Published online Apr 16, 2022. doi: 10.12998/wjcc.v10.i11.3389
Peer-review started: October 25, 2021
First decision: December 17, 2021
Revised: December 23, 2021
Accepted: February 27, 2022
Article in press: February 27, 2022
Published online: April 16, 2022
Processing time: 164 Days and 21.3 Hours
Complete response after neoadjuvant chemotherapy (rNACT) elevates the surgical outcomes of patients with breast cancer, however, non-rNACT have a higher risk of death and recurrence.
To establish novel machine learning (ML)-based predictive models for predicting probability of rNACT in breast cancer patients who intends to receive NACT.
A retrospective analysis of 487 breast cancer patients who underwent mastectomy or breast-conserving surgery and axillary lymph node dissection following neoadjuvant chemotherapy at the Hubei Cancer Hospital between January 1, 2013, and October 1, 2021. The study cohort was divided into internal training and testing datasets in a 70:30 ratio for further analysis. A total of twenty-four variables were included to develop predictive models for rNACT by multiple ML-based algorithms. A feature selection approach was used to identify optimal predictive factors. These models were evaluated by the receiver operating characteristic (ROC) curve for predictive performance.
Analysis identified several significant differences between the rNACT and non-rNACT groups, including total cholesterol, low-density lipoprotein, neutrophil-to-lymphocyte ratio, body mass index, platelet count, albumin-to-globulin ratio, platelet-to-lymphocyte ratio, and lymphocyte-to-monocyte ratio. The areas under the curve of the six models ranged from 0.81 to 0.96. Some ML-based models performed better than models using conventional statistical methods in both ROC curves. The support vector machine (SVM) model with twelve variables introduced was identified as the best predictive model.
By incorporating pretreatment serum lipids and serum inflammation markers, it is feasible to develop ML-based models for the preoperative prediction of rNACT and therefore facilitate the choice of treatment, particularly the SVM, which can improve the prediction of rNACT in patients with breast cancer.
Core Tip: For predicting response after neoadjuvant chemotherapy (rNACT), some machine learning-based models performed better than models using conventional methods, and the support vector machine model performed best. Preoperative serum lipids and serum inflammation markers have contributed to predicting rNACT in breast cancer patients. These results suggested the need to raise awareness of the importance of minimally-invasive approaches for monitoring breast cancer patients who intended to undergo neoadjuvant chemotherapy. However, the current study needs to be validated with caution and require external validation in the future.
- Citation: Ke ZR, Chen W, Li MX, Wu S, Jin LT, Wang TJ. Added value of systemic inflammation markers for monitoring response to neoadjuvant chemotherapy in breast cancer patients. World J Clin Cases 2022; 10(11): 3389-3400
- URL: https://www.wjgnet.com/2307-8960/full/v10/i11/3389.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i11.3389
Worldwide, breast cancer is a major cause of human suffering and high mortality among women[1]. Neoadjuvant chemotherapy (NACT) as a treatment for early breast cancer, can make breast conserving surgery more feasible, and may achieve more than the same chemotherapy after surgery to eradicate micrometastasis[2]. More than 65% of the patients treated with NACT have a response, and more than 15% have achieved a complete clinical response. Although some trials use the old chemotherapy regimen, more than 15% of the patients have undergone partial chemotherapy[2-4]. In other words, most patients who cannot achieve a complete pathological response after NACT may face a higher risk of death and recurrence. Therefore, it is necessary to develop a practical, convenient and efficient tool to predict the pathological response of patients with NACT breast cancer.
Machine learning (ML)-based integrated analysis is a new computer-based method, which has been widely used in medical data management in the past decade[5]. It appears at the intersection of statistics and computer science. The former attempts to learn relationships from data, while the latter emphasizes efficient computational algorithms[6,7]. Compared with traditional statistical prediction models such as logistic regression (LR), ML depends on a predetermined model. It can potentially find the interaction between variables and iteratively learn the update algorithm from the data[8,9]. Previously, several conventional predictive models have been made for predicting after NACT in breast cancer patients, including LR, GLM[10-14]. However, few reports have incorporated multiple ML-based ensemble analyses for predicting response after NACT (rNACT).
In this study, we aimed to develop a rNACT risk prediction model for breast cancer patients that utilizes pretreatment serum lipids and serum inflammation markers to stratify patients by rNACT risk on admission. We then analyzed the predictive performance of these ML-based models in a deviation cohort and then verified performance in an internal and external validation cohort.
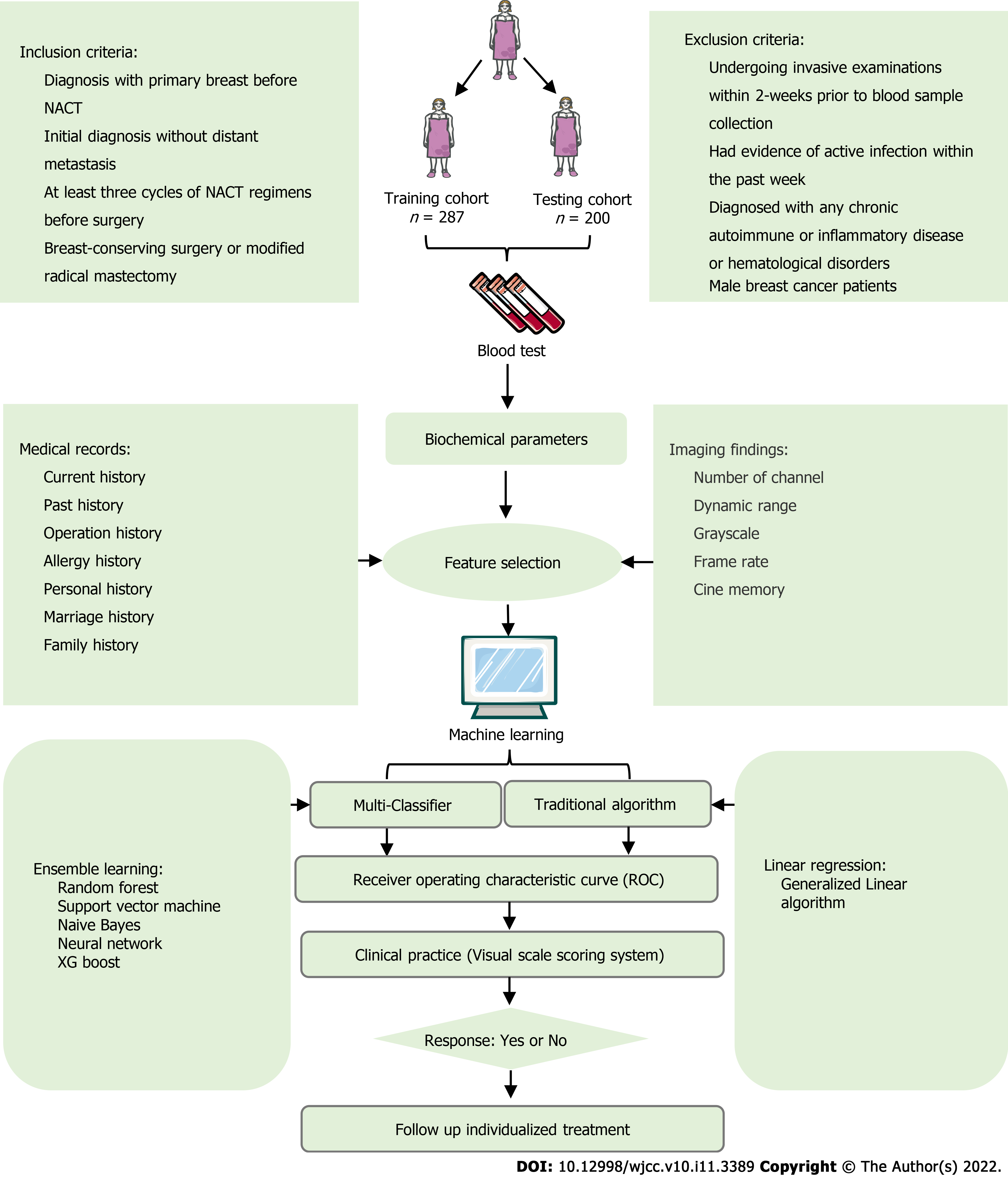
Between January 1, 2013 and October 1, 2021, we retrospectively collated data from consecutive patients who had been diagnosed with breast cancer at the Hubei Cancer Hospital. All patients had received NACT before surgery. This study was approved by the Institutional Ethics Committee of the Hubei Cancer Hospital (Reference: LLHBCH2021YN-021), in compliance with the Declaration of Helsinki. Written informed consent was obtained from all participants before any treatment. We confirmed that the data from all the patients were anonymized in this study. The inclusion and exclusion criteria were summarized in Figure 1. The study cohort was divided into internal training and testing datasets in a 70:30 ratio for further analysis.
