Published online May 26, 2022. doi: 10.12998/wjcc.v10.i15.4761
Peer-review started: December 16, 2021
First decision: January 27, 2022
Revised: February 6, 2022
Accepted: March 26, 2022
Article in press: March 26, 2022
Published online: May 26, 2022
Processing time: 158 Days and 23.3 Hours
Gastric cancer is a leading cause of cancer-related mortality worldwide. Many somatic mutations have been identified based on next-generation sequencing; they likely play a vital role in cancer treatment selection. However, next-generation sequencing has not been widely used to diagnose and treat gastric cancer in the clinic.
To test the mutant gene frequency as a guide for molecular diagnosis and personalized therapy in gastric cancer by use of next-generation sequencing.
We constructed a panel of 24 mutant genes to detect somatic nucleotide variations and copy number variations based on a next-generation sequencing technique. Our custom panel included high-mutation frequency cancer driver and tumour suppressor genes. Mutated genes were also analyzed using the cBioPortal database. The clinical annotation of important variant mutation sites was evaluated in the ClinVar database. We searched for candidate drugs for targeted therapy and immunotherapy from the OncoKB database.
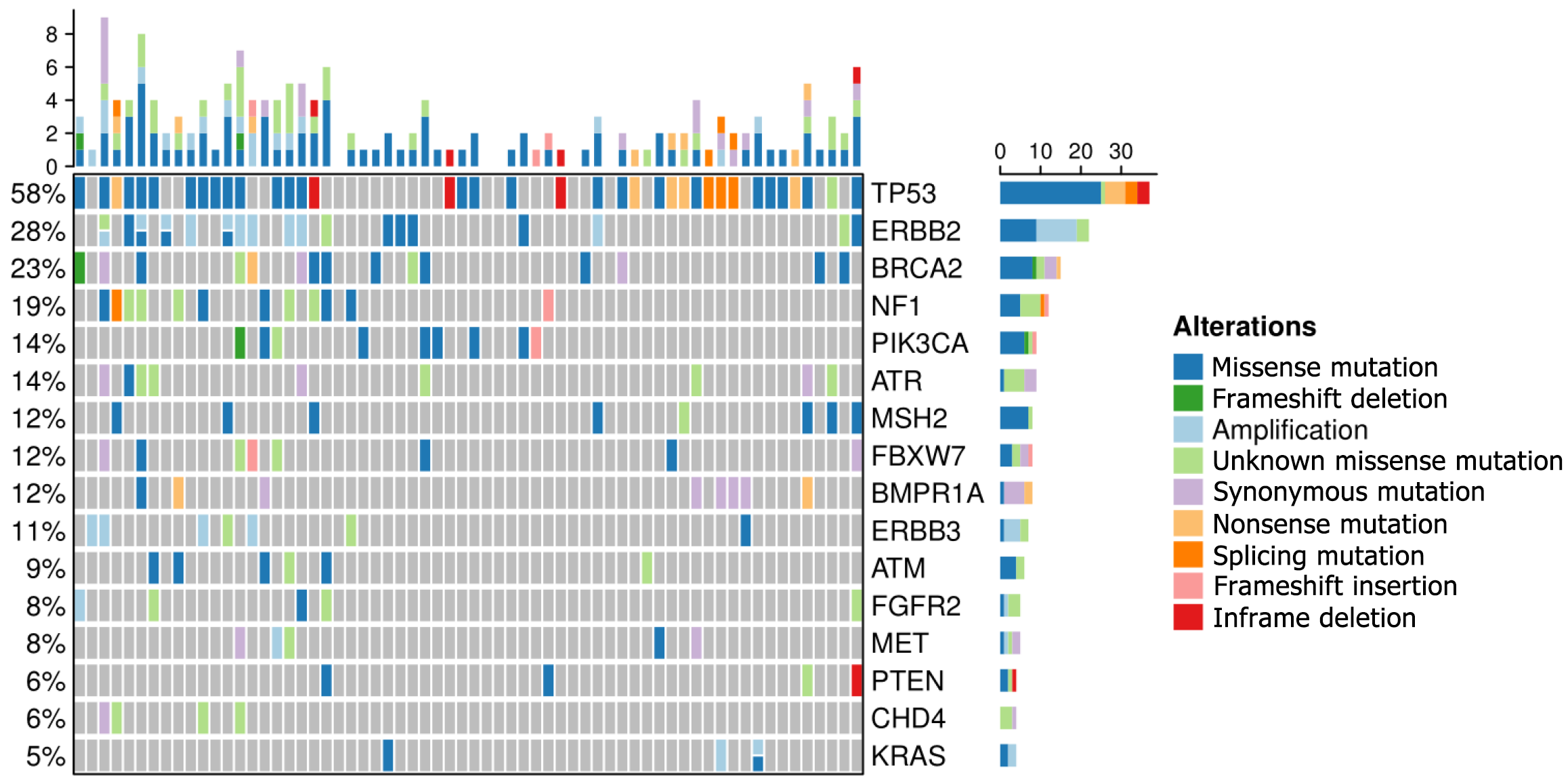
In our study, the top 16 frequently mutated genes were TP53(58%), ERBB2(28%), BRCA2 (23%), NF1 (19%), PIK3CA (14%), ATR (14%), MSH2 (12%), FBXW7 (12%), BMPR1A (12%), ERBB3 (11%), ATM (9%), FGFR2 (8%), MET (8%), PTEN (6%), CHD4 (6%), and KRAS (5%). TP53 is a commonly mutated gene in gastric cancer and has a similar frequency to that in the cBioPortal database. 33 gastric cancer patients (51.6%) with microsatellite stability and eight patients (12.5%) with microsatellite instability-high were investigated. Enrichment analyses demonstrated that high-frequency mutated genes had transmembrane receptor protein kinase activity. We discovered that BRCA2, PIK3CA, and FGFR2 gene mutations represent promising biomarkers in gastric cancer.
We developed a powerful panel of 24 genes with high frequencies of mutation that could detect common somatic mutations. The observed mutations provide potential targets for the clinical treatment of gastric cancer.
Core Tip: High frequencies of mutation might provide new insights for individualized and precise treatment by use of next-generation sequencing in gastric cancer patients. However, next-generation sequencing has not been widely used to diagnose and treat gastric cancer in clinical practice. Thus, this study analysed 24 powerful genes with high frequencies of mutation based on a next-generation sequencing technique.
- Citation: Zeng HH, Yang Z, Qiu YB, Bashir S, Li Y, Xu M. Detection of a novel panel of 24 genes with high frequencies of mutation in gastric cancer based on next-generation sequencing. World J Clin Cases 2022; 10(15): 4761-4775
- URL: https://www.wjgnet.com/2307-8960/full/v10/i15/4761.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i15.4761
Gastric cancer (GC) is a common malignancy worldwide; although the incidence and mortality of GC have declined globally over the past 50 years, it remains the fifth most frequently diagnosed cancer and the third leading cause of cancer-related death[1]. It is estimated that GC is the second most common cancer in China and ranks third in cancer-related deaths[2]. The mechanisms underlying GC metastasis and adverse prognostic factors are still unclear, mainly due to the high heterogeneity of tumours[3]. Moreover, the efficacy of traditional methods, such as surgery, radiotherapy, and chemotherapy, is limited and the majority of GC patients are already at an advanced stage when diagnosed[4]. The molecular characteristics of mutated genes can provide guidance for potential therapeutic biomarkers and have significant clinical implications for GC diagnosis and treatment[5].
The classic pathological classification of GC is mainly based on gross and histological morphology as well as cell biological characteristics, including the Borrmann, Lauren, and WHO classifications. With the development of molecular diagnostic techniques, especially next-generation sequencing (NGS), The Cancer Genome Atlas (TCGA) and the Asian Cancer Research Group (ACRG) have unprecedented insight into the molecular characteristics of GC[6,7]. Targeted therapy or immunotherapy needs to be combined with the molecular characteristics of GC. In clinical practice, both the National Comprehensive Cancer Network (NCCN) guidelines and Chinese Society of Clinical Oncology (CSCO) guidelines have affirmed the value and significance of NGS in the treatment of GC, and it has been increasingly used in clinical practice for the diagnosis of cancers, such as breast cancer and non-small-cell lung cancer[8,9]. As an essential complement to pathological diagnoses, NGS is an increasingly accessible choice for clinical doctors and researchers[10]. NGS requires only small amounts of tissue, such as fresh tissue or formalin-fixed paraffin-embedded (FFPE) tumour tissue specimens. This method can rapidly identify specific changes in DNA sequences and detect multiple targets simultaneously. NGS technology can facilitate more accurate targeted therapy and immunotherapy and improve its therapeutic effect in GC[11-13]. However, the clinical applications of molecular classification are limited because it is difficult to significantly improve therapeutic effects and select individualized treatments in GC.
