | 100 | C | 100 |
| 750 | A | G | 100 | G | 98.8 |
| 752 | C | T | 100 | T | 100 |
| 1107 | T | C | 100 | C | 99.5 |
| 2706 | A | G | 100 | G | 100 |
| 3590 | - | C | 34 | C | 31.2 |
| 4769 | A | G | 99.4 | G | 100 |
| 4883 | C | T | 91.1 | T | 80.8 |
| 5178 | C | A | 100 | A | 100 |
| 5301 | A | G | 100 | G | 100 |
| 6779 | A | G | 99.7 | G | 100 |
| 7028 | C | T | 100 | T | 99.6 |
| 8701 | A | G | 100 | G | 100 |
| 8860 | A | G | 100 | G | 100 |
| 9033 | A | G | 100 | G | 100 |
| 9180 | A | G | 99.7 | G | 100 |
| 9540 | T | C | 100 | C | 100 |
| 9554 | G | A | 100 | A | 100 |
| 10397 | AA | GG | 96.3 | GG | 98.8 |
| 10400 | C | T | 100 | T | 100 |
| 10873 | T | C | 100 | C | 100 |
| 11719 | G | A | 98.4 | A | 98 |
| 11944 | T | C | 100 | C | 98.7 |
| 12026 | A | G | 100 | G | 99.4 |
| 12705 | C | T | 100 | T | 100 |
| 14484 | T | C | 100 | C | 100 |
| 14766 | C | T | 97.8 | T | 98.4 |
| 14783 | T | C | 100 | C | 68.3 |
| 15043 | G | A | 100 | A | 98.5 |
| 15301 | G | A | 100 | A | 100 |
| 15326 | A | G | 100 | G | 100 |
| 15604 | C | T | 38.6 | T | 32 |
| 16092 | T | C | 99 | C | 99.4 |
| 16164 | A | G | 100 | G | 100 |
| 16223 | C | T | 100 | T | 99.3 |
| 16362 | T | C | 98.9 | C | 100 |
The monozygotic twins were homoplasmic for 14484T>C mutation discordant for LHON and were followed up over an 11-year period.
This study closed with the 11-year follow-up reported here.
This is the first report of a pair of monozygotic twins homoplasmic for 14484T>C mutation discordant for LHON followed up over an 11-year period. This interval between the discordant pair of twins is the longest reported to date. The Combined DNA Index System 16 STR profile confirmed the monozygosity of the twins. By using mtDNA sequencing, DHPLC and NGS, the study found that the twins have no differences in mutations and are homoplasmic at 14484T>C.
Previously, there were reports of eight sets of monozygotic and dizygotic twins having 11778 or 14484 mutations, which showed a highly variable clinical expression (Table 4). In 1987, Nikoskelainen et al[7,8] described a pair of nondiscordant 11778 LHON twins with different disease onset. In 1993, Johns et al[2] reported monozygotic twins harboring homoplasmic 11778 LHON with an onset that was 9 years apart. In 1991, Newman et al[9] disclosed two pairs of discordant monozygotic brothers and a dizygotic brother and sister for 11778 LHON, with one affected male from each pair. Harding et al[10] described three concordant 11778 and 14484 pairs of dizygous male twins. In 1997, Biousse et al[11] reported discordant monozygotic twins with heteroplasmic de novo 14484 mutation, indicating that only one twin had visual loss at the age of 17.
| Ref. | Mono/dizygotic twin (Sex) | Homo/heteroplsmy | Dis-cordant | Age at onset/onset time difference (year) | Epigenetic factors |
| 11778 | |||||
| Nikoskelainen et al[7,8], 1987 | NA (m) | Homoplasmy | No | 20/NA | NA |
| Newman et al[9], 1991 | M (m) | NA | Yes | NA | NA |
| D (m, f) | NA | Yes | NA | ||
| Johns et al[2], 1993 | M (m) | Homoplasmy | No | 34/11 | Alcohol/tobacco use |
| 14484 | |||||
| Biousse et al[11], 1997 | M (m) | Heteroplasmy | Yes | 17/ NA | None |
| Current study 2021 | M (m) | Homoplasmy | Yes | 34/ NA | Alcohol/tobacco use |
Incomplete penetrance is observed in approximately 50% of male individuals and 10% of female individuals harboring the pathogenic mutations and phenotypic expression[12]. The presence of the primary mutation is a necessary condition but not sufficient for LHON manifestation. Anatomical, hormonal or physiological factors may be contributing factors for male preponderance.