The blood samples of all patients were taken from the fasting state before chemotherapy, and the blood tests were operated by professional personnel to ensure that the blood test results were not biased. The results of the blood test are as follows: Blood routine, liver and kidney function, electrolytes, and blood lipids.
According to RECIST (version 1.1) criteria15, the efficacy of NACT is defined as follows: (1) Cardiol Res. The tumor is disappeared completely; (2) Partial response (PR). The diameter of the tumor is reduced (≥ 30%); (3) Progressive disease (PD). The diameter of the tumor was reduced (≥ 20%); and (4) Stable disease (SD). The diameter of the tumor was altered between PR and PD. Collectively, patients were considered to be responsive to NACT provided that they were evaluated as CR or PR after NACT treatment. On the contrary, patients with SD or PD were regarded as non-responsive to NACT.
Four ML-based algorithms were performed to build predictive models, we used the caret package to randomly divide the data set into two parts, 70% for model training and 30% for model testing. A total of 6 ML-based algorithms were executed to establish the predictive model. According to the principle of "two-step estimation"[16], We obtained the prediction model through variable screening and algorithm, as follows: M is the intersection of M3 and M4.The characteristic variable is marked as X and the target variable is marked as Y. The X and Y were evenly divided into two parts, namely X1, Y1, and X2, Y2. Through univariate screening, the variable quantum set M1was screened on X1 and Y1, and M2was filtered by X2 and Y2. Then, a lasso was used to fit the model again, and the filtered variables were marked asM3 and M4 Briefly, by sorting the intersection of variable sets, the optimal subset modeling is obtained. The model was evaluated by inspection, discrimination, and calibration.The receiver operating characteristic (ROC) curve was used to evaluate the recognition ability of the prediction model in the training data set and the test data set; The discrimination ability of each model was quantified by the area under the ROC curve (AUC), decision curve analysis, and clinical impact curve (CIC).
Continuous variables are expressed as mean ± SD and compared using the two-tailed t-test or the Mann-Whitney test. Categorical variables were compared using the chi-square test or Fisher's exact test. Univariate and multivariate logistic analyses were used to explore the risk factors for rNACT. Several ML-based algorithms were applied to predict rNACT, including support vector machine (SVM), random forest (RF), Naive Bayes (NB), neural network (NN), decision tree (DT), and generalized linear model (GLM)[17,18]. Among all 6 algorithms, the GLM is considered conventional methods, and the others are representative supervised ML-based algorithms. The prediction ability of the 6 models was first evaluated by the ROC curve. All analysis was performed using the Python programming language (version 3.9.2, Python Software Foundation, https://www.python.org/) and R Project for Statistical Computing (version 4.0.4, http://www.r-project.org/). Statistical analyses were performed using a two-tailed Student's t-test in PRISM software (GraphPad 6 Software) to compare the differences between rNACT and non-rNACT groups assuming equal variance. All P values were two-tailed, and P < 0.05 was considered statistically significant.
During the period of enrollment, 287 consecutive patients with breast cancer underwent mastectomy or breast-conserving surgery and axillary lymph node dissection following NACT. Besides, 201 patients were validated as external data sets for the prediction model. Demographics and baseline data were summarized in Table 1. According to the RECIST (version 1.1) criteria, rNACT was identified in 255 (88.9%) and 32 (11.1%) patients with non-rNACT in the internal whole cohort. In the external cohort, 176 (88.0%) patients were confirmed to have rNACT, and 24 (12.0%) patients represented non-rNACT. Overall, most patients with breast cancer presenting with rNACT were positively associated with pretreatment serum lipids and serum inflammation markers. No statistically significant difference was detected between two cohorts with regard to age, menopause, grade, smoking, estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) (P ≥ 0.05).