We explored a panel with 24 genes with a high frequency of mutation related to GC in the present study. We used NGS technology combined with bioinformatics methods to quickly identify differences in mutated genes and obtained valuable gene expression information. FFPE tumour tissue specimens were obtained from 64 GC patients. We analysed the correlation between TP53 gene mutations and their clinicopathological features to develop a detailed solution for important mutation sites and recommend mutation-based treatment options. In addition, we discovered promising target sites, which can provide insights into drug development in follow-up clinical research.
We recruited 64 patients with GC from the First Affiliated Hospital of Bengbu Medical College. This research was approved by the First Affiliated Hospital of Bengbu Medical College and performed in accordance with the Declaration of Helsinki. All patients provided signed informed consent for voluntary participation in the study. 64 patients had not received chemotherapy or radiotherapy before sample collection. FFPE tumour tissue specimens from GC patients who received gastrectomy or gastroendoscopic surgery were obtained from the pathology department. We used germline DNA from FFPE paracancerous tissues as a reference to detect somatic variation. We utilized a high-throughput sequencing platform and domain capture technology of the target area to test a novel 24-gene panel (Table 1).
| 24-gene panel | |||||
| ATR | ATM | AR | BRCA2 | BMPR1A | CHD4 |
| CDKN2A | ERBB2 | ERBB3 | FBXW7 | FGFR2 | KRAS |
| KDR | KIT | MET | MSH2 | MTOR | NF1 |
| PTEN | PDGFRA | PIK3CA | PTPN11 | STK11 | TP53 |
Genomic DNA (gDNA) from FFPE cancer and paracancerous tissues was extracted using a QIAGEN tissue kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer's specifications. The DNA concentration was analysed using a Qubit® 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, United States), and DNA integration was evaluated using an Agilent Bioanalyzer 2100 instrument (Agilent Technologies, Santa Clara, CA, United States). We used multimembrane polymerase chain reaction (PCR) to assess FFPE DNA integrity.
The novel 24-gene panel consisted of introns, promoters, and fusion regions. 24 mutated genes were recorded in the Catalogue of Somatic Mutations in Cancer (COSMIC) database (http://cancer.sanger.ac.uk/cosmic). The library was prepared using an Ion Ampliseq Library kit 2.0 (Life Technologies) according to the manufacturer’s specifications. We submitted 300 ng of gDNA for each tissue from each sample using an ultrasonicator (Covaris, Woburn, MA, United States). Next, small fragments of the 200 bp main band were used to perform end repair, a-tailing, and adapter ligation with the KAPA LTP Library Preparation kit according to the manufacturer’s instructions. Fragmented DNA was converted to an NGS library. After subsequent PCR amplification, the cDNA was quality-controlled and purified. The pre-library was mixed according to the proportion of the data volume. A hybridization system was configured, and magnetic beads were captured.
The library was diluted to a concentration of 100 pM using the Ion Library Equalizer kit (Life Technologies, United States) before template preparation. Enriched, templated ion sphere particles were obtained using a DA8600 Ion Proton sequencer (DAANGENE, China) equipped with an Ion P1 chip (Life Technologies, United States).
Raw NGS data analysis, including base calling, quality control, and sequence alignment to the hg19 human reference genome, was performed with Ion Reporter software (Life Technologies, Carlsbad, CA, United States). All variants were visually confirmed using the Integrative Genomics viewer (IGV). Mutations were named according to the Single Nucleotide Polymorphism database (dbSNP)(http://www.ncbi.nlm.nih.gov/snp/) and COSMIC database. The mutation population frequencies were annotated by the NHLBI Exome Sequencing Project (http://evs.gs.washington.edu/EVS/).
A “stomach adenocarcinoma” study (TCGA, Nature 2014) containing 295 samples is available in the cBioPortal database. By choosing mRNA expression Z scores relative to diploid samples (RNA Seq V2 RSEM) within two SDs from the mean (a threshold of ± 2.0), the complete data of 258 patients were downloaded from the cBioPortal database (http://www.cbioportal.org). The variant sites of mutated genes were indicated by reference to the ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar). Mutation-based targeted drug recommendation information was identified using the OncoKB database (https://github.com/oncokb). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed to explore the related enrichments and pathways using Cluster Profiler in R software.
We used R software (version 4.0.5), SPSS 22.0 (SPSS Inc., Chicago, IL, United States) and GraphPad Prism 9 (GraphPad Software Inc., La Jolla, CA, United States) for statistical analyses. Chi-square tests were employed to analyse the relationship between TP53 mutations and clinicopathological characteristics. FDR < 0.05 was used to evaluate the significance of GO terms and KEGG pathways. Differences at P < 0.05 were considered statistically significant.