Mitochondrial and nuclear DNA factors have not yet been demonstrated to have an influence on the phenotypic discordance of monozygotic twins. Moreover, an identical genome would be expected for monozygotic twins who are products of a single fertilized egg that subsequently splits into two zygotes during the zygote stage. The process to separate two cells in the zygote stage is unclear and needs further explanation[13]. Nevertheless, evidence has been found that indicates it is likely an epigenetic mechanism that affects the process of separation. External environmental factors may influence the phenotype variability.
Kirkman et al[14] reported no link between cigarette smoking and LHON. In contrast, asymptomatic relatives with heavy smoking were prone to clinical expression. The twin brothers had a similar history of cigarette smoking and alcohol consumption, but only the unaffected twin had head injury. We could not identify a strong association between smoking and the disparity in phenotypic expression of LHON in the twins in this study.
The X-linked gene and mtDNA haplogroup presumably contributes to genetically determining the expression for LHON. It is possible that the mtDNA haplogroup background drove the variability of LHON penetrance among individuals[10,15]. Hudson et al[16] described that haplogroup J2 and J1 had influences on phenotypes in Europeans with 11778G>A and 14484T>C LHON, respectively. Haplogroup B, especially sub-haplogroup B5a1, and haplogroup M likely have a larger influence in Southeast Asians with 11778G>A and 14484T>C LHON, respectively[17]. This pair of 14484T>C twins had the same haplogroup of D5a2a1, despite the occurrence of discordance.
Giordano et al[18] inferred that mitochondrial biogenesis might influence phenotypic differences of LHON expression. Although the measurement of blood mitochondrial genotypes in these discordant identical twins seem to be similar, the quantitative difference of mutant mtDNA found in the optic nerve of LHON individuals and the amount of tissue heteroplasmy may be potential factors that influence their disparity in phenotypic expression.
Additional genetic modifications and epigenetic factors are possibly associated with the occurrence of discordance for LHON during this 11-year follow-up of monozygotic twins. Future study on the effects of other DNA and mtDNA is required. For example, whole transcriptome sequencing to investigate the effects of differentially expressed genes that might modulate and play a vital role for LHON expression in identical twins should be accomplished.
The authors would like to thank the Director of the Sub-Division of Forensic Biochemistry, Institute of Forensic Medicine, Police General Hospital, Royal Thai Police for STR typing and Thai Red Cross Emerging Infectious Diseases-Health Science Centre, WHO Collaborating Centre for Research and Training on Viral Zoonoses, King Chulalongkorn Memorial Hospital for the next generation sequencing.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Genetics and heredity
Country/Territory of origin: Thailand
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Arslan N, Turkey; Jin X, China; Musoni L, Morocco; Papazafiropoulou A, Greece S-Editor: Guo XR L-Editor: A P-Editor: Chen YX
| 1. | Mackey D, Howell N. A variant of Leber hereditary optic neuropathy characterized by recovery of vision and by an unusual mitochondrial genetic etiology. Am J Hum Genet. 1992;51:1218-1228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 67] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 2. | Johns DR, Smith KH, Miller NR, Sulewski ME, Bias WB. Identical twins who are discordant for Leber's hereditary optic neuropathy. Arch Ophthalmol. 1993;111:1491-1494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 3. | Yang MJ, Tzeng CH, Tseng JY, Huang CY. Determination of twin zygosity using a commercially available STR analysis of 15 unlinked loci and the gender-determining marker amelogenin--a preliminary report. Hum Reprod. 2006;21:2175-2179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 42] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 4. | Lim KS, Naviaux RK, Wong S, Haas RH. Pitfalls in the denaturing high-performance liquid chromatography analysis of mitochondrial DNA mutation. J Mol Diagn. 2008;10:102-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 5. | Wongboonma W, Thongnoppakhun W, Auewarakul CU. A single-tube allele specific-polymerase chain reaction to detect T315I resistant mutation in chronic myeloid leukemia patients. J Hematol Oncol. 