| Variables | Dummy variables | Training cohort | Testing cohort | ||||||
| Overall (n = 287) | Yes (n = 255) | No (n = 32) | P value | Overall (n = 200) | Yes (n = 176) | No (n = 24) | P value | ||
| Age [median (IQR)],yr | 61.00 (55.00, 70.00) | 61.00 (54.00, 70.00) | 64.00 (58.75, 69.25) | 0.11 | 61.00 (55.75, 70.25) | 61.00 (55.00, 70.25) | 64.00 (57.75, 70.25) | 0.28 | |
| Menopausal (%) | Yes | 35 (12.2) | 30 (11.8) | 5 (15.6) | 0.73 | 28 (14.0) | 24 (13.6) | 4 (16.7) | 0.93 |
| No | 252 (87.8) | 225 (88.2) | 27 (84.4) | 172 (86.0) | 152 (86.4) | 20 (83.3) | |||
| T stage (%) | T1-2 | 216 (75.3) | 196 (76.9) | 20 (62.5) | 0.12 | 156 (78.0) | 140 (79.5) | 16 (66.7) | 0.24 |
| T3-4 | 71 (24.7) | 59 (23.1) | 12 (37.5) | 44 (22.0) | 36 (20.5) | 8 (33.3) | |||
| N stage (%) | N0-1 | 71 (24.7) | 58 (22.7) | 13 (40.6) | 0.04 | 52 (26.0) | 40 (22.7) | 12 (50.0) | 0.01 |
| N2-3 | 216 (75.3) | 197 (77.3) | 19 (59.4) | 148 (74.0) | 136 (77.3) | 12 (50.0) | |||
| Grade (%) | I-II | 195 (67.9) | 175 (68.6) | 20 (62.5) | 0.61 | 134 (67.0) | 118 (67.0) | 16 (66.7) | 0.11 |
| III | 92 (32.1) | 80 (31.4) | 12 (37.5) | 66 (33.0) | 58 (33.0) | 8 (33.3) | |||
| Histology (%) | IDC | 111 (38.7) | 106 (41.6) | 5 (15.6) | 0.02 | 80 (40.0) | 77 (43.8) | 3 (12.5) | 0.03 |
| ILC | 91 (31.7) | 79 (31.0) | 12 (37.5) | 61 (30.5) | 51 (29.0) | 10 (41.7) | |||
| IMC | 48 (16.7) | 41 (16.1) | 7 (21.9) | 36 (18.0) | 29 (16.5) | 7 (29.2) | |||
| Others | 37 (12.9) | 29 (11.4) | 8 (25.0) | 23 (11.5) | 19 (10.8) | 4 (16.7) | |||
| Molecular subtyping (%) | HER2-LuB | 108 (37.6) | 100 (39.2) | 8 (25.0) | < 0.01 | 76 (38.0) | 71 (40.3) | 5 (20.8) | 0.01 |
| HER2+ | 71 (24.7) | 66 (25.9) | 5 (15.6) | 53 (26.5) | 50 (28.4) | 3 (12.5) | |||
| HER2+LuB | 29 (10.1) | 24 (9.4) | 5 (15.6) | 18 (9.0) | 15 (8.5) | 3 (12.5) | |||
| LuA | 54 (18.8) | 41 (16.1) | 13 (40.6) | 37 (18.5) | 25 (14.2) | 12 (50.0) | |||
| TN | 25 (8.7) | 24 (9.4) | 1 (3.1) | 16 (8.0) | 15 (8.5) | 1 (4.2) | |||
| BMI (%) | ≤ 18 | 21 (7.3) | 21 (8.2) | 0 (0.0) | 0.22 | 11 (5.5) | 11 (6.2) | 0 (0.0) | 0.42 |
| ≥ 27 | 36 (12.5) | 31 (12.2) | 5 (15.6) | 27 (13.5) | 23 (13.1) | 4 (16.7) | |||
| 18 - 27 | 230 (80.1) | 203 (79.6) | 27 (84.4) | 162 (81.0) | 142 (80.7) | 20 (83.3) | |||
| Smoking (%) | No | 258 (89.9) | 231 (90.6) | 27 (84.4) | 0.43 | 182 (91.0) | 161 (91.5) | 21 (87.5) | 0.79 |
| Yes | 29 (10.1) | 24 (9.4) | 5 (15.6) | 18 (9.0) | 15 (8.5) | 3 (12.5) | |||
| ER (%) | Negative | 103 (35.9) | 91 (35.7) | 12 (37.5) | 0.99 | 73 (36.5) | 66 (37.5) | 7 (29.2) | 0.56 |
| Positive | 184 (64.1) | 164 (64.3) | 20 (62.5) | 127 (63.5) | 110 (62.5) | 17 (70.8) | |||
| PR (%) | Negative | 165 (57.5) | 141 (55.3) | 24 (75.0) | 0.05 | 109 (54.5) | 91 (51.7) | 18 (75.0) | 0.05 |
| Positive | 122 (42.5) | 114 (44.7) | 8 (25.0) | 91 (45.5) | 85 (48.3) | 6 (25.0) | |||
| HER2 (%) | Negative | 157 (54.7) | 140 (54.9) | 17 (53.1) | 0.99 | 110 (55.0) | 96 (54.5) | 14 (58.3) | 0.89 |
| Positive | 130 (45.3) | 115 (45.1) | 15 (46.9) | 90 (45.0) | 80 (45.5) | 10 (41.7) | |||
| PLT [median (IQR)] × 109/L | 200.00 (154.50, 268.50) | 187.00 (152.50, 245.50) | 388.00 (334.50, 454.00) | < 0.01 | 202.50 (158.75, 267.25) | 190.50 (154.00, 246.50) | 377.00 (332.50, 443.50) | < 0.01 | |
| Neutrophil [median (IQR)] × 109/L | 4.35 (3.52, 5.13) | 4.54 (3.76, 5.23) | 3.09 (2.02, 3.39) | < 0.01 | 4.44 (3.50, 5.21) | 4.60 (3.78, 5.27) | 3.09 (2.02, 3.35) | < 0.01 | |