First, we analysed 64 Chinese GC patients. The median age was 64 years. Of the patients, 78.1% were male and the primary clinical stage was II/III (79.7%). Histologically, intestinal GC accounted for 62.5%, and diffuse GC accounted for 34.4%. Subsequently, 295 patients were selected from the cBioPortal database of varying races, including Caucasian (58.3%), Asian (26.1%), and other races; 258 patients were analyzed as a control group. The majority of patients in this group were also male (60.5%). The primary clinical stage was II/III (75.2%), and the major histology was intestinal GC (65.5%). All patients’ clinical data are shown in Table 2.
| Clinical information | Custom panel, n (%) | cBioPortal database, n (%) |
| Total | 64 | 258 |
| Age median (range) | 64 (25-83) | 68 (34-90) |
| Sex | ||
| Male | 50 (78.1) | 156 (60.5) |
| Famale | 14 (21.9) | 102 (39.5) |
| T classification | ||
| T1 | 20 (31.2) | 11 (4.3) |
| T2 | 33 (51.6) | 43 (16.7) |
| T3 | 10 (15.6) | 133 (51.5) |
| T4 | 1 (1.6) | 63 (24.4) |
| Unknown | 0 (0) | 8 (3.1) |
| N classification | ||
| N0 | 24 (37.5) | 87 (33.7) |
| N1 | 18 (28.1) | 54 (20.9) |
| N2 | 6 (9.4) | 52 (20.2) |
| N3 | 16 (25.0) | 54 (20.9) |
| Unknown | 0 (0) | 11 (4.3) |
| M classification | ||
| M0 | 58 (90.6) | 238 (92.2) |
| M1 | 6 (9.4) | 18 (7) |
| Unknown | 0 (0) | 2 (0.8) |
| Clinical stage | ||
| Stage I | 10 (15.6) | 32 (12.4) |
| Stage II | 28 (43.8) | 102 (39.5) |
| Stage III | 23 (35.9) | 92 (35.7) |
| Stage IV | 3 (4.7) | 18 (7) |
| Unknown | 0 (0) | 14 (5.4) |
| Lauren class | ||
| Diffuse | 22 (34.4) | 62 (24.0) |
| Intestinal | 40 (62.5) | 169 (65.5) |
| Mixed | 2 (3.1) | 16 (6.2) |
| Unknown | 0 (0) | 11 (4.3) |
24 genes with high frequencies of mutation were detected by NGS. Our study found that the top 16 frequently mutated genes were TP53 (58%), ERBB2 (28%), BRCA2 (23%), NF1 (19%), PIK3CA (14%), ATR (14%), MSH2 (12%), FBXW7 (12%), BMPR 1A (12%), ERBB3 (11%), ATM (9%), FGFR2 (8%), MET (8%), PTEN (6%), CHD4 (6%), and KRAS (5%) (Figure 1). Most gene mutations were missense muta
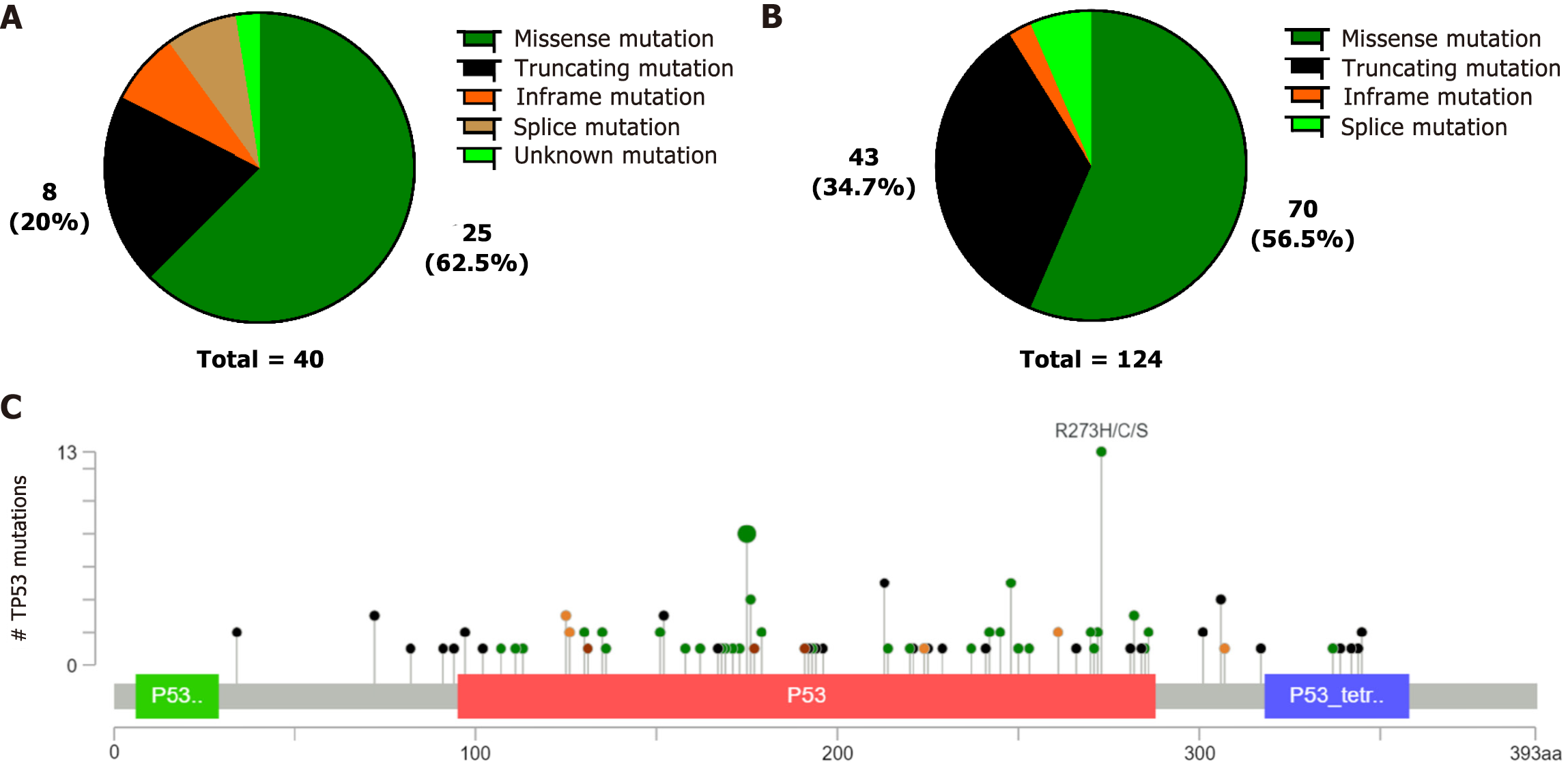
We discovered that TP53 mutations were the highest frequency in both our data and the cBioPortal database. We then investigated the relationship between TP53 mutations and clinicopathological features. Although an association was not observed between clinicopathological features and TP53 mutations (P > 0.05) in our custom panel, somatic TP53 mutations were negatively correlated with distant metastases(P = 0.040), and TP53 mutations were also observed less frequently in diffuse GC(P = 0.017) in the cBioPortal database (Table 3). The TP53 mutations were mainly missense mutations, fragment deletions, and insertions, in both our data and the cBioPortal database (Figure 3A-B). We discovered that the most common TP53 mutations were R273H/C/S, R175H/C/G, and R248Q/H in the cBioPortal database (Figure 3C). These mutation hotspots were also detected in our cohort. TP53 mutation sites are recorded as pathogenic mutations in Li-Fraumeni syndrome (LFS) and hereditary cancer predisposition in the ClinVar database.
| Clinicalinformation | Custom panel | P value | cBioPortal database | P value | ||
| Wild | Mutated | Wild | Mutated | |||
| Age | ||||||
| ≤ 65 | 16 | 19 | 0.530 | 60 | 48 | 0.878 |
| > 65 | 11 | 18 | 79 | 68 | ||
| Unknown | 0 | 0 | 2 | 1 | ||
| Sex | ||||||
| Male | 19 | 31 | 0.200 | 86 | 70 | 0.849 |
| Female | 8 | 6 | 55 | 47 | ||
| T classification | ||||||
| T1 | 10 | 10 | 0.716 | 6 | 5 | 0.451 |
| T2 | 13 | 20 | 23 | 20 | ||
| T3 | 4 | 6 | 67 | 66 | ||
| T4 | 0 | 1 | 39 | 24 | ||
| Unknown | 0 | 0 | 6 | 2 | ||
| N classification | ||||||
| N0 | 7 | 17 | 0.310 | 45 | 42 | 0.929 |
| N1 | 10 | 8 | 29 | 25 | ||
| N2 | 2 | 4 | 29 | 23 | ||
| N3 | 8 | 8 | 31 | 23 | ||
| Unknown | 0 | 0 | 7 | 4 | ||
| M classification | ||||||
| M0 | 25 | 33 | 0.645 | 127 | 111 | 0.040 |
| M1 | 2 | 4 | 14 | 4 | ||
| Unknown | 0 | 0 | 0 | 2 | ||
| clinical stage | ||||||
| Stage I | 6 | 4 | 0.414 | 17 | 15 | 0.255 |
| Stage II | 9 | 19 | 51 | 51 | ||
| Stage III | 11 | 12 | 50 | 42 | ||
| Stage IV | 1 | 2 | 14 | 4 | ||
| Unknown | 0 | 0 | 9 | 5 | ||
| Lauren class | ||||||
| Diffuse | 12 | 10 | 0.059 | 44 | 18 | 0.017 |
| Intestinal | 13 | 27 | 81 | 88 | ||
| Mixed | 2 | 0 | 10 | 6 | ||
| Unknown | 0 | 0 | 6 | 5 | ||
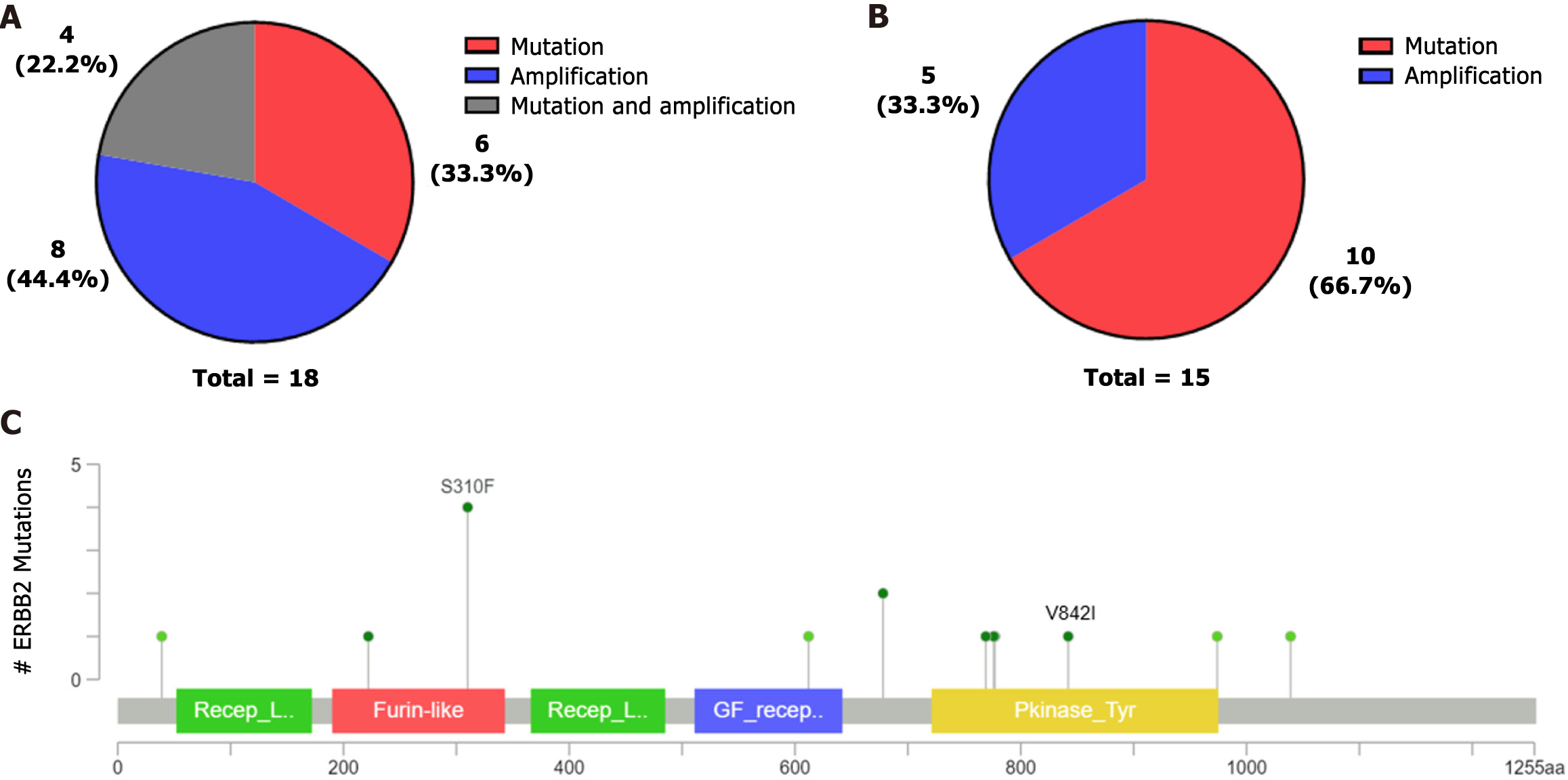
This study detected ERBB2 mutations in 18 samples. Four samples with ERBB2 point mutations also had concurrent ERBB2 amplification, and two patients had concurrent amplification with ERBB3. ERBB2 was mainly mutated in this study but mainly amplified in the cBioPortal database (Figure 4A-B). The most common ERBB2 mutation occurred at S310F in the cBioPortal database (Figure 4C), although a mutation at S310F was not detected in our study. However, four patients with the p.V842I mutation were found in our cohort. The V842I mutation site is a potential pathogenic mutation in stomach adenocarcinoma according to the ClinVar database.