2011;4:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 6. | Liu MR, Pan KF, Li ZF, Wang Y, Deng DJ, Zhang L, Lu YY. Rapid screening mitochondrial DNA mutation by using denaturing high-performance liquid chromatography. World J Gastroenterol. 2002;8:426-430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 20] [Cited by in RCA: 20] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 7. | Nikoskelainen EK, Savontaus ML, Wanne OP, Katila MJ, Nummelin KU. Leber's hereditary optic neuroretinopathy, a maternally inherited disease. A genealogic study in four pedigrees. Arch Ophthalmol. 1987;105:665-671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 104] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 8. | Nikoskelainen E, Hoyt WF, Nummelin K. Ophthalmoscopic findings in Leber's hereditary optic neuropathy. II. The fundus findings in the affected family members. Arch Ophthalmol. 1983;101:1059-1068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 91] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 9. | Newman NJ, Lott MT, Wallace DC. The clinical characteristics of pedigrees of LHON with the 11778 mutation. Am J Ophthalmol 1991; 111: 750-762. [RCA] [DOI] [Full Text] [Cited by in Crossref: 258] [Cited by in RCA: 266] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 10. | Harding AE, Sweeney MG, Govan GG, Riordan-Eva P. Pedigree analysis in Leber hereditary optic neuropathy families with a pathogenic mtDNA mutation. Am J Hum Genet. 1995;57:77-86. [PubMed] |
| 11. | Biousse V, Brown MD, Newman NJ, Allen JC, Rosenfeld J, Meola G, Wallace DC. De novo 14484 mitochondrial DNA mutation in monozygotic twins discordant for Leber's hereditary optic neuropathy. Neurology. 1997;49:1136-1138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 39] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 12. | Seedorff T. The inheritance of Leber's disease. A genealogical follow-up study. Acta Ophthalmol (Copenh). 1985;63:135-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 42] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 13. | Hall JG. Twinning. Lancet. 2003;362:735-743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 449] [Cited by in RCA: 399] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 14. | Kirkman MA, Yu-Wai-Man P, Korsten A, Leonhardt M, Dimitriadis K, De Coo IF, Klopstock T, Chinnery PF. Gene-environment interactions in Leber hereditary optic neuropathy. Brain. 2009;132:2317-2326. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 276] [Cited by in RCA: 255] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 15. | Bu XD, Rotter JI. X chromosome-linked and mitochondrial gene control of Leber hereditary optic neuropathy: evidence from segregation analysis for dependence on X chromosome inactivation. Proc Natl Acad Sci U S A. 1991;88:8198-8202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 123] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 16. | Hudson G, Carelli V, Spruijt L, Gerards M, Mowbray C, Achilli A, Pyle A, Elson J, Howell N, La Morgia C, Valentino ML, Huoponen K, Savontaus ML, Nikoskelainen E, Sadun AA, Salomao SR, Belfort R Jr, Griffiths P, Yu-Wai-Man P, de Coo RF, Horvath R, Zeviani M, Smeets HJ, Torroni A, Chinnery PF. Clinical expression of Leber hereditary optic neuropathy is affected by the mitochondrial DNA-haplogroup background. Am J Hum Genet. 2007;81:228-233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 278] [Cited by in RCA: 284] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 17. | Kaewsutthi S, Phasukkijwatana N, Joyjinda Y, Chuenkongkaew W, Kunhapan B, Tun AW, Suktitipat B, Lertrit P. Mitochondrial haplogroup background may influence Southeast Asian G11778A Leber hereditary optic neuropathy. Invest Ophthalmol Vis Sci. 2011;52:4742-4748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 18. | Giordano C, Iommarini L, Giordano L, Maresca A, Pisano A, Valentino ML, Caporali L, Liguori R, Deceglie S, Roberti M, Fanelli F, Fracasso F, Ross-Cisneros FN, D'Adamo P, Hudson G, Pyle A, Yu-Wai-Man P, Chinnery PF, Zeviani M, Salomao SR, Berezovsky A, Belfort R Jr, Ventura DF, Moraes M, Moraes Filho M, Barboni P, Sadun F, De Negri A, Sadun AA, Tancredi A, Mancini M, d'Amati G, Loguercio Polosa P, Cantatore P, Carelli V. Efficient mitochondrial biogenesis drives incomplete penetrance in Leber's hereditary optic neuropathy. Brain. 2014;137:335-353. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 215] [Cited by in RCA: 224] [Article Influence: 20.4] [Reference Citation Analysis (0)] |