| MONO [median (IQR)] × 109/L | 0.40 (0.25, 0.57) | 0.38 (0.24, 0.50) | 0.88 (0.80, 0.94) | < 0.01 | 0.41 (0.25, 0.58) | 0.37 (0.23, 0.49) | 0.86 (0.75, 0.91) | < 0.01 | |
| Lymphocyte [median (IQR)] × 109/L | 3.09 (2.92, 3.30) | 3.13 (2.96, 3.32) | 1.64 (1.14, 2.16) | < 0.01 | 3.08 (2.92, 3.30) | 3.12 (2.98, 3.32) | 1.67 (1.14, 2.16) | < 0.01 | |
| NLR [median (IQR)] | 1.43 (1.20, 1.67) | 1.42 (1.20, 1.64) | 1.56 (1.21, 2.68) | 0.03 | 1.48 (1.20, 1.70) | 1.47 (1.20, 1.67) | 1.56 (1.22, 2.75) | < 0.01 | |
| LMR [median (IQR)] | 7.62 (5.59, 12.96) | 8.51 (6.18, 13.31) | 1.86 (1.51, 2.47) | < 0.01 | 7.59 (5.60, 13.07) | 8.53 (6.26, 13.46) | 1.87 (1.59, 2.52) | < 0.01 | |
| PLR [median (IQR)] | 64.45 (49.59, 85.43) | 61.99 (47.81, 78.50) | 237.76 (189.53, 342.96) | < 0.01 | 65.33 (50.30, 85.89) | 62.36 (49.02, 78.10) | 226.10 (189.53, 305.51) | < 0.01 | |
| ALB [median (IQR)],g/L | 47.00 (39.00, 55.00) | 48.00 (41.00, 56.00) | 32.50 (27.50, 37.00) | < 0.01 | 47.00 (39.00, 55.25) | 49.00 (41.00, 56.00) | 31.50 (27.50, 37.00) | < 0.01 | |
| GLB [median (IQR)],g/L | 23.00 (20.00, 26.00) | 22.00 (20.00, 25.00) | 35.00 (30.75, 41.00) | < 0.01 | 23.00 (20.00, 26.00) | 23.00 (20.00, 25.00) | 35.50 (30.75, 41.25) | < 0.01 | |
| A/G [median (IQR)] | 2.11 (1.76, 2.53) | 2.18 (1.88, 2.57) | 0.88 (0.82, 1.03) | < 0.01 | 2.11 (1.71, 2.50) | 2.18 (1.85, 2.55) | 0.88 (0.82, 1.03) | < 0.01 | |
| LDL [median (IQR)],mmol/L | 3.00 (2.88, 3.10) | 2.97 (2.87, 3.08) | 3.13 (3.05, 3.24) | < 0.01 | 3.00 (2.90, 3.08) | 2.97 (2.88, 3.07) | 3.13 (3.04, 3.22) | < 0.01 | |
| HDL [median (IQR)],mmol/L | 1.26 (1.19, 1.33) | 1.29 (1.20, 1.34) | 1.17 (1.08, 1.20) | < 0.01 | 1.25 (1.18, 1.32) | 1.28 (1.20, 1.33) | 1.15 (1.08, 1.19) | 0.02 | |
| TC [median (IQR)], mmol/L | 0.52 (0.49, 0.56) | 0.52 (0.48, 0.56) | 0.56 (0.52, 0.58) | < 0.01 | 0.52 (0.48, 0.56) | 0.52 (0.48, 0.56) | 0.56 (0.52, 0.57) | < 0.01 | |
| TG [median (IQR)], mmol/L | 1.82 (1.61, 2.15) | 1.78 (1.59, 2.04) | 2.34 (2.27, 2.39) | < 0.01 | 1.85 (1.60, 2.15) | 1.79 (1.59, 2.04) | 2.34 (2.27, 2.39) | < 0.01 | |
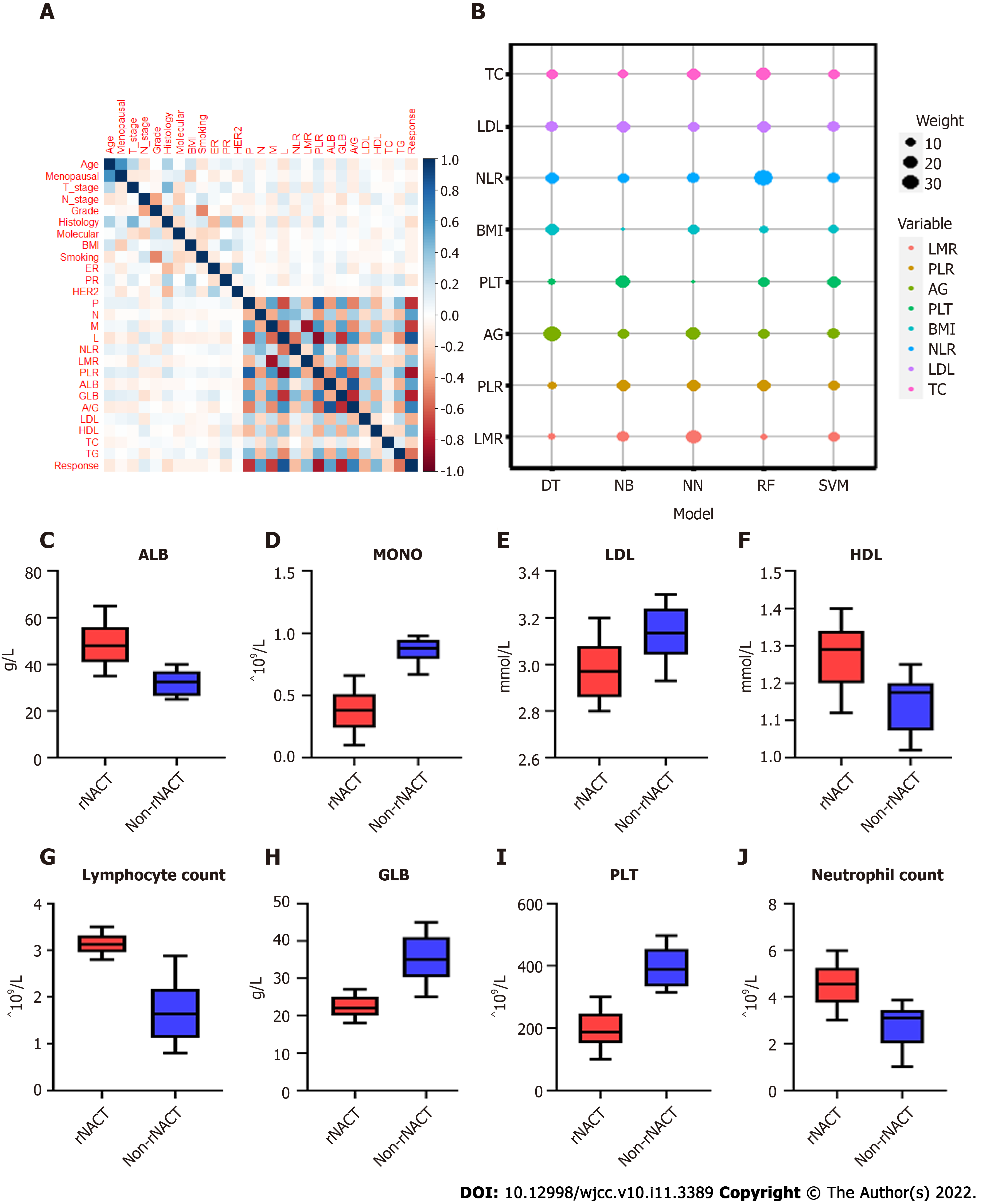
By feature selection, the twenty-four variables for each algorithm were screened by their predictive importance. As depicted in Figure 2A, only twelve of the candidate features were eventually chosen for modeling, among which eight features had a positive association with rNACT, including PLT, monocyte count (MONO), neutrophil-to-lymphocyte ratio (NLR), lymphocyte-to-monocyte ratio (LMR), platelet-to-lymphocyte ratio (PLR), low-density lipoprotein (LDL), A/G, and total cholesterol (TC). Four features were negatively correlated with rNACT, including high-density lipoprotein (HDL), triglyceride (TG), BMI, and age. The weight of the top eight variables was shown in Figure 2B. The pretreatment serum lipids and serum inflammation markers also showed significant differences between rNACT and non-rNACT groups (Figure 2C-J). Multivariable logistic analysis using raw data of the candidate features proved that the features selected by stepwise analysis exhibited similar risk implications (Supplementary Table 1). Based on our results, NLR (OR: 1.02, 95%CI: 0.78-1.26), LMR (OR: 1.44, 95%CI: 1.32-1.56), PLR (OR:2.54, 95%CI: 1.81-6.94), PLT (OR:1.87, 95%CI: 1.76-1.98), LDL (OR:1.01, 95%CI: 0.89-1.13), BMI (OR:1.23, 95%CI: 0.78-1.68), A/G (OR:1.69, 95%CI: 1.24-2.14), TC (OR:0.71, 95%CI: 0.26-1.16), and TG (OR:0.42, 95%CI: 0.17-0.68) were positively correlated with rNACT.