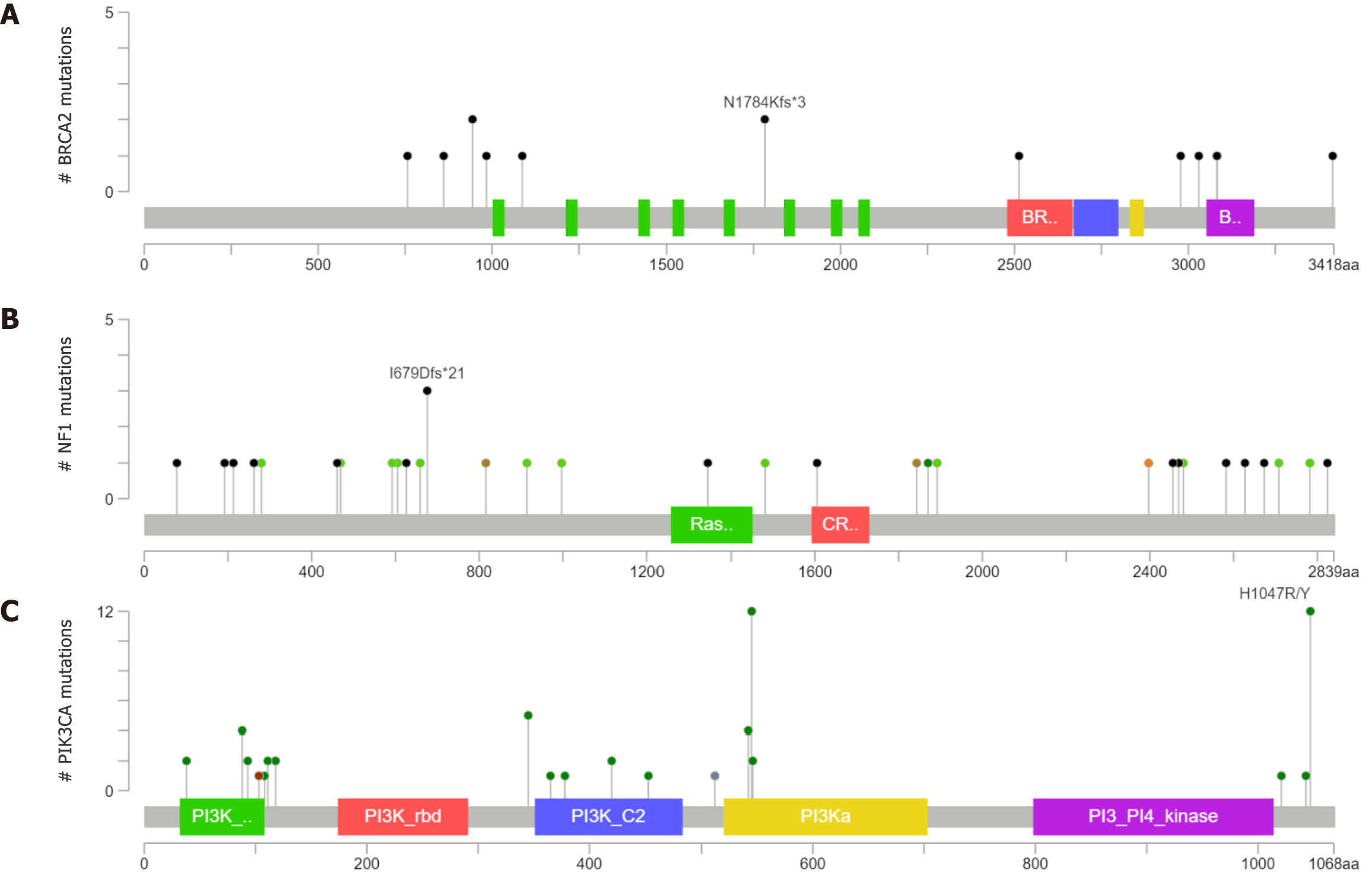
BRCA2 mutations were found in 15 patients in our cohort; 3 mutations were synonymous and 12 were nonsynonymous. The nonsynonymous mutations included missense mutations and frameshift mutations. Two samples had BRCA2 at N1784Kfs*3 in the cBioPortal database (Figure 5A), but we did not find any BRCA2 mutations at N1784Kfs*3 in our panel. NF1 mutations were found in 13 patients in our cohort. NF1 was detected at the I679Dfs*21 location in three samples in the cBioPortal database (Figure 5B), but none of the patients in our study had a mutation at this particular location. The PIK3CA mutation was found in nine patients in our cohort. The mutation hotspots of PIK3CA were E542K, E545K, and H1047R, which was consistent with the cBioPortal database (Figure 5C). Furthermore, we found the E545K mutation in four patients and H1047R in one patient. E545K and H1047R are recorded as likely pathogenic mutations in stomach adenocarcinoma according to the ClinVar database.
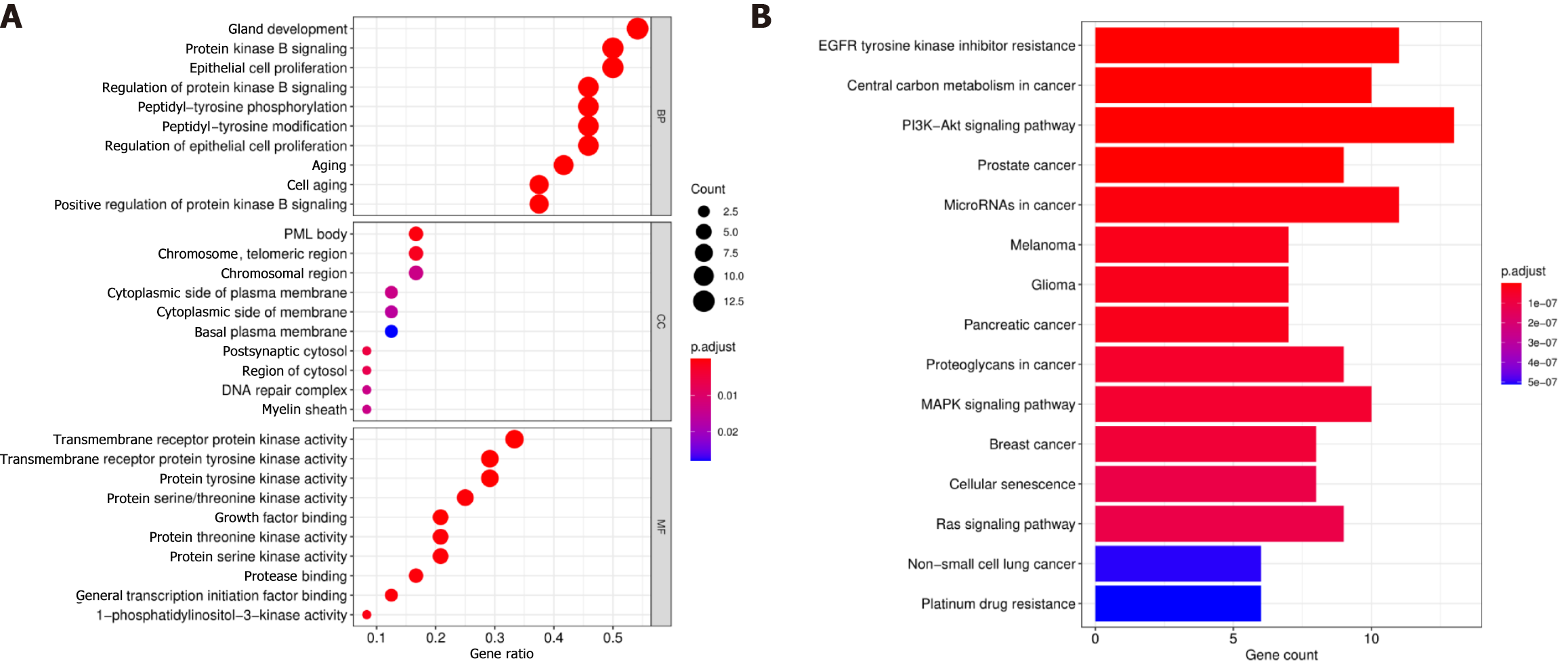
We performed an enrichment analysis to identify the biological function of the 24 differentially mutated genes associated with GC. The GO enrichment analysis included biological processes, cellular components, and molecular functions. The related biological processes primarily consisted of protein kinase B signalling, epithelial cell proliferation, and protein kinase B signalling regulation; the cellular components included the chromosome region and cytoplasmic side of the plasma membrane; and the enriched molecular functions consisted of transmembrane receptor protein kinase activity. Thus, the GO enrichment analysis demonstrated that the biological function of these mutations might be related to the tumour interaction with transmembrane receptor protein kinase activity (Figure 6A). The KEGG pathway analysis indicated that these genes were enriched in tumour-related signalling pathways, including the PI3K−Akt, MAPK, and Ras signalling pathways and platinum drug resistance (Figure 6B). In summary, these results indicated that the function of these mutated genes was highly associated with the occurrence of GC.