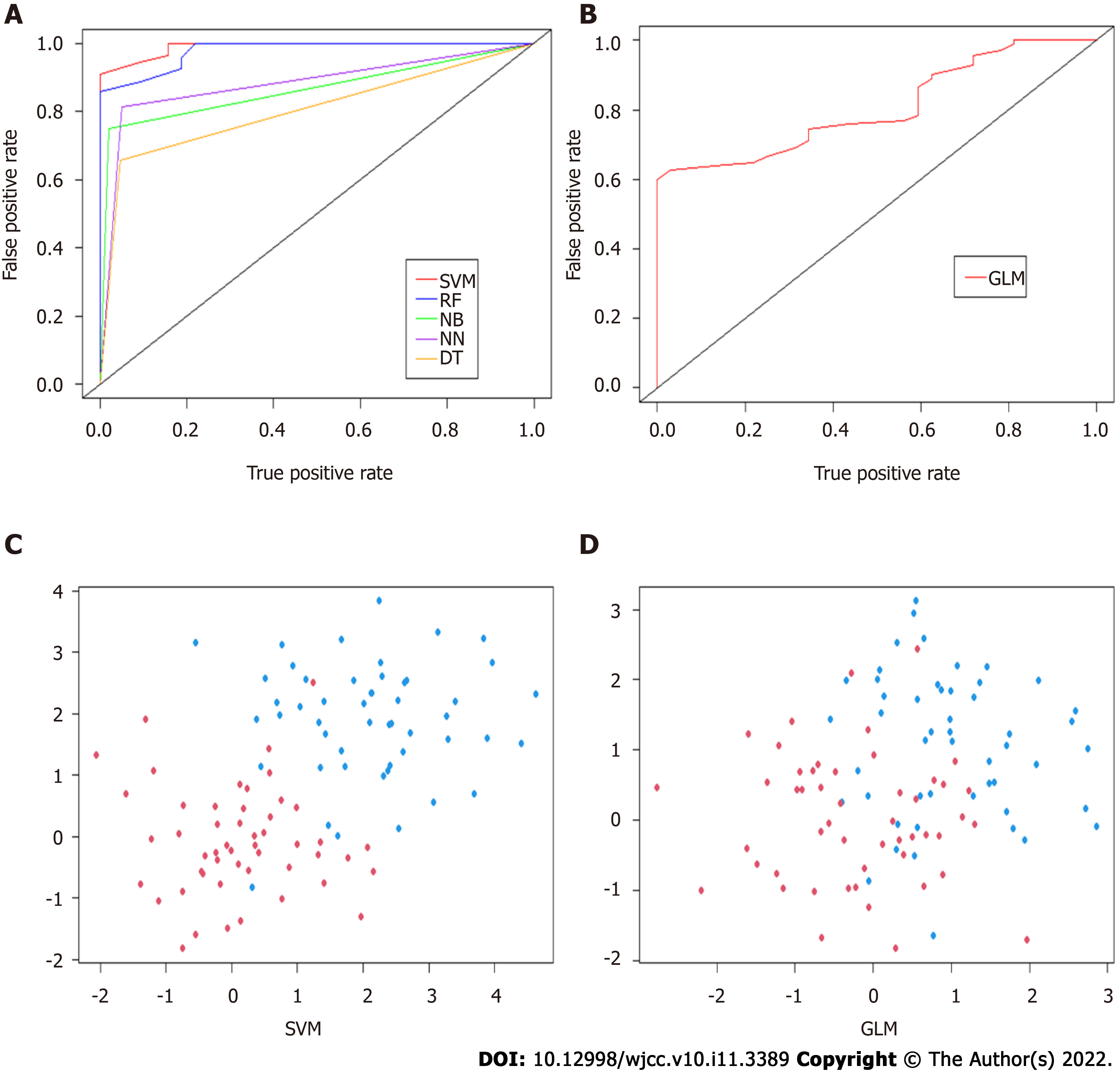
A total of twelve preoperative variables were used to develop predictive models for rNACT based on six algorithms. The predictive performance of all models was shown in Figure 3A and B and Table 2. The best performance was observed in the SVM model (AUC = 0.96, 95%CI: 0.91-1.01), which performed similarly to RF model (AUC = 0.94, 95%CI: 0.87-1.01), superior than NB model (AUC = 0.86, 95%CI: 0.79-0.93), NN model (AUC = 0.88, 95%CI: 0.82-0.94), DT model (AUC = 0.83, 95%CI: 0.77-0.89), and GLM (AUC = 0.81, 95%CI: 0.71-0.91). All ML-based models were better than conventional model. Furthermore, the optimal model SVM showed superior to the traditional linear model in discrimination (Figure 3C and D).
| Model | AUC (95%CI) | Sensitivity | Specificity | PPV | NPV | Kappa | Brier |
| SVM | 0.96 (0.91-1.01) | 96.58 | 45.28 | 88.63 | 75.00 | 0.68 | 0.06 |
| RF | 0.94 (0.87-1.01) | 94.44 | 68.75 | 86.67 | 68.75 | 0.65 | 0.07 |
| NB | 0.86 (0.79-0.93) | 96.36 | 35.80 | 83.14 | 75.00 | 0.62 | 0.07 |
| NN | 0.88 (0.82-0.94) | 93.15 | 25.00 | 93.15 | 53.15 | 0.62 | 0.07 |
| DT | 0.83 (0.77-0.89) | 94.50 | 24.14 | 74.12 | 65.63 | 0.59 | 0.07 |
| GLM | 0.81 (0.71-0.91) | 95.70 | 23.76 | 69.80 | 75.00 | 0.57 | 0.08 |
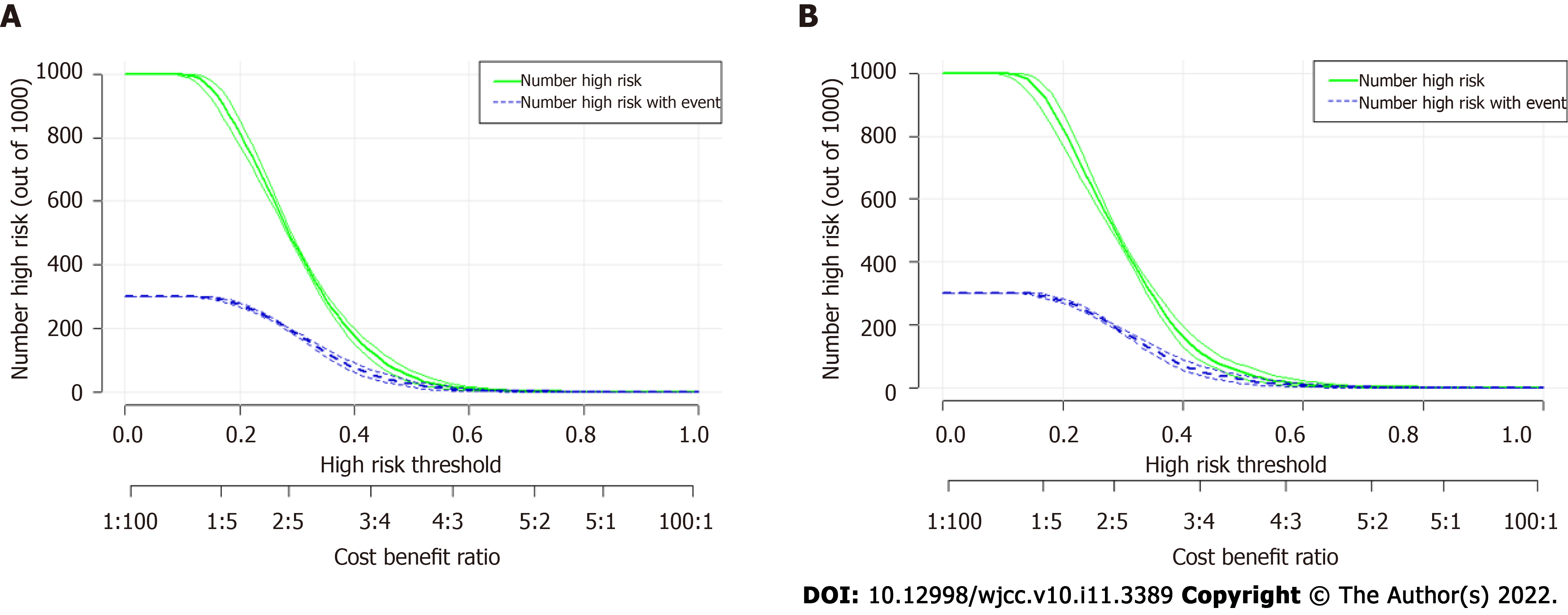
To further validate the performance of the SVM model, we also adopted CIC to evaluate the prediction efficiency, as illustrated in Figure 4A, the CIC demonstrated that the stratification of rNACT could be distinguished in the training cohorts. These results were also parallel to risk factors of rNACT delineated in the validation cohorts (Figure 4B), indicating that the selected features were highly relevant to rNACT.
Reliable markers of chemosensitivity help select patients who most benefit from NACT[19]. Previous studies on the candidate predictors of NACT efficacy in breast cancer patients are discordant, suggesting that the potential predictors to predict efficacy is insufficient[20-23]. In addition, whilst many studies report the predictive outcomes of breast cancer patients who have received NACT, however, relatively few have investigated the individual contribution of multiple models to accuracy, especially prediction efficiency[24-27]. Whilst this study indicates that ML-based predictive algorithms should be included in NACT risk assessments in breast cancer patients, it also highlights the importance of conducting newly predictive models for clinical management.
Supervised ML algorithms have been a dominant method in the data mining field[17]. In recent years, ML-based algorithms were widely used for the evaluation of disease prognosis[28-30]. In this study, extensive variables were made to identify those predictive that applied more than one supervised ML algorithm on rNACT prediction. Based on the ML algorithm, we employed a variety of statistical, probabilistic, and optimization methods to learn from experience and detect useful patterns from large, unstructured, and complex datasets. To sum up, we extracted the data from the patient’s medical records as much as possible. With the help of different algorithms, such as automated text categorisation[31], network intrusion detection[32], optimizing manufacturing process[33], etc., we finally obtained meaningful candidate variables. Given the growing applicability and effectiveness of supervised ML algorithms on predictive disease modeling. Interestingly, we found that the SVM algorithm is applied most robust in predicting rNACT, which denotes superior performance than the conventional linear prediction model. Besides, the remaining machine prediction models are better than GLM. Therefore, our research demonstrated that, compared with the traditional model, machine learning modeling prediction rNACT could obtain better prediction performance.