To evaluate the clinical value and predict the response to targeted therapy in these gene mutations, we searched for targeted drugs using the OncoKB database (https://github.com/oncokb). The OncoKB database, a comprehensive and precise oncology database, can classify mutated genes and provide clinical evidence levels. There are four OncoKB therapeutic levels of evidence: level 1, FDA-approved drugs; level 2, standard care; level 3, clinical evidence; and level 4, biological evidence. We discovered that many gene mutations in our data were recognized as standard care biomarkers corresponding to FDA-approved drugs (Table 4). Ten patients showed ERBB2 amplification in our data; these patients would apparently benefit from FDA-approved ERBB2 inhibitors, including trastuzumab and pertuzumab. For patients with an ERBB2 mutation at V842I, which is level 3B in breast cancer, neratinib can be used. Alpelisib plus fulvestrant targets the PIK3CA alteration at the C420R, E542, E545, Q546, and H1047 sites. BRCA2 alteration is classified as level 3B in peritoneal serous carcinoma and breast cancer, and it responds to olaparib and talazoparib. The KRAS mutation at G12C is considered level 3B and represents a predictive target for adagrasib and sotorasib in non-small-cell lung cancer. The NF1, FGFR2, PTEN, MET, KRAS, KIT, MTOR, CDKN2A, and PDGFRA mutations are currently level 4 mutations. Large clinical studies are needed to explore the efficacy of these targets in GC. The results can provide clinical actionability for targeted therapy.
| Gene | Variation type | Variable area | Associated drug | Level of evidence |
| ERBB2 | CNV | Amplification | Trastuzumab, Pembrolizumab | 1 |
| NF1 | Mutation | Oncogenic variants | Trametinib, Cobimetinib | 4 |
| PIK3CA | Mutation | C420R, E542, E545, Q546, H1047 | Alpelisib + Fulvestrant | 3B |
| BRCA2 | Mutation | Oncogenic variants | Olaparib, Talazoparib, Niraparib, Rucaparib | 3B |
| FGFR2 | Fusions/Mutation | Fusions, Oncogenic variants | Infigratinib, Erdafitinib, Debio1347, AZD4547 | 4 |
| PTEN | Mutation | Oncogenic variants | GSK2636771, AZD8186 | 4 |
| MET | CNV, exon 14-skipping | Exon 14-skipping, Amplification | Crizotinib | 4 |
| KRAS | Mutation/CNV | G12C, Oncogenic Mutations | Adagrasib, Sotorasib, Trametinb, Cobimetinib, Binimetinib | 3B, 4 |
| KIT | Mutation/CNV | Exon 8, 9, 11, 13, 14, 17, 18 | Imatinib, Sunitinib, Regorafenib | 4 |
| MTOR | Mutation | Oncogenic variants | Everolimus, Temsirolimus | 4 |
| CDKN2A | Mutation | Oncogenic variants | Palbociclib, Ribociclib, Abemaciclib | 4 |
| PDGFRA | Mutation/CNV | Exon 12, 14, 18 | Imatinib, Sunitinib, Regorafenib | 4 |
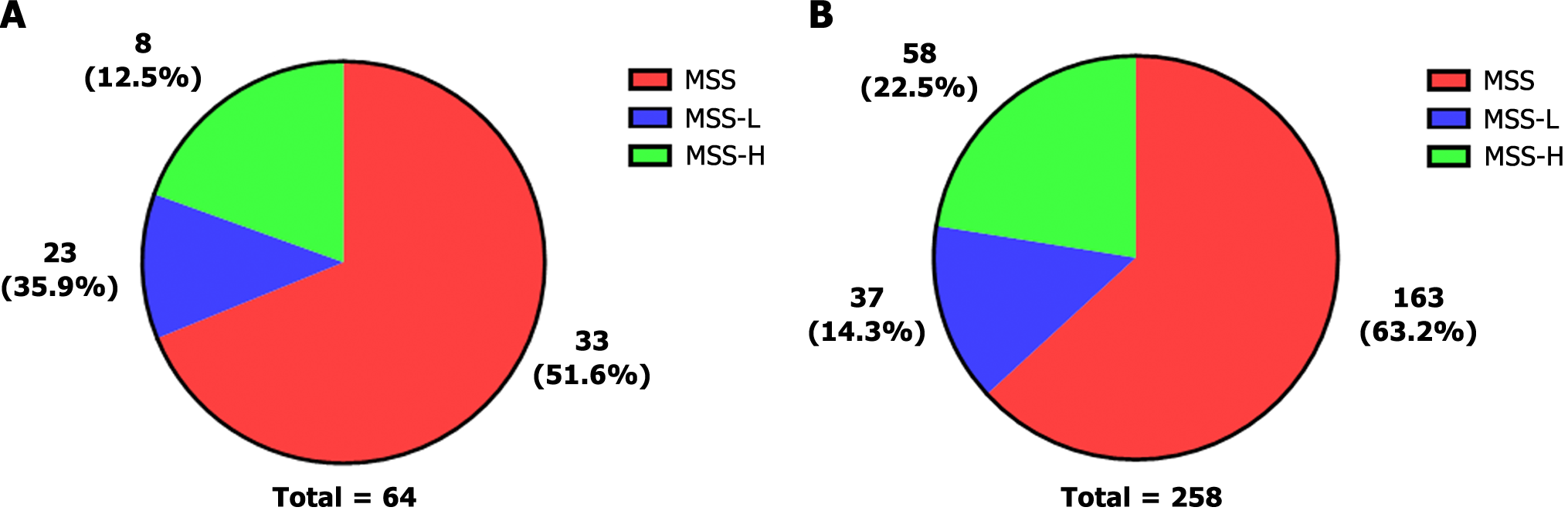
Microsatellite instability (MSI) is a biomarker of sensitivity to immune checkpoint inhibitors. Microsatellites are short tandem repeats throughout the human genome. Tumour tissue has altered microsatellite length due to the insertion or deletion of repeated units; this tissue promotes the appearance of new microsatellite alleles compared to normal tissue. We obtained the results of MSI testing and divided MSI into three levels. MSI = 0 was defined as microsatellite stability (MSS); 0 < MSI < 0.3 was defined as microsatellite instability-low (MSI-L); and MSI ≥ 0.3 was defined as microsatellite instability high (MSI-H).
MSS was the primary type in the present study, accounting for 51.6% of patients. The percent of MSI-L was 35.9%, and the percent of MSI-H was 12.5% (Figure 7A). In the cBioPortal database, MSS was also the dominant type. However, the proportion of MSI-H was higher than that observed in our data (Figure 7B). The mismatch repair (MMR) system repairs defects during DNA replication. MSI can thus result from germline mutations in certain MMR genes. MSH2 is a DNA mismatch repair gene that is associated with Lynch syndrome; we found MSH2 mutations in nine patients. We found the T8M mutation in four patients. This mutation is recorded as a pathogenic mutation in Lynch syndrome and hereditary cancer predisposition according to the ClinVar database. Overall, these results suggest that MSI plays a significant role in the heterogeneity of GC and the selection of treatment options.