Inflammation is associated with the development and malignant progression of most cancers[34]. Inflammatory blood markers have emerged as potential prognostic factors in various cancers, such as NLR, LMR, and PLR. Activated inflammatory cells are sources of reactive oxygen species and reactive nitrogen intermediates that can promote cancer initiation[35]. In breast cancer, pretreatment NLR values are associated with patient prognosis[36]. Similarly, our study indicated that NLR, LMR, and PLR values can be reliably used to predict breast patient responses to NACT treatment, which can effectively stratify patients based upon their likelihood of achieving rNACT. Besides, we also found that pretreatment abnormal A/G ratio, which might be attributable to rNACT. Indeed, a low pretreatment A/G ratio is associated with poor prognosis in human cancers[37]. The importance of lipids in tumor progression, invasion, and metastasis has been described in the previous studies[38]. High triglycerides and low levels of HDL are observed to promote tumor growth[39]. In the present study, we observed that LDL, TC, and BMI were highly associated with rNACT, consistent with previous studies[37,38]. Collectively, clinicians can more effectively weigh the relative costs and benefits of pretreatment serum lipids and serum inflammation markers to ensure that they act in the optimal choice of breast cancer patients.
There are multiple strengths to this study. First, our observations were limited to retrospective studies from a single-center, these findings need further multi-institutional validation with larger sample size. Second, our nomograms were merely validated via an internal training set, external verification using independent patient set is necessary. Third, this is a retrospective study that could not completely avoid missing data and measurement biases, more candidate useful biomarkers may be needed to develop predictive models in the future.
In summary, for predicting rNACT, some ML-based models performed better than models using conventional methods, and the SVM model performed best. Preoperative serum lipids and serum inflammation markers have contributed to predicting rNACT in breast cancer patients. These results suggested the need to raise awareness of the importance of minimally-invasive approaches for monitoring breast cancer patients who intended to undergo NACT. However, the current study needs to be validated with caution and require external validation in the future.
Complete response after neoadjuvant chemotherapy (rNACT) elevates the surgical outcomes of patients with breast cancer, however, non-rNACT have a higher risk of death and recurrence.
In this study, we aimed to develop an rNACT risk prediction model for breast cancer patients that utilizes pretreatment serum lipids and serum inflammation markers to stratify patients by rNACT risk on admission. We then analyzed the predictive performance of these ML-based models in a deviation cohort and then verified performance in an internal and external validation cohort.
In this study, we aimed to develop an rNACT risk prediction model for breast cancer patients that utilizes pretreatment serum lipids and serum inflammation markers to stratify patients by rNACT risk on admission. We then analyzed the predictive performance of these ML-based models in a deviation cohort and then verified performance in an internal and external validation cohort.
A retrospective analysis of 487 breast cancer patients who underwent mastectomy or breast-conserving surgery and axillary lymph node dissection following NACT at the Hubei Cancer Hospital between January 1, 2013, and October 1, 2021. The study cohort was divided into internal training and testing datasets in a 70:30 ratio for further analysis. A total of twenty-four variables were included to develop predictive models for rNACT by multiple ML-based algorithms. A feature selection approach was used to identify optimal predictive factors. These models were evaluated by the receiver operating characteristic (ROC) curve for predictive performance.
Analysis identified several significant differences between the rNACT and non-rNACT groups, including total cholesterol, low-density lipoprotein, neutrophil-to-lymphocyte ratio, body mass index, platelet count, albumin-to-globulin ratio (A/G), platelet-to-lymphocyte ratio, and lymphocyte-to-monocyte ratio. The areas under the curve of the six models ranged from 0.81 to 0.96. Some ML-based models performed better than models using conventional statistical methods in both ROC curves. The support vector machine (SVM) model with twelve variables introduced was identified as the best predictive model.
By incorporating pretreatment serum lipids and serum inflammation markers, it is feasible to develop ML-based models for the preoperative prediction of rNACT and therefore facilitate the choice of treatment, particularly the SVM, which can improve the prediction of rNACT in patients with breast cancer.
For predicting rNACT, some ML-based models performed better than models using conventional methods, and the SVM model performed best. Preoperative serum lipids and serum inflammation markers have contributed to predicting rNACT in breast cancer patients. These results suggested the need to raise awareness of the importance of minimally-invasive approaches for monitoring breast cancer patients who intended to undergo NACT. However, the current study needs to be validated with caution and require external validation in the future.
The authors gratefully acknowledge all of our participants for sharing their medical records. The authors also wish to thank the staff members at Hubei Cancer Hospital for their assistance with data collection.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Fang S, China; Nath J, India S-Editor: Xing YX L-Editor: A P-Editor: Xing YX
| 1. | Coughlin SS. Epidemiology of Breast Cancer in Women. Adv Exp Med Biol. 2019;1152:9-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 202] [Article Influence: 33.7] [Reference Citation Analysis (2)] |
| 2. | Early Breast Cancer Trialists' Collaborative Group (EBCTCG). Long-term outcomes for neoadjuvant versus adjuvant chemotherapy in early breast cancer: meta-analysis of individual patient data from ten randomised trials. Lancet Oncol. 2018;19:27-39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 593] [Cited by in RCA: 734] [Article Influence: 104.9] [Reference Citation Analysis (0)] |
| 3. | Wolmark N, Wang J, Mamounas E, Bryant J, Fisher B. Preoperative chemotherapy in patients with operable breast cancer: nine-year results from National Surgical Adjuvant Breast and Bowel Project B-18. J Natl Cancer Inst Monogr. 2001;96-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 924] [Cited by in RCA: 918] [Article Influence: 39.9] [Reference Citation Analysis (0)] |
| 4. | Powles TJ, Hickish TF, Makris A, Ashley SE, O'Brien ME, Tidy VA, Casey S, Nash AG, Sacks N, Cosgrove D. Randomized trial of chemoendocrine therapy started before or after surgery for treatment of primary breast cancer. J Clin Oncol. 1995;13:547-552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 267] [Cited by in RCA: 248] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 5. | Thrall JH, Li X, Li Q, Cruz C, Do S, Dreyer K, Brink J. Artificial Intelligence and Machine Learning in Radiology: Opportunities, Challenges, Pitfalls, and Criteria for Success. J Am Coll Radiol. 2018;15:504-508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 313] [Article Influence: 44.7] [Reference Citation Analysis (0)] |
| 6. | Deo RC. Machine Learning in Medicine. Circulation. 2015;132:1920-1930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1155] [Cited by in RCA: 1961] [Article Influence: 217.9] [Reference Citation Analysis (6)] |
| 7. | O'Mahony C, Jichi F, Pavlou M, Monserrat L, Anastasakis A, Rapezzi C, Biagini E, Gimeno JR, Limongelli G, McKenna WJ, Omar RZ, Elliott PM; Hypertrophic Cardiomyopathy Outcomes Investigators. A novel clinical risk prediction model for sudden cardiac death in hypertrophic cardiomyopathy (HCM risk-SCD). Eur Heart J. 2014;35:2010-2020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 630] [Cited by in RCA: 843] [Article Influence: 70.3] [Reference Citation Analysis (0)] |
| 8. | Waljee AK, Higgins PD. Machine learning in medicine: a primer for physicians. Am J Gastroenterol. 2010;105:1224-1226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 103] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 9. | Austin PC, Merlo J. Intermediate and advanced topics in multilevel logistic regression analysis. Stat Med. 2017;36:3257-3277. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 257] [Cited by in RCA: 402] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 10. | Davis J Jr, Hoskin TL, Day CN, Wickre M, Piltin MA, Caudle AS, Boughey JC. Performance and Clinical Utility of Models Predicting Eradication of Nodal Disease in Patients with Clinically Node-Positive Breast Cancer Treated with Neoadjuvant Chemotherapy by Tumor Biology. Ann Surg Oncol. 2020;27:4678-4686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 11. | Zhang J, Xiao L, Pu S, Liu Y, He J, Wang K. Can We Reliably Identify the Pathological Outcomes of Neoadjuvant Chemotherapy in Patients with Breast Cancer? Ann Surg Oncol. 2021;28:2632-2645. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 12. | Kim WH, Kim HJ, Park HY, Park JY, Chae YS, Lee SM, Cho SH, Shin KM, Lee SY. Axillary Pathologic Complete Response to Neoadjuvant Chemotherapy in Clinically Node-Positive Breast Cancer Patients: A Predictive Model Integrating the Imaging Characteristics of Ultrasound Restaging with Known Clinicopathologic Characteristics. Ultrasound Med Biol. 2019;45:702-709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 13. | Hwang HW, Jung H, Hyeon J, Park YH, Ahn JS, Im YH, Nam SJ, Kim SW, Lee JE, Yu JH, Lee SK, Choi M, Cho SY, Cho EY. A nomogram to predict pathologic complete response (pCR) and the value of tumor-infiltrating lymphocytes (TILs) for prediction of response to neoadjuvant chemotherapy (NAC) in breast cancer patients. Breast Cancer Res Treat. 2019;173:255-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 98] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 14. | Matsuda N, Hayashi N, Ohde S, Yagata H, Kajiura Y, Yoshida A, Suzuki K, Nakamura S, Tsunoda H, Yamauchi H. A nomogram for predicting locoregional recurrence in primary breast cancer patients who received breast-conserving surgery after neoadjuvant chemotherapy. J Surg Oncol. 2014;109:764-769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 15. | Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15860] [Cited by in RCA: 21630] [Article Influence: 1351.9] [Reference Citation Analysis (1)] |
| 16. | Fan J, Lv J. A Selective Overview of Variable Selection in High Dimensional Feature Space. Stat Sin. 2010;20:101-148. [PubMed] |