NGS is a robust technology platform in the field of oncology that can help doctors accurately and thoroughly understand the gene mutation status of patients in a short period of time and thereby assist in guiding selection of therapeutic strategies[14]. In this study, 24 mutated genes were identified as GC-related genes with a high frequency of mutation; they can be used to provide patients with personalized and accurate treatment. Our results showed that somatic nucleotide variations were detected in all 64 patients. Thus, our study identified an efficient custom panel of 24 mutant genes that can quickly provide common drug targets with a relatively low detection cost.
Our study found a higher mutation frequency of TP53, ERBB2, BRCA2, NF1, and PIK3CA in GC. TP53 is the most common gene mutation associated with GC, which is consistent with previously reported results[15]. TP53 is closely associated with biological processes, including the effects of the cell cycle, DNA repair, energy metabolism, and antioxidant defence[16,17]. Our study discovered TP53 mutation hotspots, including R175, R248, and R273. A related clinical study also confirmed that TP53 point mutations in tumours mainly occurred at the same conserved sites. Recent research indicated that GC patients with these hotspot mutations had poor prognoses[18]. A study demonstrated that the prodrug APR-246 could represent a new cancer treatment strategy for mutant p53 because it can promote tumour cell death[19]. Lee et al.found that adavosertib (WEE1 inhibitor) combined with paclitaxel showed high tumour reduction in GC patients with TP53 mutations[20].
ERBB2 hotspot mutations are found in the extracellular domain (ECD; primarily S310F) in the public database. However, we identified an ERBB2 mutation at V842I, which is commonly found in colorectal cancer[21].We discovered that 15 patients carried BRCA1/2 mutations. The current study also demonstrated that GC or GEJ tumour patients carried BRCA1/2 mutations[22], which may be related to tumorigenesis, including GC development. A molecular analysis of BRCA1/2 mutations clearly revealed distinct categories of tumours[23]. A recent study showed that NF1 has prognostic relevance to clinical outcomes in GC patients[24] and leads to the dysregulation of the RAS/MAPK pathway[25]. Our study demonstrated that PIK3CA mutations were highly frequent and occurred at the E542K and H1047R sites. A previous study showed that overexpression of the PIK3CA gene due to H1047R or E542K point mutations can induce gain-of-function activation of the PI3K-AKT-mTOR pathway and cause phenotypic changes in human cancers, such as enhanced proliferation and invasion[26]. These genes could become potential prognostic markers and biomarkers for the treatment of GC patients.
In addition, we found different phenomena in two separate cohorts. In our database, KRAS and CDKN2A had lower mutation frequencies and BMPR1A and MSH2 had higher mutation frequencies than those in the cBioPortal database. This discrepancy may be due to tumour heterogeneity, sample size, and/or patient race. We found three patients with KRAS mutations and two with KRAS amplification. Fu et al[27] demonstrated that Chinese GC patients with KRAS mutations at the G12V site had shorter OS than patients with wild-type KRAS. BMPR1A and MSH2 are GC susceptibility genes; therefore, this finding has potential implications for screening and managing these gene carriers.
We then explored promising drugs for potential therapeutic targets. ERBB2 is also known as human epidermal growth factor receptor-2 and belongs to the family of epidermal growth factor (EGF) receptor tyrosine kinases[28]. EGF receptor tyrosine kinases can act on downstream MAPKs and the PI3K pathway to promote cancer development. Trastuzumab, combined with chemotherapy and pembrolizumab, has shown promising antitumour activity in patients with advanced GC[29]; therefore, the OncokB database recommends these drugs as level 1. A recent clinical study found synergistic antitumour activity of an Fc-optimized anti-HER2 drug (margetuximab) with an anti-PD-1 checkpoint blocking drug (pembrolizumab)[30]. In addition, several new ERBB2 tyrosine kinase inhibitors have been developed, including tucatinib, poziotinib, and pirotonib[31]. Tyrosine kinase inhibitors (TKIs) directly act on intra cellular protein tyrosine kinases by competitively binding to the tyrosine kinase domain and inhibiting tyrosine kinase phosphorylation. TKIs thus play an important role in personalized therapy of GC.
PIK3CA, a downstream signalling pathway of EGFR and ERBB2, has been frequently identified as a mechanism of drug resistance. A phase I multicentre clinical trial evaluated the safety of the PI3K inhibitor BYL719 in combination with the HSP90 inhibitor AUY922 for treating advanced GC. Currently, the FDA has approved PARP inhibitors for treating ovarian, breast, prostate, and pancreatic cancers that carry BRCA mutations. The detection of embryonic BRCA1/2 mutations may help promote the development of PARP inhibitors in gastroesophageal cancer. We then determined that the BRCA2 mutation was associated with genetic susceptibility for cancer. The BRCA2 mutation at I2490T is likely associated with pathogenic mutations in hereditary breast-ovarian cancer syndrome according to the ClinVar database.
FGFR2 is a member of the fibroblast growth factor receptor (FGFR) family and is located at 10q26. Our study found that approximately 8% of patients had FGFR2 mutations. Prior research has indicated that approximately 5%-10% of GC patients show FGFR2 amplification, which is associated with a poor prognosis[32]. The FIGHT study demonstrated that bemarituzumab combined with chemotherapy significantly improved progression-free survival and overall survival in fibroblast growth factor receptor 2b-positive (FGFR2b+) advanced GC[33]. Research has shown that the CDK4/6 inhibitor PD-0332991 could be a promising therapeutic agent for those with CDKN2A mutations, which occur at high frequency in GC[34].
Numerous studies have shown that MSI is caused by defects in MMR genes, which are closely related to the occurrence of tumours. MSI has been shown to play an essential role in the development of GC. MSI is a highly mutated phenotype caused by short repeat polymorphisms and single nucleotide substitutions in MMR DNA. The hMSH2 gene is one of the critical proteins in MMR. A clinical study suggested that MSI-H/dMMR-positive tumour patients who received preoperative chemotherapy combined with surgery had a poorer prognosis than patients who underwent surgery alone. This result indicated that identifying the MSI/MMR status might help screen patients with GC who need preoperative chemotherapy[35]. More importantly, MSI-H/dMMR has been recognized as one of the critical biomarkers in immunotherapy for GC[36]. The Keynote-062 study showed that pembrolizumab combined with chemo therapy could prolong the survival of GC patients with MSI-H[37]. The results suggested that immunotherapy should play a significant role in GC.
Our research found that the MSH2 mutation probability was 18.75% in our panel but 2.7% in the cBioPortal database. This difference might be due to the selected population and sample quantity. In addition, it could be related to race. A study reporting clinical risk factors found that male sex, advanced age, MLH1 or MSH2 mutations, and a family history of Lynch syndrome were associated with an increased risk of GC[38]. These findings suggest that an individualized, risk-stratified surveillance approach may be appropriate for individuals carrying Lynch syndrome-associated mutations to reduce GC risk.
Taken together, these findings suggest that this novel panel of 24 genes with high frequencies of mutation might provide new insights for individualized and precise treatment in GC patients. Nevertheless, there are some limitations to our study. First, we only compared our data with that of data from the cBioPortal database; thus, a multi-database analysis is required for further validation. Second, the use of NGS could lead to few clinically meaningful responses. Patients and their doctors may jointly decide to order a gene panel at no additional cost from the public health care system.In addition, the frequently mutated genes need to be further explored to determine their functions and underlying mechanisms. Therefore, large cohorts in multicentre prospective clinical studies are needed to validate these prognostic mutation genes and their frequency to guide clinical targeted treatment.
A novel panel of 24 gastric cancer-related genes with a high frequency of mutation can be used to quickly detect mutated genes based on high-throughput sequencing. Mutant gene information, including insertions, deletions, copy number changes, and structural variations, can be efficiently captured. The observed mutations can provide potential targets for the clinical treatment of gastric cancer.
Next-generation sequencing (NGS) technology combined with bioinformatics methods provides quick identification of differences in mutated genes and valuable gene expression information. However, NGS has not been widely used to diagnose and treat gastric cancer in clinical practice.
The Cancer Genome Atlas (TCGA) and the Asian Cancer Research Group (ACRG) have provided unprecedented insight into the molecular characteristics of gastric cancer. We constructed a panel of 24 mutant genes to identify promising target sites.