| 17. | Uddin S, Khan A, Hossain ME, Moni MA. Comparing different supervised machine learning algorithms for disease prediction. BMC Med Inform Decis Mak. 2019;19:281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 288] [Cited by in RCA: 555] [Article Influence: 92.5] [Reference Citation Analysis (0)] |
| 18. | Christodoulou E, Ma J, Collins GS, Steyerberg EW, Verbakel JY, Van Calster B. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J Clin Epidemiol. 2019;110:12-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 935] [Cited by in RCA: 1015] [Article Influence: 169.2] [Reference Citation Analysis (0)] |
| 19. | Pu S, Wang K, Liu Y, Liao X, Chen H, He J, Zhang J. Nomogram-derived prediction of pathologic complete response (pCR) in breast cancer patients treated with neoadjuvant chemotherapy (NCT). BMC Cancer. 2020;20:1120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 20. | Fayanju OM, Ren Y, Thomas SM, Greenup RA, Plichta JK, Rosenberger LH, Tamirisa N, Force J, Boughey JC, Hyslop T, Hwang ES. The Clinical Significance of Breast-only and Node-only Pathologic Complete Response (pCR) After Neoadjuvant Chemotherapy (NACT): A Review of 20,000 Breast Cancer Patients in the National Cancer Data Base (NCDB). Ann Surg. 2018;268:591-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 152] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 21. | Corbeau I, Jacot W, Guiu S. Neutrophil to Lymphocyte Ratio as Prognostic and Predictive Factor in Breast Cancer Patients: A Systematic Review. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 22. | Machireddy A, Thibault G, Tudorica A, Afzal A, Mishal M, Kemmer K, Naik A, Troxell M, Goranson E, Oh K, Roy N, Jafarian N, Holtorf M, Huang W, Song X. Early Prediction of Breast Cancer Therapy Response using Multiresolution Fractal Analysis of DCE-MRI Parametric Maps. Tomography. 2019;5:90-98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 23. | Cho N, Im SA, Park IA, Lee KH, Li M, Han W, Noh DY, Moon WK. Breast cancer: early prediction of response to neoadjuvant chemotherapy using parametric response maps for MR imaging. Radiology. 2014;272:385-396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 70] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 24. | Gu J, Polley EC, Denis M, Carter JM, Pruthi S, Gregory AV, Boughey JC, Fazzio RT, Fatemi M, Alizad A. Early assessment of shear wave elastography parameters foresees the response to neoadjuvant chemotherapy in patients with invasive breast cancer. Breast Cancer Res. 2021;23:52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 25. | Zhang J, Sun M, Chang E, Lu CY, Chen HM, Wu SY. Pathologic response as predictor of recurrence, metastasis, and survival in breast cancer patients receiving neoadjuvant chemotherapy and total mastectomy. Am J Cancer Res. 2020;10:3415-3427. [PubMed] |
| 26. | Iwasa H, Kubota K, Hamada N, Nogami M, Nishioka A. Early prediction of response to neoadjuvant chemotherapy in patients with breast cancer using diffusion-weighted imaging and gray-scale ultrasonography. Oncol Rep. 2014;31:1555-1560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 27. | Maier AM, Heil J, Harcos A, Sinn HP, Rauch G, Uhlmann L, Gomez C, Stieber A, Funk A, Barr RG, Hennigs A, Riedel F, Schäfgen B, Hug S, Marmé F, Sohn C, Golatta M. Prediction of pathological complete response in breast cancer patients during neoadjuvant chemotherapy: Is shear wave elastography a useful tool in clinical routine? Eur J Radiol. 2020;128:109025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 28. | Heo J, Yoon JG, Park H, Kim YD, Nam HS, Heo JH. Machine Learning-Based Model for Prediction of Outcomes in Acute Stroke. Stroke. 2019;50:1263-1265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 340] [Article Influence: 68.0] [Reference Citation Analysis (0)] |
| 29. | Kalafi EY, Nor NAM, Taib NA, Ganggayah MD, Town C, Dhillon SK. Machine Learning and Deep Learning Approaches in Breast Cancer Survival Prediction Using Clinical Data. Folia Biol (Praha). 2019;65:212-220. [PubMed] |
| 30. | Peiffer-Smadja N, Rawson TM, Ahmad R, Buchard A, Georgiou P, Lescure FX, Birgand G, Holmes AH. Machine learning for clinical decision support in infectious diseases: a narrative review of current applications. Clin Microbiol Infect. 2020;26:584-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 259] [Article Influence: 43.2] [Reference Citation Analysis (0)] |
| 31. | Lee Y, Ragguett RM, Mansur RB, Boutilier JJ, Rosenblat JD, Trevizol A, Brietzke E, Lin K, Pan Z, Subramaniapillai M, Chan TCY, Fus D, Park C, Musial N, Zuckerman H, Chen VC, Ho R, Rong C, McIntyre RS. Applications of machine learning algorithms to predict therapeutic outcomes in depression: A meta-analysis and systematic review. J Affect Disord. 2018;241:519-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 180] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 32. | Dutta V, Choraś M, Pawlicki M, Kozik R. A Deep Learning Ensemble for Network Anomaly and Cyber-Attack Detection. Sensors (Basel). 2020;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 33. | Sadeghi Aghili SA, Fatahi Valilai O, Haji A, Khalilzadeh M. Dynamic mutual manufacturing and transportation routing service selection for cloud manufacturing with multi-period service-demand matching. PeerJ Comput Sci. 2021;7:e461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 34. | Todoric J, Antonucci L, Karin M. Targeting Inflammation in Cancer Prevention and Therapy. Cancer Prev Res (Phila). 2016;9:895-905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 281] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 35. | Singh R, Mishra MK, Aggarwal H. Inflammation, Immunity, and Cancer. Mediators Inflamm. 2017;2017:6027305. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 160] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 36. | Guo W, Lu X, Liu Q, Zhang T, Li P, Qiao W, Deng M. Prognostic value of neutrophil-to-lymphocyte ratio and platelet-to-lymphocyte ratio for breast cancer patients: An updated meta-analysis of 17079 individuals. Cancer Med. 2019;8:4135-4148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 119] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 37. | Lv GY, An L, Sun XD, Hu YL, Sun DW. Pretreatment albumin to globulin ratio can serve as a prognostic marker in human cancers: a meta-analysis. Clin Chim Acta. 2018;476:81-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 88] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 38. | Hashemi SA, Bathaie SZ, Mohagheghi MA. Crocetin and crocin decreased cholesterol and triglyceride content of both breast cancer tumors and cell lines. Avicenna J Phytomed. 2020;10:384-397. [PubMed] |
| 39. | Lofterød T, Mortensen ES, Nalwoga H, Wilsgaard T, Frydenberg H, Risberg T, Eggen AE, McTiernan A, Aziz S, Wist EA, Stensvold A, Reitan JB, Akslen LA, Thune I. Impact of pre-diagnostic triglycerides and HDL-cholesterol on breast cancer recurrence and survival by breast cancer subtypes. BMC Cancer. 2018;18:654. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 66] [Article Influence: 9.4] [Reference Citation Analysis (0)] |