We used germline DNA from FFPE tumour tissue specimens to explore 24 genes with a high frequency of mutation, including ATR, ATM, AR, BRCA2, BMPR1A, CHD4, CDKN2A, ERBB2, ERBB3, FBXW7, FGFR2, KRAS, KDR, KIT, MET, MSH2, MTOR, NF1, PTEN, PDGFRA, PIK3CA, PTPN11, STK11, and TP53.
24 genes with a high frequency of mutation were compared using the cBioPortal database. Then, the clinical annotation of important variant mutation sites was evaluated with the ClinVar database. Finally, candidate drugs for targeted therapy and immunotherapy were identified from the OncoKB database.
TP53, ERBB2, BRCA2, NF1, and PIK3CA had a higher mutation frequency in our data and the cBioPortal database. MSS was the primary type of MSI. BRCA2, PIK3CA, and FGFR2 gene mutations were identified as promising biomarkers in gastric cancer.
In the present study, the 24 mutated genes were detected in all 64 patients. This novel panel of 24 genes with high frequencies of mutation might provide new insights for individualized and precise treatment of gastric cancer patients.
An efficient custom panel of 24 mutant genes can rapidly identify common drug targets with a relatively low detection cost. Furthermore, it can offer effective personalized treatment for gastric cancer patients.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kawabata H, Japan; Koganti S, United States S-Editor: Wu YXJ L-Editor: A P-Editor: Chen YX
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55615] [Article Influence: 7945.0] [Reference Citation Analysis (131)] |
| 2. | Cao M, Li H, Sun D, Chen W. Cancer burden of major cancers in China: A need for sustainable actions. Cancer Commun (Lond). 2020;40:205-210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 226] [Cited by in RCA: 305] [Article Influence: 61.0] [Reference Citation Analysis (0)] |
| 3. | Ho SWT, Tan P. Dissection of gastric cancer heterogeneity for precision oncology. Cancer Sci. 2019;110:3405-3414. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 4. | Thrift AP, El-Serag HB. Burden of Gastric Cancer. Clin Gastroenterol Hepatol. 2020;18:534-542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 432] [Cited by in RCA: 951] [Article Influence: 190.2] [Reference Citation Analysis (1)] |
| 5. | Matsuoka T, Yashiro M. Biomarkers of gastric cancer: Current topics and future perspective. World J Gastroenterol. 2018;24:2818-2832. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 230] [Cited by in RCA: 308] [Article Influence: 44.0] [Reference Citation Analysis (7)] |
| 6. | Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5015] [Cited by in RCA: 4815] [Article Influence: 437.7] [Reference Citation Analysis (2)] |
| 7. | Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, Ye XS, Do IG, Liu S, Gong L, Fu J, Jin JG, Choi MG, Sohn TS, Lee JH, Bae JM, Kim ST, Park SH, Sohn I, Jung SH, Tan P, Chen R, Hardwick J, Kang WK, Ayers M, Hongyue D, Reinhard C, Loboda A, Kim S, Aggarwal A. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1071] [Cited by in RCA: 1565] [Article Influence: 156.5] [Reference Citation Analysis (0)] |
| 8. | Mosele F, Remon J, Mateo J, Westphalen CB, Barlesi F, Lolkema MP, Normanno N, Scarpa A, Robson M, Meric-Bernstam F, Wagle N, Stenzinger A, Bonastre J, Bayle A, Michiels S, Bièche I, Rouleau E, Jezdic S, Douillard JY, Reis-Filho JS, Dienstmann R, André F. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020;31:1491-1505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 569] [Cited by in RCA: 757] [Article Influence: 151.4] [Reference Citation Analysis (0)] |
| 9. | Marotti JD, de Abreu FB, Wells WA, Tsongalis GJ. Triple-Negative Breast Cancer: Next-Generation Sequencing for Target Identification. Am J Pathol. 2017;187:2133-2138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 10. | Arnold M, Abnet CC, Neale RE, Vignat J, Giovannucci EL, McGlynn KA, Bray F. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterology. 2020;159:335-349.e15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 857] [Cited by in RCA: 1206] [Article Influence: 241.2] [Reference Citation Analysis (0)] |
| 11. | Tan KT, Yeh CN, Chang YC, Cheng JH, Fang WL, Yeh YC, Wang YC, Hsu DS, Wu CE, Lai JI, Chang PM, Chen MH, Lu ML, Chen SJ, Chao Y, Hsiao M. PRKDC: new biomarker and drug target for checkpoint blockade immunotherapy. J Immunother Cancer. 2020;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 12. | Jin Y, Chen DL, Wang F, Yang CP, Chen XX, You JQ, Huang JS, Shao Y, Zhu DQ, Ouyang YM, Luo HY, Wang ZQ, Wang FH, Li YH, Xu RH, Zhang DS. The predicting role of circulating tumor DNA landscape in gastric cancer patients treated with immune checkpoint inhibitors. Mol Cancer. 2020;19:154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 13. | Kim HS, Kim JH, Jang HJ, Han B, Zang DY. Pathological and Prognostic Impacts of FGFR2 Overexpression in Gastric Cancer: A Meta-Analysis. J Cancer. 2019;10:20-27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 14. | Xiao W, Ren L, Chen Z, Fang LT, Zhao Y, Lack J, Guan M, Zhu B, Jaeger E, Kerrigan L, Blomquist TM, Hung T, Sultan M, Idler K, Lu C, Scherer A, Kusko R, Moos M, Xiao C, Sherry ST, Abaan OD, Chen W, Chen X, Nordlund J, Liljedahl U, Maestro R, Polano M, Drabek J, Vojta P, Kõks S, Reimann E, Madala BS, Mercer T, Miller C, Jacob H, Truong T, Moshrefi A, Natarajan A, Granat A, Schroth GP, Kalamegham R, Peters E, Petitjean V, Walton A, Shen TW, Talsania K, Vera CJ, Langenbach K, de Mars M, Hipp JA, Willey JC, Wang J, Shetty J, Kriga Y, Raziuddin A, Tran B, Zheng Y, Yu Y, Cam M, Jailwala P, Nguyen C, Meerzaman D, Chen Q, Yan C, Ernest B, Mehra U, Jensen RV, Jones W, Li JL, Papas BN, Pirooznia M, Chen YC, Seifuddin F, Li Z, Liu X, Resch W, Wu L, Yavas G, Miles C, Ning B, Tong W, Mason CE, Donaldson E, Lababidi S, Staudt LM, Tezak Z, Hong H, Wang C, Shi L. Toward best practice in cancer mutation detection with whole-genome and whole-exome sequencing. Nat Biotechnol. 2021;39:1141-1150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 87] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 15. | Wahl AF, Donaldson KL, Fairchild C, Lee FY, Foster SA, Demers GW, Galloway DA. Loss of normal p53 function confers sensitization to Taxol by increasing G2/M arrest and apoptosis. Nat Med. 1996;2:72-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 458] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 16. | Wasylishen AR, Lozano G. Attenuating the p53 Pathway in Human Cancers: Many Means to the Same End. Cold Spring Harb Perspect Med. 2016;6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 103] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 17. | Sabapathy K, Lane DP. Therapeutic targeting of p53: all mutants are equal, but some mutants are more equal than others. Nat Rev Clin Oncol. 2018;15:13-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 359] [Article Influence: 44.9] [Reference Citation Analysis (0)] |
| 18. | Moon S, Balch C, Park S, Lee J, Sung J, Nam S. Systematic Inspection of the Clinical Relevance of TP53 Missense Mutations in Gastric Cancer. IEEE/ACM Trans Comput Biol Bioinform. 2019;16:1693-1701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 19. | Haffo L, Lu J, Bykov VJN, Martin SS, Ren X, Coppo L, Wiman KG, Holmgren A. Inhibition of the glutaredoxin and thioredoxin systems and ribonucleotide reductase by mutant p53-targeting compound APR-246. Sci Rep. 2018;8:12671. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 20. | Lee J, Kim ST, Kim K, Lee H, Kozarewa I, Mortimer PGS, Odegaard JI, Harrington EA, Lee J, Lee T, Oh SY, Kang JH, Kim JH, Kim Y, Ji JH, Kim YS, Lee KE, Kim J, Sohn TS, An JY, Choi MG, Lee JH, Bae JM, Kim S, Kim JJ, Min YW, Min BH, Kim NKD, Luke S, Kim YH, Hong JY, Park SH, Park JO, Park YS, Lim HY, Talasaz A, Hollingsworth SJ, Kim KM, Kang WK. Tumor Genomic Profiling Guides Patients with Metastatic Gastric Cancer to Targeted Treatment: The VIKTORY Umbrella Trial. Cancer Discov. 2019;9:1388-1405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 151] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 21. | Mishra R, Hanker AB, Garrett JT. Genomic alterations of ERBB receptors in cancer: clinical implications. Oncotarget. 2017;8:114371-114392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 90] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 22. | Avanesyan AA, Sokolenko AP, Ivantsov AO, Kleshchev MA, Maydin MA, Bizin IV, Raskin GA, Shelekhova KV, Gorodnova TV, Bessonov AA, Anisimova EI, Volynshchikova OA, Romanko AA, Ni VI, Broyde RV, Tkachenko OB, Whitehead AJ, Scherbakov AM, Imyanitov EN. Gastric Cancer in BRCA1 Germline Mutation Carriers: Results of Endoscopic Screening and Molecular Analysis of Tumor Tissues. Pathobiology. 2020;87:367-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 23. | Jonsson P, Bandlamudi C, Cheng ML, Srinivasan P, Chavan SS, Friedman ND, Rosen EY, Richards AL, Bouvier N, Selcuklu SD, Bielski CM, Abida W, Mandelker D, Birsoy O, Zhang L, Zehir A, Donoghue MTA, Baselga J, Offit K, Scher HI, O'Reilly EM, Stadler ZK, Schultz N, Socci ND, Viale A, Ladanyi M, Robson ME, Hyman DM, Berger MF, Solit DB, Taylor BS. Tumour lineage shapes BRCA-mediated phenotypes. Nature. 2019;571:576-579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 302] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 24. | Liu D, Zhang Y, Li Y, Fan K. Neurofibromatosis type-1 is a prognostic indicator in human gastric carcinoma. Oncotarget. 2017;8:82910-82919. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 25. | Philpott C, Tovell H, Frayling IM, Cooper DN, Upadhyaya M. The NF1 somatic mutational landscape in sporadic human cancers. Hum Genomics. 2017;11:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 219] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 26. | Zhang Y, Kwok-Shing Ng P, Kucherlapati M, Chen F, Liu Y, Tsang YH, de Velasco G, Jeong KJ, Akbani R, Hadjipanayis A, Pantazi A, Bristow CA, Lee E, Mahadeshwar HS, Tang J, Zhang J, Yang L, Seth S, Lee S, Ren X, Song X, Sun H, Seidman J, Luquette LJ, Xi R, Chin L, Protopopov A, Westbrook TF, Shelley CS, Choueiri TK, Ittmann M, Van Waes C, Weinstein JN, Liang H, Henske EP, Godwin AK, Park PJ, Kucherlapati R, Scott KL, Mills GB, Kwiatkowski DJ, Creighton CJ. A Pan-Cancer Proteogenomic Atlas of PI3K/AKT/mTOR Pathway Alterations. Cancer Cell. 2017;31:820-832.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 335] [Cited by in RCA: 446] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 27. | Fu XH, Chen ZT, Wang WH, Fan XJ, Huang Y, Wu XB, Huang JL, Wang JX, Lin HJ, Tan XL, Wang L, Wang JP. KRAS G12V Mutation is an Adverse Prognostic Factor of Chinese Gastric Cancer Patients. J Cancer. 2019;10:821-828. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 28. | Han S, Park S, An J, Yang JY, Chung JW, Kim YJ, Kim KO, Park DK, Kwon KA, Lee WK, Nam S, Kim JH. HER2 as a potential biomarker of lymph node metastasis in undifferentiated early gastric cancer. Sci Rep. 2020;10:5270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 29. | Chung HC, Bang YJ, S Fuchs C, Qin SK, Satoh T, Shitara K, Tabernero J, Van Cutsem E, Alsina M, Cao ZA, Lu J, Bhagia P, Shih CS, Janjigian YY. First-line pembrolizumab/placebo plus trastuzumab and chemotherapy in HER2-positive advanced gastric cancer: KEYNOTE-811. Future Oncol. 2021;17:491-501. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 129] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 30. | Rugo HS, Im SA, Cardoso F, Cortés J, Curigliano G, Musolino A, Pegram MD, Wright GS, Saura C, Escrivá-de-Romaní S, De Laurentiis M, Levy C, Brown-Glaberman U, Ferrero JM, de Boer M, Kim SB, Petráková K, Yardley DA, Freedman O, Jakobsen EH, Kaufman B, Yerushalmi R, Fasching PA, Nordstrom JL, Bonvini E, Koenig S, Edlich S, Hong S, Rock EP, Gradishar WJ; SOPHIA Study Group. Efficacy of Margetuximab vs Trastuzumab in Patients With Pretreated ERBB2-Positive Advanced Breast Cancer: A Phase 3 Randomized Clinical Trial. JAMA Oncol. 2021;7:573-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 245] [Article Influence: 61.3] [Reference Citation Analysis (0)] |
| 31. | Meric-Bernstam F, Johnson AM, Dumbrava EEI, Raghav K, Balaji K, Bhatt M, Murthy RK, Rodon J, Piha-Paul SA. Advances in HER2-Targeted Therapy: Novel Agents and Opportunities Beyond Breast and Gastric Cancer. Clin Cancer Res. 2019;25:2033-2041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 235] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 32. | Su X, Zhan P, Gavine PR, Morgan S, Womack C, Ni X, Shen D, Bang YJ, Im SA, Ho Kim W, Jung EJ, Grabsch HI, Kilgour E. FGFR2 amplification has prognostic significance in gastric cancer: results from a large international multicentre study. Br J Cancer. 2014;110:967-975. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 149] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 33. | Catenacci DVT, Rasco D, Lee J, Rha SY, Lee KW, Bang YJ, Bendell J, Enzinger P, Marina N, Xiang H, Deng W, Powers J, Wainberg ZA. Phase I Escalation and Expansion Study of Bemarituzumab (FPA144) in Patients With Advanced Solid Tumors and FGFR2b-Selected Gastroesophageal Adenocarcinoma. J Clin Oncol. 2020;38:2418-2426. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 34. | Huang S, Ye H, Guo W, Dong X, Wu N, Zhang X, Huang Z. CDK4/6 inhibitor suppresses gastric cancer with CDKN2A mutation. Int J Clin Exp Med. 2015;8:11692-11700. [PubMed] |
| 35. | Smyth EC, Wotherspoon A, Peckitt C, Gonzalez D, Hulkki-Wilson S, Eltahir Z, Fassan M, Rugge M, Valeri N, Okines A, Hewish M, Allum W, Stenning S, Nankivell M, Langley R, Cunningham D. Mismatch Repair Deficiency, Microsatellite Instability, and Survival: An Exploratory Analysis of the Medical Research Council Adjuvant Gastric Infusional Chemotherapy (MAGIC) Trial. JAMA Oncol. 2017;3:1197-1203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 409] [Cited by in RCA: 388] [Article Influence: 48.5] [Reference Citation Analysis (0)] |
| 36. | Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, Wong F, Azad NS, Rucki AA, Laheru D, Donehower R, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Greten TF, Duffy AG, Ciombor KK, Eyring AD, Lam BH, Joe A, Kang SP, Holdhoff M, Danilova L, Cope L, Meyer C, Zhou S, Goldberg RM, Armstrong DK, Bever KM, Fader AN, Taube J, Housseau F, Spetzler D, Xiao N, Pardoll DM, Papadopoulos N, Kinzler KW, Eshleman JR, Vogelstein B, Anders RA, Diaz LA Jr. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3799] [Cited by in RCA: 4909] [Article Influence: 613.6] [Reference Citation Analysis (0)] |
| 37. | Shitara K, Van Cutsem E, Bang YJ, Fuchs C, Wyrwicz L, Lee KW, Kudaba I, Garrido M, Chung HC, Lee J, Castro HR, Mansoor W, Braghiroli MI, Karaseva N, Caglevic C, Villanueva L, Goekkurt E, Satake H, Enzinger P, Alsina M, Benson A, Chao J, Ko AH, Wainberg ZA, Kher U, Shah S, Kang SP, Tabernero J. Efficacy and Safety of Pembrolizumab or Pembrolizumab Plus Chemotherapy vs Chemotherapy Alone for Patients With First-line, Advanced Gastric Cancer: The KEYNOTE-062 Phase 3 Randomized Clinical Trial. JAMA Oncol. 2020;6:1571-1580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 821] [Cited by in RCA: 845] [Article Influence: 169.0] [Reference Citation Analysis (1)] |
| 38. | Syngal S, Brand RE, Church JM, Giardiello FM, Hampel HL, Burt RW; American College of Gastroenterology. ACG clinical guideline: Genetic testing and management of hereditary gastrointestinal cancer syndromes. Am J Gastroenterol. 2015;110:223-62; quiz 263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 957] [Cited by in RCA: 1086] [Article Influence: 108.6] [Reference Citation Analysis (0)] |